In this issue of Blood, Boccalatte and colleagues report on innovative mass spectrometry–driven proteomic studies designed to determine the effect of the NPM-ALK oncogenic tyrosine kinase on the phosphoproteomic-signaling pathway of ALCL.1
The t(2;5)(p23;q35) chromosomal aberration resulting in over-expression of a chimeric oncogene, nucleophosmin-anaplastic lymphoma kinase (NPM-ALK),2 is the most common translocation found in anaplastic large cell lymphoma (ALCL). The constitutive activation of the oncogenic ALK tyrosine kinase induces signaling events that lead to the neoplastic phenotype of ALCL. Numerous signaling pathways downstream of NPM-ALK have been identified including phospholipase Cγ (PLCγ), phosphatidylinositol-3-kinase (PI3K), RAC-serine/threonine-protein kinase (AKT), signal transducer and activator of transcription 3 (STAT3), and the nonreceptor protein kinase (SRC).3 However, most of these studies have been using hypothesis-driven approaches that lead to the identification of the role of a single protein or pathway. Furthermore, analysis of the activation state of a protein as implied by phosphorylation status or sites of phosphorylations are limited to those for which phosphorylation-specific antibodies exist.
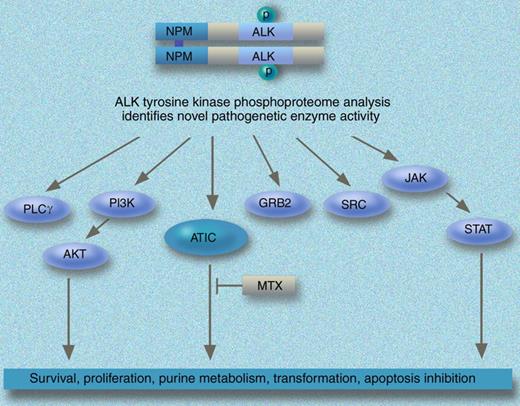
Mass spectrometry–based quantitative phosphoproteomic analysis identifies ATIC as a novel mediator of NPM-ALK. Professional illustration by Marie Dauenheimer.
Mass spectrometry–based quantitative phosphoproteomic analysis identifies ATIC as a novel mediator of NPM-ALK. Professional illustration by Marie Dauenheimer.
Large scale unbiased approaches limited to the transcriptional signature have been used to identify “ALK” target genes, however, the translation of these data sets to signaling proteins induced by a tyrosine kinase may have limited biologic relevance. Advances in liquid chromatography–tandem mass spectrometry–driven proteomics have shifted the paradigm of translational cancer research that enables fast and reliable large scale study of proteins involved in many disease models.4 It follows that identifying the ALK-regulated proteome5,6 and phosphoproteome would lead to key insights into the signaling pathways important for the pathogenesis of ALCL.
Using quantitative proteomic-based approaches coupled with an enrichment strategy that allow the identification of not just the phosphorylated peptides/proteins but specific phosphorylation sites within the peptides,7 the authors present a large dataset of phosphoproteins in 6 cell lines derived from ALCLs and alterations that occur in response to NPM-ALK shRNA or ALK kinase inhibitors. A common ALK phosphotyrosine signature, which included both up-regulation of phoshoproteins and down-regulation of other phosphoproteins, was identified using complementary approaches including small molecular ALK inhibitor (CEP14083) and shRNA. The power of this technology is demonstrated by the observation that of the proteins that are exclusively identified in ALK+cells, 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase (ATIC) is phosphorylated at Y104. The authors provide functional validation of the role of ATIC and demonstrate that its phosphorylation is regulated by NPM/ALK. The biologic relevance of ALK-mediated ATIC activity is highlighted by the observation that the transformylase and total enzyme activity of ATIC is reduced by exposure to methotrexate and could confer resistance to the drug.
These studies not only add to the complexity of downstream signaling pathways induced by ALK but raise the potential for other approaches that enable “activity profiling” of other posttranslational mechanisms to study ALK signaling. These large scale proteomic approaches will have clinical implications in other ALK-deregulated cancers, such as lung cancer8 and neuroblastoma.9
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal