Abstract
The outcome of classical Hodgkin lymphoma (cHL) patients may be related to the tumor microenvironment, which in turn may be influenced by Epstein-Barr virus (EBV) infection. To characterize the cHL microenvironment, a set of 63 cHL tissue samples was profiled using DNA microarrays. Their gene expression profile differed from that of histiocyte T cell–rich B-cell lymphoma (H/TCRBCL) samples that were used as controls, mainly due to high expression of PDCD1/PD-1 in H/TCRBCL. EBV+ cHL tissues could be distinguished from EBV− samples by a gene signature characteristic of Th1 and antiviral responses. Samples from cHL patients with favorable outcome overexpressed genes specific for B cells and genes involved in apoptotic pathways. An independent set of 146 cHL samples was analyzed using immunohistochemistry. It showed a significant adverse value in case of high percentage of either TIA-1+-reactive cells or topoisomerase-2+ tumor cells, whereas high numbers of BCL11A+, FOXP3+, or CD20+ reactive cells had a favorable influence. Our results suggest an antitumoral role for B cells in the cHL microenvironment and a stronger stromal influence of the PD1 pathway in H/TCRBCL than cHL. The observation of Th1/ antiviral response in EBV+ cHL tissues provides a basis for novel treatment strategies.
Introduction
Current therapeutic approaches are often effective for the treatment of classical Hodgkin lymphoma (cHL). Nonetheless, approximately 40% of patients are refractory to initial treatment and/or relapse after achieving complete remission.1,2 Various prognostic systems are currently used to predict the outcome of cHL patients, including the International Prognostic Score (IPS), which is considered the “gold standard” for advanced-stage patients.3 However, the IPS has limited applicability for early-stage patients. Other prognostic systems have been developed for patients with localized disease,4 but there is no consensus about a model to be used in the routine management of all cHL patients. Identification of biologic markers that could help to select patients at high risk of treatment failure remains a crucial challenge
As suggested by the correlation between the clinical course of cHL patients and the plasma levels of particular cytokines,5 the severity of the disease may result from cell signaling networks operating within neoplastic tissues. cHL lesions are characterized by the presence of a minority of malignant cells (usually < 5%), designated as Reed-Sternberg (RS) cells, which reside in a mixture of reactive cells composed of T and B cells, macrophages, plasma cells, and granulocytes.6 Reactive cells are thought to favor the proliferation of RS cells through cytokines and chemokines acting as paracrine factors.7
An aberrant immune response in the vicinity of RS cells is supposed to account for the maintenance of an immunosuppressive environment. It has been initially proposed that a local Th2 reaction predominates,7 but more recently, it was suggested that T regulatory (Treg) cells and PD1+ T cells also interact with RS cells,8-10 which produce the Treg attractant galectin-18 and the PD-1 ligand, PDL-1.10 On the other hand, the observation of numerous CXCR3+ lymphocytes in some HL tumors has raised the possibility of an occasional Th1-predominant immune response.11
The influence of Epstein-Barr virus (EBV) infection on the microenvironment remains unclear. EBV is present in RS cells of 40% to 60% of cHL patients and was shown to contribute to their survival.12 EBV-infected RS cells were shown to stimulate the stromal production of particular chemokines such as CXCL1011 and to produce CCL20, capable of attracting Treg cells.13 It has been suggested that immunologic reactions against EBV can occur in the peripheral blood of some cHL patients.14 However, no comprehensive characterization of intratumoral immunologic alterations induced by EBV+ RS cells has been described so far.
Several retrospective studies using immunohistochemistry (IHC) have attempted to define adverse prognostic markers associated with RS cells, such as high expression of BCL215 or topoisomerase-IIα (topo-IIα),16 and/or loss of HGAL17 or HLA class II molecules.18 Other IHC reports have highlighted the characteristics of nonmalignant immune cells that may predict unfavorable outcome, in particular a low infiltration of intratumoral FOXP3+ Treg cells in combination with a high percentage of either granzyme B+ or TIA-1+ lymphocytes.19-22
In contrast to immunohistochemistry (IHC), gene expression profiling has been rarely used to analyze cHL tissues. A recent series of 23 cHL cases was focused on EBV-induced alterations.13 Only 2 prognostic studies have been published so far. In 2002, we reported the first study that suggested the existence of correlations between gene expression profiles and prognosis,23 although the number of cases (n = 21) was too small to draw a final conclusion. More recently, a gene profiling analysis of 29 cHL cases focused on patients with advanced disease.24 In the present study, we have analyzed a large series of cHL tissues from patients with various clinical characteristics, and we have identified new prognostic factors related to the microenvironment.
Methods
Patients
A set of 63 pretreatment cHL tissue samples was profiled using Affymetrix DNA microarrays (Santa Clara, CA). They were collected at the time of diagnosis from 63 different cHL patients (including 10 children younger than 15 years) who underwent lymph node surgical biopsy in several French hematologic centers (Marseilles, Lille, Dijon, Nancy, Paris) belonging to the network of the Groupe d'Etude des Lymphomas de l'Adulte (GELA). Each patient (or parents, for children) gave written informed consent in accordance with the Declaration of Helsinki with the approval of ethics committees from all participating institutions. Samples were snap-frozen in liquid nitrogen within 30 minutes of removal. All cases were de novo reviewed by 2 hematopathologists (B.C. and L.X.) before analysis. They were classified according to the WHO classification6 as nodular sclerosis (NS; n = 42), mixed cellularity (MC; n = 17), and lymphocyte depleted (n = 1). Three samples that displayed predominant features of interfollicular involvement were considered as unclassified to avoid any bias in the comparison between NS and MC subtypes. EBV status of cHL samples was assigned according to the IHC detection of latent membrane protein-1 (LMP1) and/or in situ hybridization of EBER as negative (n = 35), positive (n = 18), or undetermined due to technical reasons (n = 10). Clinical characteristics of these patients are detailed in Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article. Briefly, cHL patients presented with either Ann Arbor stage I-II (n = 30) or Ann Arbor stage III-IV (n = 29) and were treated with relatively uniform and standard first-line anthracyclin-based chemotherapy regimens (mostly ABVD [doxorubicin, bleomycin, vinblastine, dacarbazine] for adults). Forty-one patients (31 adults and 10 children) had a favorable outcome defined by sustained complete remission for a minimal follow-up of 50 months, and 21 adult patients had an unfavorable outcome, characterized by either failure to achieve complete remission, progressive disease, or relapse during follow-up. These latter criteria were used to define events in univariate Cox analysis. The outcome of one patient was considered as not valuable due to the occurrence of sarcoidosis during follow-up. Control samples profiled with DNA microarrays included tissues from histiocyte/T cell–rich B-cell lymphoma (H/TCRBCL, n = 5) and from benign lymphadenitis (n = 5). All cHL and H/TCRBCL tissue specimens contained more than 80% of neoplastic areas, as assessed using frozen sections adjacent to the profiled samples. Three HL-derived cell lines (L-428, KM-H2, HDLM-2), the fibroblastic cell line HFFB, and the endothelial cell line HUVEC were also profiled as controls.
Gene profiling data were validated and/or completed by analyzing an independent set of 146 cHL patients (different from those analyzed by DNA microarrays) using tissue microarrays (TMAs) and IHC on paraffin tissue samples. Clinical characteristics of these patients are detailed in Table 1. An additional group of 13 H/TCRBCL samples was used for IHC validation experiments on whole paraffin sections.
Clinical characteristics of TMA cHL patients and correlations with bcl11A immunohistochemical expression on reactive cells
| Characteristic . | n (%) . | BCL11A IHC score . | P . | |
|---|---|---|---|---|
| ≤ 2 . | > 2 . | |||
| Age | ||||
| < 45 y | 121 (84) | 49 | 59 | NS |
| ≥ 45 y | 23 (16) | 11 | 10 | |
| Sex | ||||
| Male | 82 (56.2) | 40 | 36 | NS |
| Female | 64 (43.8) | 21 | 34 | |
| Histologic subtype | ||||
| Nodular sclerosis | 106 (78.5) | 40 | 55 | |
| Mixed cellularity | 26 (19.3) | 14 | 11 | |
| Lymphocyte depletion | 2 (1.5) | 2 | 0 | |
| Unclassified | 1 (0.7) | 0 | 0 | |
| EBV tumor status | ||||
| Negative | 115 (79.3) | 51 | 56 | NS |
| Positive | 30 (20.7) | 10 | 14 | |
| Ann Arbor stage | ||||
| 1 | 9 (6.2) | 0 | 8 | .024 |
| 2 | 77 (52.7) | 31 | 38 | |
| 3 | 29 (19.9) | 13 | 13 | |
| 4 | 31 (21.2) | 17 | 11 | |
| B symptoms | ||||
| Absent | 69 (58.5) | 21 | 43 | NS |
| Present | 49 (41.5) | 23 | 20 | |
| Bulky tumor | ||||
| Absent | 70 (59.8) | 20 | 42 | .002 |
| Present | 47 (40.2) | 28 | 15 | |
| Extranodal extension | ||||
| Absent | 98 (83.8) | 34 | 55 | NS |
| Present | 19 (16.2) | 11 | 7 | |
| No. of nodal sites involved | ||||
| < 3 | 67 (52.3) | 23 | 37 | NS |
| ≥ 3 | 61 (47.7) | 27 | 27 | |
| Inguinal involvement | ||||
| Absent | 88 (90.7) | 30 | 49 | NS |
| Present | 9 (9.3) | 7 | 2 | |
| ESR | ||||
| < 50 | 64 (52) | 25 | 37 | NS |
| ≥ 50 | 59 (48) | 24 | 25 | |
| Hemoglobin | ||||
| < 10.5 g/dL | 29 (21.5) | 14 | 12 | NS |
| ≥ 10.5 g/dL | 106 (78.5) | 40 | 56 | |
| White cell count | ||||
| ≤ 15 × 109/L or less | 110 (80.3) | 43 | 57 | NS |
| > 15 × 109/L | 27 (19.7) | 12 | 11 | |
| Lymphocyte count | ||||
| < 0.6 × 109/L or < 8% | 22 (17.5) | 12 | 5 | .033 |
| ≥ 0.6 × 109/L or ≥ 8% | 104 (82.5) | 39 | 60 | |
| Albumin | ||||
| < 40 g/L | 56 (53.8) | 28 | 20 | .037 |
| ≥ 40 g/L | 48 (46.2) | 16 | 30 | |
| International Prognostic Score | ||||
| IPS ≤ 2 | 60 (47.6) | 16 | 39 | .003 |
| IPS > 2 | 66 (52.4) | 35 | 25 | |
| Outcome | ||||
| Favorable | 74 (51) | 8 | 53 | < .001 |
| Unfavorable | 71 (49) | 53 | 17 | |
| Characteristic . | n (%) . | BCL11A IHC score . | P . | |
|---|---|---|---|---|
| ≤ 2 . | > 2 . | |||
| Age | ||||
| < 45 y | 121 (84) | 49 | 59 | NS |
| ≥ 45 y | 23 (16) | 11 | 10 | |
| Sex | ||||
| Male | 82 (56.2) | 40 | 36 | NS |
| Female | 64 (43.8) | 21 | 34 | |
| Histologic subtype | ||||
| Nodular sclerosis | 106 (78.5) | 40 | 55 | |
| Mixed cellularity | 26 (19.3) | 14 | 11 | |
| Lymphocyte depletion | 2 (1.5) | 2 | 0 | |
| Unclassified | 1 (0.7) | 0 | 0 | |
| EBV tumor status | ||||
| Negative | 115 (79.3) | 51 | 56 | NS |
| Positive | 30 (20.7) | 10 | 14 | |
| Ann Arbor stage | ||||
| 1 | 9 (6.2) | 0 | 8 | .024 |
| 2 | 77 (52.7) | 31 | 38 | |
| 3 | 29 (19.9) | 13 | 13 | |
| 4 | 31 (21.2) | 17 | 11 | |
| B symptoms | ||||
| Absent | 69 (58.5) | 21 | 43 | NS |
| Present | 49 (41.5) | 23 | 20 | |
| Bulky tumor | ||||
| Absent | 70 (59.8) | 20 | 42 | .002 |
| Present | 47 (40.2) | 28 | 15 | |
| Extranodal extension | ||||
| Absent | 98 (83.8) | 34 | 55 | NS |
| Present | 19 (16.2) | 11 | 7 | |
| No. of nodal sites involved | ||||
| < 3 | 67 (52.3) | 23 | 37 | NS |
| ≥ 3 | 61 (47.7) | 27 | 27 | |
| Inguinal involvement | ||||
| Absent | 88 (90.7) | 30 | 49 | NS |
| Present | 9 (9.3) | 7 | 2 | |
| ESR | ||||
| < 50 | 64 (52) | 25 | 37 | NS |
| ≥ 50 | 59 (48) | 24 | 25 | |
| Hemoglobin | ||||
| < 10.5 g/dL | 29 (21.5) | 14 | 12 | NS |
| ≥ 10.5 g/dL | 106 (78.5) | 40 | 56 | |
| White cell count | ||||
| ≤ 15 × 109/L or less | 110 (80.3) | 43 | 57 | NS |
| > 15 × 109/L | 27 (19.7) | 12 | 11 | |
| Lymphocyte count | ||||
| < 0.6 × 109/L or < 8% | 22 (17.5) | 12 | 5 | .033 |
| ≥ 0.6 × 109/L or ≥ 8% | 104 (82.5) | 39 | 60 | |
| Albumin | ||||
| < 40 g/L | 56 (53.8) | 28 | 20 | .037 |
| ≥ 40 g/L | 48 (46.2) | 16 | 30 | |
| International Prognostic Score | ||||
| IPS ≤ 2 | 60 (47.6) | 16 | 39 | .003 |
| IPS > 2 | 66 (52.4) | 35 | 25 | |
| Outcome | ||||
| Favorable | 74 (51) | 8 | 53 | < .001 |
| Unfavorable | 71 (49) | 53 | 17 | |
Gene expression analysis
Details of gene expression analysis are given in Document S1. Briefly, total RNA was extracted from frozen samples as described.23 Affymetrix U133 A2.0 human oligonucleotide microarrays were used for gene expression profiling. Preparation of cRNA, hybridizations, washes, and detection were completed as recommended by the supplier.
Gene expression data were analyzed by the Robust Multichip Average method in R using Bioconductor and associated packages.25 Unsupervised hierarchic clustering was done using the Cluster program.26 Results were displayed using the TreeView program.26 To identify the gene clusters responsible for the resulting subdivision of samples, we used the method of quality-threshold (QT) clustering27 and selected the gene clusters with minimal number of 50 genes and minimal correlation of 0.75. Two types of supervised analysis were used. For the comparison of samples based on a continuous variable (clinical outcome), we used univariate Cox analysis. For the comparison of samples based on a categoric variable, we used a discriminating score with confidence levels being estimated by 100 random permutations of samples.28,29 Distinction between subgroups was assessed by linear discriminant analysis (LDA).30
Gene set enrichment analysis (GSEA)31 was performed to assess by supervised analysis the differential expression of potentially relevant previously defined gene sets. Three genes lists were tested: the biologic processes from the Gene Ontology database (http://www.geneontology.org32 ), the functional/pathway gene sets C2 from the Molecular Signatures Database (http://www.broad.mit.edu/gsea/msigdb/index.jsp33 ), and the T cell–related gene sets previously described elsewhere.34 To assess significance of GSEA results, 100 permutations were done. The minimum size of tested gene sets was 10 genes. We report those gene sets found to be significant at 5% risk with a false discovery rate of less than 25%. All microarray data have been deposited with Gene Expression Omnibus (GEO) under accession number GSE13996.35
RQ-PCR analysis
As a validation of DNA microarray data, we performed reverse quantitative polymerase chain reaction (RQ-PCR) analysis of 2 selected genes (IFNG and IFIH1 [MDA5]) that we found differentially expressed between EBV+ and EBV− cHL subgroups. Expression levels were measured in 7 cHL samples (4 EBV+ and 3 EBV−) previously profiled by DNA microarrays. GAPDH was used as the endogenous RNA and cDNA quantity control. Briefly, 2 μg total RNA was subjected to reverse transcription using oligo(dT)15 primers and Moloney murine leukemia virus (m-MLV) reverse transcriptase (Invitrogen, Carlsbad, CA). One-fiftieth of this cDNA was used as the template for amplification of IFNG, IFIH1, and GAPDH transcripts, using a TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA) on an ABI PRISM 7900 HT Fast Real Time PCR System (Applied Biosystems). Primers were designed according to the Sequence Detection System Software 2.1 (Applied Biosystems) and were referenced by the supplier (Applied Biosystems): Hs00174143_m1 for IFNG, Hs00223420_m1 for IFIH1, and Hs99999905_m1 for GAPDH. Analysis of the quantitative real-time PCR curves was performed by the RQ analysis software (Applied Biosystems). Normal peripheral blood lymphocytes were used as a reference sample. Amplification of the GAPDH gene was used as a normalization control for each sample.
Immunohistochemistry
We assessed the protein expression of 17 markers in an independent set of 146 cHL samples using IHC as described.36 The selection was based on antibody availability and on other criteria: IHC was used as a direct validation process for both BCL11A and BFAR markers, whose genes belonged to the molecular prognostic gene signature provided by DNA microarrays. Other IHC markers were specific for cell populations suspected to influence the molecular signature, such as CD20 for B cells and BDCA2 for plasmacytoid dendritic cells (pDCs). Other markers were reported to have prognostic value in cHL, such as BCL2,15 FOXP-3,19-22 TIA-1,19,20 granzyme B,19-21 STAT1,20 and topo-IIα.16 The remaining antibodies were markers of reactive cells that may interact with RS cells, such as tryptase (mast cells), PS100 (dendritic cells), CD8, CD57, and PD-1 (T-cell subsets). SMAD4 was analyzed because SMAD4-deficient stromal T cells are known to produce abundant Th2 cytokines that can stimulate the growth of tumor cells.37 Briefly, areas containing malignant RS cells surrounded by a significant amount of reactive background were marked in the paraffin blocks from each cHL case; 1-mm-diameter cylinders from 3 different areas were then included in the TMA. Primary antibodies were as follows: BCL11A (kindly gifted from the Leukemia Research Fund Monoclonal Antibody Facility, University of Oxford, Oxford, United Kingdom), BCL2 (Dako, Glostrup, Denmark), LMP-1 (Dako), CD57 (Novocastra, Newcastle upon Tyne, United Kingdom), FOXP-3 (eBioscience, San Diego, CA), tryptase (Dako), TIA-1 (Immunotech, Marseilles, France), granzyme B (Novocastra), PS100 (Immunotech), BFAR (kind gift from Maryla Krajewska, Burnham Institute for Medical Research, La Jolla CA), CD20 (Dako), CD8 (Dako), STAT1 (Cell Signaling, Beverly, MA), BDCA2 (dendritics, Lyon, France), topo-IIα (Dako), SMAD4 (Novocastra), and PD-1.38 Dilutions were as recommended by the respective suppliers, except for BCL11A (1/50), BFAR (1/100), and PD-1 (1/50). The PD-1 antibody was also used to validate the supervised H/TCRBCL signature on a group of 13 H/TCRBCL paraffin tissue samples analyzed on whole paraffin sections. After staining, slides were independently evaluated by 2 hematopathologists (L.X. and B.C.). Because the intensity of each marker varied between cases due to the use of different tissue fixatives, only the count of positively stained cells was estimated. The mean of the counts obtained at high power (×400) magnification from 3 different representative TMA cores per case was calculated by each reader in each case. The mean of the counts from each reader was then calculated to assess the IHC score used for statistical analysis. The scores were defined as 0 (no staining), 1 (< 10% of positive cells), 2 (10%-50% of positive cells), or 3 (50%-100% of positive cells). When relatively balanced numbers of patients were obtained in each class group, all possible cutoffs were tested (0 vs 1-3, 0-1 vs 2-3, and 0-2 vs 3); and the cutoff with the strongest effect on the hazard ratio was eventually retained. When necessary, RS cells and reactive cells were separately counted. Pictures were obtained using a DMD 108 microvideo system (Leica, Wetzlar, Germany).
Statistical analysis
The association between 2 categoric variables was examined using Fisher exact or χ2 tests. The primary end point was event-free survival (EFS), which was defined by the time interval between the diagnosis of the disease and the date of relapse or death (any cause). Overall survival was defined by the time between the diagnosis and the date of death (any cause). Event-free patients were right censored at the date of the last follow up. Survival curves were derived from Kaplan-Meier estimates and compared by the log-rank test. Significant changes of relative risks of event were explored using Cox proportional hazard models in univariate and multivariate analysis. Multivariate models were built using a backward stepwise selection of variables to minimize the Akaike information criterion. The stepwise procedure started from the variables selected with a univariate P value less than .1 and a percentage of missing values less than 15%. All results are presented with their 95% confidence intervals. Statistical tests were 2 sided at the 5% level of significance. All the statistical analyses were done using the “survival” package from R.2.7.0 statistical software (R Foundation for Statistical Computing, Vienna, Austria).
Results
Unsupervised clustering reveals molecular heterogeneity of cHL
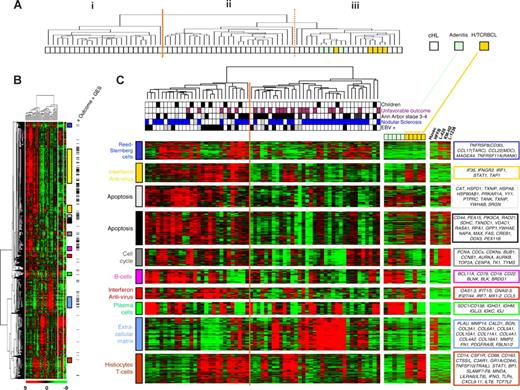
Unsupervised hierarchic clustering of the complete data including 63 cHL, 5 H/TCRBCL, and 5 benign lymphadenitis tissue samples identified 3 major clusters of samples (hereafter designated I, II, and III; Figure 1A). All H/TCRBCL and adenitis samples were grouped in cluster III. As shown by the length of dendrogram branches, these control samples were more homogeneous at the transcriptional level than cHL samples, although that could be due to their comparatively small number.
Global gene expression profiling of cHL samples and control samples. (A) The hierarchic clustering of all tissue samples analyzed using cDNA microarrays. The resulting dendrogram includes 63 lymph node tissue samples from cHL patients (white boxes), 5 from histiocyte/T cell–rich B-cell lymphoma (H/TCRBCL) patients (orange boxes), and 5 from benign lymphadenitis patients (green boxes). This clustering is based on the mRNA expression levels of 10 394 probe sets retained after a filtering process that removed probe sets with low and poorly measured expression as defined by an expression value inferior to 100 units in all 73 samples. Three major clusters of samples are observed and designated as I, II, and III. The heterogeneity of cHL samples is obvious. H/TCRBCL and benign lymphadenitis samples are grouped in cluster III and seem more homogeneous at the transcriptional level than cHL samples, although that could be due to their comparatively small number. (B) Hierarchic clustering restricted to cHL samples (n = 63) and based on 6229 probe sets with significant variation in mRNA expression levels across these samples. Each row represents a gene and each column represents a sample. The 2 separated color matrixes on the right correspond to the expression profile of control tissue samples (5 benign lymphadenitis, 5 H/TCRBCL samples, and 5 cell lines from left to right). Because these control samples are not considered in the clustering of cHL samples, genes are in the same order than in the major left matrix. The expression level of each gene in a single sample is relative to its median abundance across the 63 cHL tissue samples and is depicted according to a color scale (log2 scale) shown at the bottom. Red and green indicate expression levels, respectively, above and below the median. The magnitude of deviation from the median is represented by the color saturation. The dendrogram of cHL tissue samples (above matrix) represent overall similarities in gene expression profiles, whereas colored bars on the right indicate the locations of 11 gene clusters of interest. Black dashes on the right represent the 501 probe sets associated with clinical outcome according to the univariate Cox analysis (“Outcome” GES). A zoomed view of panel B is shown in panel C, highlighting the dendrogram and gene clusters. In the dendrogram (C, top) of cHL tissue samples, 2 large groups are evidenced by clustering and delimited by an orange vertical line. Below the dendrogram, relevant characteristics of cHL tissue samples are represented according to a color ladder (gray when unclassifiable or unavailable): age (adult, white; children, black), clinical outcome (favorable, white; unfavorable, purple), Ann Arbor stage (I-II, white; III-V, black), histologic type (nodular sclerosis, blue; mixed cellularity, white), and EBV tumoral status (negative, white; positive, black). The control samples are colored as in panel A. The 5 cell lines are HUVEC (endothelial cells), HFFB (fibroblastic cells), L-428, KM-H2, and L-1236 (Reed-Sternberg cells) (from left to right). Expanded views of selected gene clusters corresponding to relevant cell types/function are named from top to bottom: “Reed-Sternberg cells” (dark blue bar), “interferon pathway and antiviral response (1)” (orange bar), “apoptosis” (2 clusters; light gray and black bars), “cell cycle” (dark gray bar), “B cells” (pink bar), “interferon pathway and antiviral response (2)” (red bar), “plasma cells” (green bar), “extracellular matrix” (light blue bar), and “histiocytes/T cells/innate immune response” (brown bar). The “cell metabolism” cluster (yellow bar in B) is not zoomed in panel C. The most relevant genes included in these clusters are indicated on the right by their EntrezGene symbol.39
Global gene expression profiling of cHL samples and control samples. (A) The hierarchic clustering of all tissue samples analyzed using cDNA microarrays. The resulting dendrogram includes 63 lymph node tissue samples from cHL patients (white boxes), 5 from histiocyte/T cell–rich B-cell lymphoma (H/TCRBCL) patients (orange boxes), and 5 from benign lymphadenitis patients (green boxes). This clustering is based on the mRNA expression levels of 10 394 probe sets retained after a filtering process that removed probe sets with low and poorly measured expression as defined by an expression value inferior to 100 units in all 73 samples. Three major clusters of samples are observed and designated as I, II, and III. The heterogeneity of cHL samples is obvious. H/TCRBCL and benign lymphadenitis samples are grouped in cluster III and seem more homogeneous at the transcriptional level than cHL samples, although that could be due to their comparatively small number. (B) Hierarchic clustering restricted to cHL samples (n = 63) and based on 6229 probe sets with significant variation in mRNA expression levels across these samples. Each row represents a gene and each column represents a sample. The 2 separated color matrixes on the right correspond to the expression profile of control tissue samples (5 benign lymphadenitis, 5 H/TCRBCL samples, and 5 cell lines from left to right). Because these control samples are not considered in the clustering of cHL samples, genes are in the same order than in the major left matrix. The expression level of each gene in a single sample is relative to its median abundance across the 63 cHL tissue samples and is depicted according to a color scale (log2 scale) shown at the bottom. Red and green indicate expression levels, respectively, above and below the median. The magnitude of deviation from the median is represented by the color saturation. The dendrogram of cHL tissue samples (above matrix) represent overall similarities in gene expression profiles, whereas colored bars on the right indicate the locations of 11 gene clusters of interest. Black dashes on the right represent the 501 probe sets associated with clinical outcome according to the univariate Cox analysis (“Outcome” GES). A zoomed view of panel B is shown in panel C, highlighting the dendrogram and gene clusters. In the dendrogram (C, top) of cHL tissue samples, 2 large groups are evidenced by clustering and delimited by an orange vertical line. Below the dendrogram, relevant characteristics of cHL tissue samples are represented according to a color ladder (gray when unclassifiable or unavailable): age (adult, white; children, black), clinical outcome (favorable, white; unfavorable, purple), Ann Arbor stage (I-II, white; III-V, black), histologic type (nodular sclerosis, blue; mixed cellularity, white), and EBV tumoral status (negative, white; positive, black). The control samples are colored as in panel A. The 5 cell lines are HUVEC (endothelial cells), HFFB (fibroblastic cells), L-428, KM-H2, and L-1236 (Reed-Sternberg cells) (from left to right). Expanded views of selected gene clusters corresponding to relevant cell types/function are named from top to bottom: “Reed-Sternberg cells” (dark blue bar), “interferon pathway and antiviral response (1)” (orange bar), “apoptosis” (2 clusters; light gray and black bars), “cell cycle” (dark gray bar), “B cells” (pink bar), “interferon pathway and antiviral response (2)” (red bar), “plasma cells” (green bar), “extracellular matrix” (light blue bar), and “histiocytes/T cells/innate immune response” (brown bar). The “cell metabolism” cluster (yellow bar in B) is not zoomed in panel C. The most relevant genes included in these clusters are indicated on the right by their EntrezGene symbol.39
Hierarchic clustering of the 63 cHL and the 6229 probe sets with significant variation in expression level across the samples (SD > 0.5) is shown in Figure 1B and C. Samples were distributed in 2 main groups (Figure 1C) and this distribution was correlated with clinical outcome, because among the 28 patients of the left group, 23 (82%) had favorable outcome, whereas among the 35 patients of the right group, only 18 (51%) had favorable outcome (P = .03, Fisher exact test). No significant correlation was found between this distribution and the age of patients (adults vs children), Ann Arbor stage (I-II vs III-IV), EBV status (positive vs negative), or histologic type (MC vs NS).
Gene clustering revealed groups of coordinately expressed genes. Quality threshold (QT) clustering identified 17 gene clusters (correlation 0.75), some of which represented expression signatures corresponding to defined biologic processes or cell types (see colored bars on the right of Figure 1B and zooms in Figure 1C). They included a “cell-cycle” cluster (84 probe sets) that was globally overexpressed in cell lines compared with tissues and included genes encoding PCNA, cyclins, CDCs and CDKNs, STK6, AURKB, TOP2A, CENPA, TK1, and TYMS; an “interferon pathway and antiviral response” cluster (236 probe sets) that was enriched in genes involved in response to viral infections such as IFI35, IFNGR2, IRF1, STAT1, and TAP1. This cluster was overexpressed in most EBV+ cHL samples, as well as in H/TCRBCL samples. The largest cluster designated “cell metabolism” (1141 probe sets) included many genes involved in oxidative phosphorylation, protein synthesis, protein folding, and protein ubiquitination. Two “apoptosis” clusters (52 and 161 probe sets) were also evidenced, including FAS, DDX3, MAX, TXNIP, HSPD1, HSPA8, HSP90AB1, and TANK. Other gene clusters reflected specific cell types present in cHL tissues. The “plasma cell” cluster (81 probe sets) was overexpressed in adenitis compared with H/TCRBCL and included genes coding for immunoglobulins and syndecan-1 (SDC1/CD138). The “B-cell” cluster (62 probe sets) was overexpressed in adenitis samples; it included genes involved in B-cell function such as BCL11A, CD19, CD22, CD79A/B, BANK1, VPREB3, STAP1 (BRDG1), BLNK, and BLK. The “extracellular matrix” cluster (323 probe sets) contained genes related to extracellular matrix remodeling such as collagen genes, MMPs, CALD1, PLAU, PDGFRs, FN1, SPARC, and DCN. As expected, this cluster was strongly expressed in both HFFB and HUVEC cell lines (derived from fibroblasts and endothelial cells, respectively), but was not expressed in RS cell lines. The “histiocytes/T cells/innate immune response” cluster (222 probe sets) was overexpressed in H/TCRBCL samples and in most EBV+ cHL samples. It was enriched in markers associated with either histiocytes or T cells, or involved in innate immune response such as CD14, CD68, CSF1R, CD163, CTSS/L, C3AR1, FCGR1(CD64), FCGR3A (CD16a), STAT1, TNFSF10 (TRAIL), MNDA, LILRA/Bs, GBP1, CXCL9-11, SLAMF7-8, TLR1/2/4/5/8, TCF7L2, and LILRA6 (ILT8). Finally, 2 other gene clusters of interest were just below the correlation threshold of QT clustering. They included a “Reed-Sternberg cell” cluster (97 probe sets; correlation, 0.58), characterized by overexpression of genes encoding known RS cell markers such as TNFRSF8 (CD30), CCL17 (TARC), CCL22 (MDC), MAGEA4, and TNFRSF11A (RANK). As expected, this latter cluster was strongly expressed in HL cell lines but was not expressed in non-Hodgkin cell lines and tissues. The last gene cluster was an additional “interferon pathway and antiviral response” cluster (60 probe sets; correlation, 0.61), which was enriched in genes involved in response to viral infections such as OAS1-2-3, CCL5/RANTES, IFIT1, IFIT5, GNAI2-3, IFI44, IFI27, IRF7, and MX1-2. This latter cluster was overexpressed in most EBV+ cHL samples.
Visual inspection revealed that a few of these independent gene clusters were responsible for much of the subdivision of cHL samples (Figure 1C).
The clinical outcome of cHL patients is correlated with a specific gene expression signature
In search for a gene expression signature associated with the clinical outcome of cHL patients, we applied supervised Cox analysis to adult patients. With a 5% FDR, 501 probe sets were associated with outcome, corresponding to 450 unique genes comprising 47 genes (52 probe sets) associated with unfavorable outcome and 403 genes (449 probe sets) associated with favorable outcome (Table S2). The resulting LDA-based classification of adult samples is shown in Figure 2A with a correct classification of 90% (P < .001, Fisher exact test).
Supervised classification of cHL samples based on the “clinical outcome” signature and on the “EBV status” signature. (A) The classification of 52 informative cHL adult samples using the molecular signature of 501 probe sets correlated with clinical outcome. Thirty-one samples were from patients with favorable outcome (FO) and 21 samples from patients with unfavorable outcome (UO). Expression data and clinical features (top panel) are depicted as in Figure 1. Probe sets are ordered from top to bottom by their decreasing discriminating score. Tumor samples are ordered from left to right according to their LDA function score. The solid orange line indicates the threshold 0 that separates the 2 predicted classes of samples referred to as “predicted FO group” (at the left of the line) and “predicted UO group” (right to the line). The bottom panel represents the LDA posterior probability for each sample to belong to the FO group (y-axis), with the samples (x-axis) ranked according to their LDA function score. The observed clinical outcome is indicated by colored dots: green indicates FO; red, UO. There was a statistically significant correlation between the observed clinical outcome and the predicted clinical outcome based on the corresponding gene signature, as shown by the resulting cross table. With a representation similar to panel A, panel B shows the molecular classification of 53 informative cHL samples using the 425 probe sets identified as differentially expressed between the 18 EBV+ samples and the 35 EBV− samples (top panel). The LDA posterior probability (y-axis) is the probability for each sample to belong to the EBV− group (bottom panel). The observed EBV status is indicated by colored dots: green indicates negative; red, positive. The statistical correlation between the observed EBV status and the predicted EBV status was highly significant, as shown by the resulting cross table.
Supervised classification of cHL samples based on the “clinical outcome” signature and on the “EBV status” signature. (A) The classification of 52 informative cHL adult samples using the molecular signature of 501 probe sets correlated with clinical outcome. Thirty-one samples were from patients with favorable outcome (FO) and 21 samples from patients with unfavorable outcome (UO). Expression data and clinical features (top panel) are depicted as in Figure 1. Probe sets are ordered from top to bottom by their decreasing discriminating score. Tumor samples are ordered from left to right according to their LDA function score. The solid orange line indicates the threshold 0 that separates the 2 predicted classes of samples referred to as “predicted FO group” (at the left of the line) and “predicted UO group” (right to the line). The bottom panel represents the LDA posterior probability for each sample to belong to the FO group (y-axis), with the samples (x-axis) ranked according to their LDA function score. The observed clinical outcome is indicated by colored dots: green indicates FO; red, UO. There was a statistically significant correlation between the observed clinical outcome and the predicted clinical outcome based on the corresponding gene signature, as shown by the resulting cross table. With a representation similar to panel A, panel B shows the molecular classification of 53 informative cHL samples using the 425 probe sets identified as differentially expressed between the 18 EBV+ samples and the 35 EBV− samples (top panel). The LDA posterior probability (y-axis) is the probability for each sample to belong to the EBV− group (bottom panel). The observed EBV status is indicated by colored dots: green indicates negative; red, positive. The statistical correlation between the observed EBV status and the predicted EBV status was highly significant, as shown by the resulting cross table.
As shown in Figure 1B, many of these 501 probe sets were included in several gene clusters of interest described in the previous section. On the one hand, 23% of probe sets of the “B-cell” cluster, such as BCL11A, BANK1, STAP1 (BRDG1), BLNK, FCER2, CD24, and CCL21, were associated with favorable outcome, as were 19% of those of an “apoptosis” cluster with genes such as YWHAB, CASP8, PTPRC, and TANK, and 11% of those of the “cell metabolism” cluster. On the other hand, 9% of probe sets of the “extracellular matrix” cluster with genes such as collagen genes (COL1A1/4A1/4A2/5A1/18A1), THBS1/2, FN1, EDNRA, ITGB5, and LAMA4 were associated with unfavorable outcome. These results were further corroborated by GSEA analysis. Indeed, among the gene lists significantly overrepresented in favorable outcome samples (P < .001) were lists associated with B cells (“Sig_BCR_Signalling_Pathway,” “St_B_Cell_Antigen_Receptor”), with apoptosis (“Apoptosis_Kegg,” “St_FAS_Signaling_Pathway,” “Apoptosis_Genmapp,” “Death Pathway,” “Apoptosis”), and with cell metabolism (“mRNA.Processing,” “RNA.Splicing,” “RNA.Processing,” “Protein.Amino.Acid.Dephosphorylation,” “Oxidative_Phosphorylation”). Conversely, among the gene lists significantly overrepresented in unfavorable outcome samples (P < .001) were lists associated with stroma remodeling (“Skeletal.Development,” “Nervous.System.Development”).
To validate this signature at the protein level and to extend the analysis to other potential prognostic markers, we performed IHC on an independent series of 146 cHL samples. Examples of IHC staining for BCL11A, CD20, FOXP3, and TIA-1 are shown in Figures 3, 4.
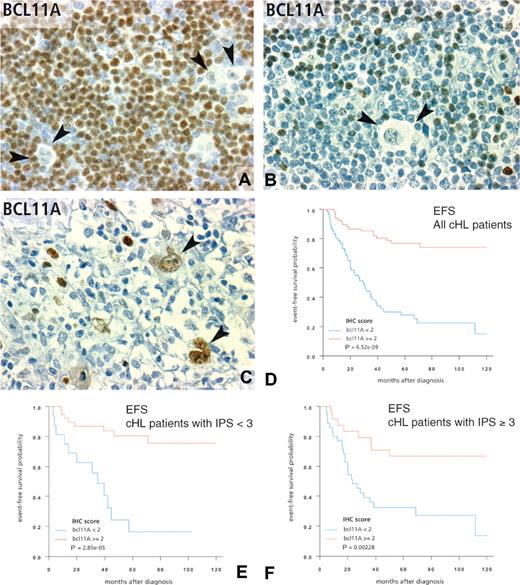
Immunohistochemical patterns of Bcl11A expression in classical Hodgkin lymphoma and correlations with survival. (A,B) The usual pattern of BCL11A positivity in cHL tissues. The immunostaining is located mainly in the nucleus of reactive small lymphocytes surrounding Reed-Sternberg cells (arrows). Variable numbers of positive plasmacytoid dendritic cells were also observed. The number of positive reactive cells scored as high/score 3 or low/score 1 (A and B, respectively) was strongly correlated with EFS in the whole population of 143 informative patients (different from those analyzed by DNA microarrays), as shown by Kaplan-Meier estimates (D). Among these patients, Bcl11A positivity has also a significant prognostic value in the subgroup of patients with an international prognostic score of 2 or less (E) and in patients with an IPS more than 2 (F) as shown by Kaplan-Meier estimates of EFS. Panel C illustrates an infrequent pattern of Bcl11A expression in cHL tissues, with a positive signal located in neoplastic Reed-Sternberg cells ( ). This latter pattern was not statistically correlated with survival. Panels A-C correspond to a 40×/0.75 objective lens (×400 magnification).
). This latter pattern was not statistically correlated with survival. Panels A-C correspond to a 40×/0.75 objective lens (×400 magnification).
Immunohistochemical patterns of Bcl11A expression in classical Hodgkin lymphoma and correlations with survival. (A,B) The usual pattern of BCL11A positivity in cHL tissues. The immunostaining is located mainly in the nucleus of reactive small lymphocytes surrounding Reed-Sternberg cells (arrows). Variable numbers of positive plasmacytoid dendritic cells were also observed. The number of positive reactive cells scored as high/score 3 or low/score 1 (A and B, respectively) was strongly correlated with EFS in the whole population of 143 informative patients (different from those analyzed by DNA microarrays), as shown by Kaplan-Meier estimates (D). Among these patients, Bcl11A positivity has also a significant prognostic value in the subgroup of patients with an international prognostic score of 2 or less (E) and in patients with an IPS more than 2 (F) as shown by Kaplan-Meier estimates of EFS. Panel C illustrates an infrequent pattern of Bcl11A expression in cHL tissues, with a positive signal located in neoplastic Reed-Sternberg cells ( ). This latter pattern was not statistically correlated with survival. Panels A-C correspond to a 40×/0.75 objective lens (×400 magnification).
). This latter pattern was not statistically correlated with survival. Panels A-C correspond to a 40×/0.75 objective lens (×400 magnification).
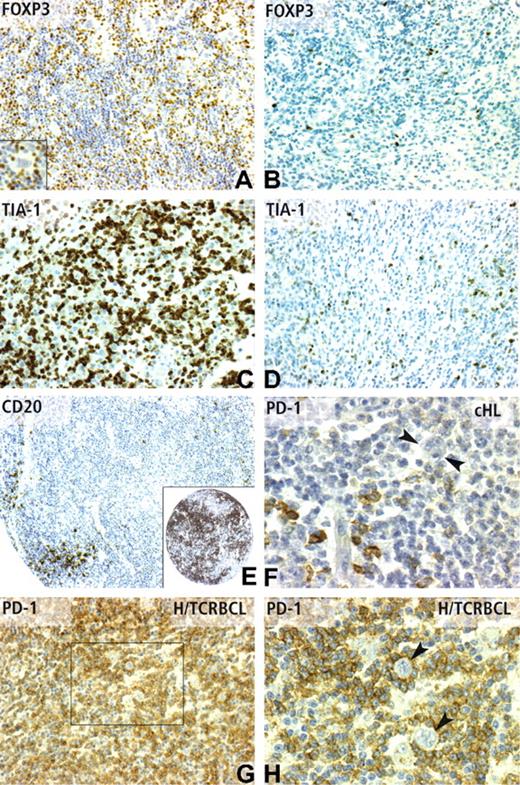
Immunohistochemical patterns of CD20, FOXP3 TIA-1, and PD-1 expression. (A) FOXP3 nuclear positivity in numerous reactive lymphocytes (IHC score 3), but not in Reed-Sternberg cells (inset) in a cHL case with favorable outcome. In contrast, only rare scattered FOXP3-positive lymphocytes (IHC score 1) are observed in a cHL case with unfavorable outcome (B). TIA-1 positivity in cHL cases with poor outcome was frequently observed as high (IHC score 3) (C), whereas cHL cases with favorable outcome often display a low level (IHC score 1) of TIA-1 expression (D). (E) CD20 reactivity on a TMA core from a case of cHL with unfavorable outcome, which contained only scarce positive reactive lymphocytes (IHC score 1), compared with the strong CD20 positivity (IHC score 3) of a case of cHL with favorable outcome (inset). PD-1 immunostaining was detected in a minority of reactive lymphocytes in most cHL cases (F), whereas Reed-Sternberg cells are negative (F,  ). As expected from cDNA microarray data, the PD-1 antibody strongly stained the vast majority of reactive T lymphocytes in H/TCRBCL cases (G), whereas neoplastic B cells were negative, as evidenced by high-power view (H,
). As expected from cDNA microarray data, the PD-1 antibody strongly stained the vast majority of reactive T lymphocytes in H/TCRBCL cases (G), whereas neoplastic B cells were negative, as evidenced by high-power view (H,  ). Panels A-D and G were obtained using a 20×/0.6 objective lens (×200 magnification). Panel E was obtained using a 10×/0.32 objective lens (×100 magnification). Panels F and H correspond to a 40×/0.75 lens (×400 magnification).
). Panels A-D and G were obtained using a 20×/0.6 objective lens (×200 magnification). Panel E was obtained using a 10×/0.32 objective lens (×100 magnification). Panels F and H correspond to a 40×/0.75 lens (×400 magnification).
Immunohistochemical patterns of CD20, FOXP3 TIA-1, and PD-1 expression. (A) FOXP3 nuclear positivity in numerous reactive lymphocytes (IHC score 3), but not in Reed-Sternberg cells (inset) in a cHL case with favorable outcome. In contrast, only rare scattered FOXP3-positive lymphocytes (IHC score 1) are observed in a cHL case with unfavorable outcome (B). TIA-1 positivity in cHL cases with poor outcome was frequently observed as high (IHC score 3) (C), whereas cHL cases with favorable outcome often display a low level (IHC score 1) of TIA-1 expression (D). (E) CD20 reactivity on a TMA core from a case of cHL with unfavorable outcome, which contained only scarce positive reactive lymphocytes (IHC score 1), compared with the strong CD20 positivity (IHC score 3) of a case of cHL with favorable outcome (inset). PD-1 immunostaining was detected in a minority of reactive lymphocytes in most cHL cases (F), whereas Reed-Sternberg cells are negative (F,  ). As expected from cDNA microarray data, the PD-1 antibody strongly stained the vast majority of reactive T lymphocytes in H/TCRBCL cases (G), whereas neoplastic B cells were negative, as evidenced by high-power view (H,
). As expected from cDNA microarray data, the PD-1 antibody strongly stained the vast majority of reactive T lymphocytes in H/TCRBCL cases (G), whereas neoplastic B cells were negative, as evidenced by high-power view (H,  ). Panels A-D and G were obtained using a 20×/0.6 objective lens (×200 magnification). Panel E was obtained using a 10×/0.32 objective lens (×100 magnification). Panels F and H correspond to a 40×/0.75 lens (×400 magnification).
). Panels A-D and G were obtained using a 20×/0.6 objective lens (×200 magnification). Panel E was obtained using a 10×/0.32 objective lens (×100 magnification). Panels F and H correspond to a 40×/0.75 lens (×400 magnification).
As expected from initial reports,40,41 BCL11A staining in cHL samples was evidenced in a fraction of reactive cells corresponding to small B lymphocytes and pDCs, and more rarely in tumor cells (Figure 3). We searched for a correlation between IHC results and clinical outcome of cHL patients (results detailed in Tables 2,3). Univariate Cox proportional hazards regression model showed an adverse value on overall survival (OS) in case of high percentage of either TIA-1+-reactive cells or topo-IIa tumor cells, whereas high numbers of CD20+ or BCL11A+-reactive cells had a favorable influence on OS (Table 2). With respect to EFS, univariate analysis retained high counts of FOXP3+-, CD20+-, or BCL11A+-reactive cells as significant features of favorable prognosis, whereas high counts of topo-IIa+ neoplastic cells were correlated with poor outcome (Table 3). The significant value of both TIA-1 and FOXP3 was dependent on the choice of the cutoff, whereas the 2 different cutoffs tested for BCL11A were both effective (data not shown). No prognostic value on either OS or EFS was found for IHC detection of BCL2, LMP-1, PD-1, CD57, tryptase, granzyme B, BFAR, PS100, CD8, STAT1, SMAD4, and BDCA2.
Overall survival analysis
| Variable . | Class/cutoff . | n (%) . | Univariate analysis* . | Multivariate analysis† . | ||
|---|---|---|---|---|---|---|
| Hazard ratio of event risk . | P . | Hazard ratio of event risk . | P . | |||
| Bcl2‡ | IHC score > 0 | 53 (39.8) | 1.64 (0.843-3.188) | .15 | ||
| EBV‡ | IHC score > 0 | 29 (20.4) | 0.627 (0.243-1.618) | .33 | ||
| PD1 | IHC score > 1 | 8 (18.2) | 0.447 (0.055-3.625) | .45 | ||
| CD57 | IHC score > 1 | 42 (35.3) | 1.09 (0.497-2.373) | .84 | ||
| FOXP3 | IHC score > 1 | 97 (73.5) | 0.908 (0.415-1.987) | .81 | ||
| Tryptase | IHC score > 1 | 116 (83.5) | 1.86 (0.566-6.088) | .31 | ||
| TIA-1 | IHC score > 2 | 34 (24.5) | 2.4 (1.202-4.786) | .013 | 1.91 (0.841-4.331) | .12 |
| Granzyme B | IHC score > 1 | 40 (36.4) | 1.31 (0.618-2.779) | .48 | ||
| PS100 | IHC score > 1 | 77 (53.8) | 1.28 (0.651-2.522) | .47 | ||
| TopoIIa‡ | IHC score > 1 | 66 (49.3) | 2.85 (1.358-5.961) | .006 | 4.11 (1.663-10.136) | .002 |
| BFAR‡ | IHC score > 1 | 84 (70.6) | 0.798 (0.356-1.792) | .59 | ||
| Smad4 | IHC score > 0 | 20 (29.4) | 0.799 (0.287-2.224) | .67 | ||
| Bcl11A | IHC score > 1 | 68 (53.1) | 0.104 (0.04-0.268) | 2.90 × 10−6 | 0.103 (0.032-0.329) | .001 |
| CD20 | IHC score > 1 | 76 (53.9) | 0.322 (0.157-0.658) | .002 | 0.325 (0.135-0.781) | .012 |
| CD8 | IHC score > 1 | 54 (45.4) | 1.28 (0.601-2.728) | .52 | ||
| Stat1 | IHC score > 1 | 33 (42.9) | 1.13 (0.48-2.672) | .78 | ||
| BDCA2 | IHC score > 1 | 24 (35.3) | 1.28 (0.405-4.051) | .67 | ||
| Age | ≥ 45 y | 21 (14.9) | 2.56 (1.197-5.477) | .015 | 6.47 (2.321-18.019) | .001 |
| Sex | Female | 64 (44.8) | 0.44 (0.211-0.918) | .029 | 0.364 (0.147-0.904) | .03 |
| Ann Arbor | III-IV | 59 (41.3) | 1.39 (0.716-2.708) | .33 | ||
| B symptoms | Present | 47 (40.5) | 2.87 (1.301-6.331) | .009 | ||
| Bulky tumor | Present | 47 (40.5) | 5.25 (2.292-12.012) | 8.80 × 10−5 | ||
| Extranodal disease | Present | 19 (16.4) | 1.51 (0.569-4.014) | .41 | ||
| No. of nodal sites | ≥ 3 | 61 (48) | 1.11 (0.528-2.325) | .79 | ||
| Inguinal involvement | Present | 9 (9.4) | 2.74 (0.926-8.094) | .069 | ||
| ESR | ≥ 50 | 59 (48.4) | 1.2 (0.57-2.518) | .63 | ||
| Hemoglobin | ≥ 10.5 g/dL | 105 (78.4) | 0.795 (0.357-1.771) | .57 | ||
| White cell count | > 15 × 109/L | 27 (19.9) | 2.54 (1.222-5.269) | .013 | 2.79 (1.212-6.418) | .016 |
| Lymphocyte count | ≥ 0.6 × 109/L or ≥ 8% | 103 (82.4) | 0.533 (0.236-1.204) | .13 | ||
| Albumin level | ≥ 40 g/L | 47 (45.6) | 0.326 (0.131-0.812) | .016 | ||
| Variable . | Class/cutoff . | n (%) . | Univariate analysis* . | Multivariate analysis† . | ||
|---|---|---|---|---|---|---|
| Hazard ratio of event risk . | P . | Hazard ratio of event risk . | P . | |||
| Bcl2‡ | IHC score > 0 | 53 (39.8) | 1.64 (0.843-3.188) | .15 | ||
| EBV‡ | IHC score > 0 | 29 (20.4) | 0.627 (0.243-1.618) | .33 | ||
| PD1 | IHC score > 1 | 8 (18.2) | 0.447 (0.055-3.625) | .45 | ||
| CD57 | IHC score > 1 | 42 (35.3) | 1.09 (0.497-2.373) | .84 | ||
| FOXP3 | IHC score > 1 | 97 (73.5) | 0.908 (0.415-1.987) | .81 | ||
| Tryptase | IHC score > 1 | 116 (83.5) | 1.86 (0.566-6.088) | .31 | ||
| TIA-1 | IHC score > 2 | 34 (24.5) | 2.4 (1.202-4.786) | .013 | 1.91 (0.841-4.331) | .12 |
| Granzyme B | IHC score > 1 | 40 (36.4) | 1.31 (0.618-2.779) | .48 | ||
| PS100 | IHC score > 1 | 77 (53.8) | 1.28 (0.651-2.522) | .47 | ||
| TopoIIa‡ | IHC score > 1 | 66 (49.3) | 2.85 (1.358-5.961) | .006 | 4.11 (1.663-10.136) | .002 |
| BFAR‡ | IHC score > 1 | 84 (70.6) | 0.798 (0.356-1.792) | .59 | ||
| Smad4 | IHC score > 0 | 20 (29.4) | 0.799 (0.287-2.224) | .67 | ||
| Bcl11A | IHC score > 1 | 68 (53.1) | 0.104 (0.04-0.268) | 2.90 × 10−6 | 0.103 (0.032-0.329) | .001 |
| CD20 | IHC score > 1 | 76 (53.9) | 0.322 (0.157-0.658) | .002 | 0.325 (0.135-0.781) | .012 |
| CD8 | IHC score > 1 | 54 (45.4) | 1.28 (0.601-2.728) | .52 | ||
| Stat1 | IHC score > 1 | 33 (42.9) | 1.13 (0.48-2.672) | .78 | ||
| BDCA2 | IHC score > 1 | 24 (35.3) | 1.28 (0.405-4.051) | .67 | ||
| Age | ≥ 45 y | 21 (14.9) | 2.56 (1.197-5.477) | .015 | 6.47 (2.321-18.019) | .001 |
| Sex | Female | 64 (44.8) | 0.44 (0.211-0.918) | .029 | 0.364 (0.147-0.904) | .03 |
| Ann Arbor | III-IV | 59 (41.3) | 1.39 (0.716-2.708) | .33 | ||
| B symptoms | Present | 47 (40.5) | 2.87 (1.301-6.331) | .009 | ||
| Bulky tumor | Present | 47 (40.5) | 5.25 (2.292-12.012) | 8.80 × 10−5 | ||
| Extranodal disease | Present | 19 (16.4) | 1.51 (0.569-4.014) | .41 | ||
| No. of nodal sites | ≥ 3 | 61 (48) | 1.11 (0.528-2.325) | .79 | ||
| Inguinal involvement | Present | 9 (9.4) | 2.74 (0.926-8.094) | .069 | ||
| ESR | ≥ 50 | 59 (48.4) | 1.2 (0.57-2.518) | .63 | ||
| Hemoglobin | ≥ 10.5 g/dL | 105 (78.4) | 0.795 (0.357-1.771) | .57 | ||
| White cell count | > 15 × 109/L | 27 (19.9) | 2.54 (1.222-5.269) | .013 | 2.79 (1.212-6.418) | .016 |
| Lymphocyte count | ≥ 0.6 × 109/L or ≥ 8% | 103 (82.4) | 0.533 (0.236-1.204) | .13 | ||
| Albumin level | ≥ 40 g/L | 47 (45.6) | 0.326 (0.131-0.812) | .016 | ||
Univariate Cox proportional hazards regression model.
Cox proportional hazards regression model after backward stepwise variables selection procedure.
Bcl2, EBV, topoisomerase-2, and BFAR immunostainings were analyzed on Reed-Sternberg cells, whereas the results mentioned for all other antibodies were analyzed on reactive cells.
Event-free survival analysis
| Variable . | Class/cutoff . | n (%) . | Univariate analysis* . | Multivariate analysis† . | ||
|---|---|---|---|---|---|---|
| Hazard ratio of event risk . | P . | Hazard ratio of event risk . | P . | |||
| Bcl2‡ | IHC score > 0 | 53 (39.8) | 1.22 (0.726-2.034) | .46 | ||
| EBV‡ | IHC score > 0 | 29 (20.4) | 0.668 (0.338-1.319) | .24 | ||
| PD1 | IHC score > 1 | 8 (18.2) | 0.627 (0.179-2.193) | .46 | ||
| CD57 | IHC score > 1 | 42 (35.3) | 1.42 (0.81-2.493) | .22 | ||
| FOXP3 | IHC score > 1 | 97 (73.5) | 0.555 (0.32-0.966) | .037 | ||
| Tryptase | IHC score > 1 | 75 (54) | 0.892 (0.537-1.482) | .66 | ||
| TIA-1 | IHC score > 2 | 34 (24.5) | 1.16 (0.654-2.063) | .61 | ||
| Granzyme B | IHC score > 1 | 40 (36.4) | 1.25 (0.707-2.201) | .45 | ||
| PS100 | IHC score > 1 | 77 (53.8) | 0.808 (0.49-1.33) | .4 | ||
| TopoIIa‡ | IHC score > 1 | 66 (49.3) | 1.75 (1.048-2.918) | .032 | ||
| BFAR‡ | IHC score > 1 | 84 (70.6) | 0.756 (0.416-1.372) | .36 | ||
| Smad4 | IHC score > 0 | 20 (29.4) | 0.742 (0.345-1.597) | .45 | ||
| Bcl11A | IHC score > 1 | 68 (53.1) | 0.216 (0.122-0.38) | 1.10 × 10−7 | 0.206 (0.107-0.394) | 1.80 × 10−6 |
| CD20 | IHC score > 1 | 76 (53.9) | 0.532 (0.318-0.89) | .016 | ||
| CD8 | IHC score >1 | 54 (45.4) | 0.792 (0.451-1.39) | .42 | ||
| Stat1 | IHC score > 1 | 33 (42.9) | 0.746 (0.378-1.472) | .4 | ||
| BDCA2 | IHC score > 1 | 24 (35.3) | 1.31 (0.625-2.745) | .47 | ||
| Age | ≥ 45 y | 21 (14.9) | 1.46 (0.758-2.808) | .26 | ||
| Sex | Female | 64 (44.8) | 0.71 (0.426-1.185) | .19 | ||
| Ann Arbor | III-IV | 59 (41.3) | 1.19 (0.719-1.976) | .5 | ||
| B symptom | Present | 47 (40.5) | 1.57 (0.901-2.738) | .11 | ||
| Bulky tumor | Present | 47 (40.5) | 2.35 (1.348-4.104) | .003 | ||
| Extranodal disease | Present | 19 (16.4) | 1.2 (0.56-2.589) | .63 | ||
| No. of nodal sites | ≥ 3 | 61 (48) | 1.18 (0.677-2.054) | .56 | ||
| Inguinal involvement | Present | 9 (9.4) | 1.78 (0.69-4.594) | .23 | ||
| ESR | ≥ 50 | 59 (48.4) | 0.747 (0.428-1.302) | .3 | ||
| Hemoglobin | ≥ 10.5 g/dL | 105 (78.4) | 0.783 (0.428-1.432) | .43 | ||
| White cell count | > 15 × 109/L | 27 (19.9) | 1.98 (1.107-3.527) | .021 | 1.8 (0.912-3.559) | .09 |
| Lymphocyte count | ≥ 0.6 × 109/L or ≥ 8% | 103 (82.4) | 0.886 (0.455-1.725) | .72 | ||
| Albumin level | ≥ 40 g/L | 47 (45.6) | 0.337 (0.169-0.668) | .002 | ||
| Variable . | Class/cutoff . | n (%) . | Univariate analysis* . | Multivariate analysis† . | ||
|---|---|---|---|---|---|---|
| Hazard ratio of event risk . | P . | Hazard ratio of event risk . | P . | |||
| Bcl2‡ | IHC score > 0 | 53 (39.8) | 1.22 (0.726-2.034) | .46 | ||
| EBV‡ | IHC score > 0 | 29 (20.4) | 0.668 (0.338-1.319) | .24 | ||
| PD1 | IHC score > 1 | 8 (18.2) | 0.627 (0.179-2.193) | .46 | ||
| CD57 | IHC score > 1 | 42 (35.3) | 1.42 (0.81-2.493) | .22 | ||
| FOXP3 | IHC score > 1 | 97 (73.5) | 0.555 (0.32-0.966) | .037 | ||
| Tryptase | IHC score > 1 | 75 (54) | 0.892 (0.537-1.482) | .66 | ||
| TIA-1 | IHC score > 2 | 34 (24.5) | 1.16 (0.654-2.063) | .61 | ||
| Granzyme B | IHC score > 1 | 40 (36.4) | 1.25 (0.707-2.201) | .45 | ||
| PS100 | IHC score > 1 | 77 (53.8) | 0.808 (0.49-1.33) | .4 | ||
| TopoIIa‡ | IHC score > 1 | 66 (49.3) | 1.75 (1.048-2.918) | .032 | ||
| BFAR‡ | IHC score > 1 | 84 (70.6) | 0.756 (0.416-1.372) | .36 | ||
| Smad4 | IHC score > 0 | 20 (29.4) | 0.742 (0.345-1.597) | .45 | ||
| Bcl11A | IHC score > 1 | 68 (53.1) | 0.216 (0.122-0.38) | 1.10 × 10−7 | 0.206 (0.107-0.394) | 1.80 × 10−6 |
| CD20 | IHC score > 1 | 76 (53.9) | 0.532 (0.318-0.89) | .016 | ||
| CD8 | IHC score >1 | 54 (45.4) | 0.792 (0.451-1.39) | .42 | ||
| Stat1 | IHC score > 1 | 33 (42.9) | 0.746 (0.378-1.472) | .4 | ||
| BDCA2 | IHC score > 1 | 24 (35.3) | 1.31 (0.625-2.745) | .47 | ||
| Age | ≥ 45 y | 21 (14.9) | 1.46 (0.758-2.808) | .26 | ||
| Sex | Female | 64 (44.8) | 0.71 (0.426-1.185) | .19 | ||
| Ann Arbor | III-IV | 59 (41.3) | 1.19 (0.719-1.976) | .5 | ||
| B symptom | Present | 47 (40.5) | 1.57 (0.901-2.738) | .11 | ||
| Bulky tumor | Present | 47 (40.5) | 2.35 (1.348-4.104) | .003 | ||
| Extranodal disease | Present | 19 (16.4) | 1.2 (0.56-2.589) | .63 | ||
| No. of nodal sites | ≥ 3 | 61 (48) | 1.18 (0.677-2.054) | .56 | ||
| Inguinal involvement | Present | 9 (9.4) | 1.78 (0.69-4.594) | .23 | ||
| ESR | ≥ 50 | 59 (48.4) | 0.747 (0.428-1.302) | .3 | ||
| Hemoglobin | ≥ 10.5 g/dL | 105 (78.4) | 0.783 (0.428-1.432) | .43 | ||
| White cell count | > 15 × 109/L | 27 (19.9) | 1.98 (1.107-3.527) | .021 | 1.8 (0.912-3.559) | .09 |
| Lymphocyte count | ≥ 0.6 × 109/L or ≥ 8% | 103 (82.4) | 0.886 (0.455-1.725) | .72 | ||
| Albumin level | ≥ 40 g/L | 47 (45.6) | 0.337 (0.169-0.668) | .002 | ||
Univariate Cox proportional hazards regression model.
Cox proportional hazards regression model after backward stepwise variables selection procedure.
Bcl2, EBV, Topoisomerase-2, and BFAR immunostainings were analyzed on Reed-Sternberg cells, whereas the results mentioned for all other antibodies were analyzed on reactive cells.
Next, we applied univariate analysis to the prognostic impact of cHL classical prognostic parameters (Tables 2,3). The prognostic influence of IHC markers, adjusted for classical prognostic factors, was assessed in multivariate analysis by the Cox proportional hazards model. TIA-1, CD20, BCL11A, topo-IIa, age older than 45 years, female sex, and high leukocyte count remained as informative parameters after multivariate analysis of OS (Table 2). Multivariate analysis of EFS led to a final model including only BCL11A and leukocyte count as informative predictors of EFS (Table 3). BLC11A was the most powerful predictive marker for EFS (Figure 3D), was correlated with the IPS score (Table 1), and also had a predictive value within the groups of patients with IPS less than 3 and IPS more than 3 (Figure 3E,F).
The signature of EBV+ cHL tissues is enriched in genes characteristic of Th1 and antiviral responses
We next searched for a signature of EBV+ samples that could indicate a specific immune reaction within cHL tissues.
Supervised analysis was applied to identify genes differentially expressed between EBV+ (n = 18) and EBV− (n = 35) cHL samples. With a 0.01% risk, 425 probe sets corresponding to 357 unique genes/ESTs were identified (Table S3), including 422 probe sets (354 unique genes) overexpressed in EBV+ samples and 3 probe sets (3 unique genes) overexpressed in EBV− samples. The resulting LDA-based classification of samples is shown in Figure 2B with a correct classification rate of 96% (P < .001, Fisher exact test).
The genes overrepresented in the EBV+ signature belonged to several significant functional categories including “innate immune response” (GSEA, P = .018), “immune response” (GSEA, P = .005), and “defense response” (GSEA, P = .007). EBV+ cHL had higher levels of expression of IFNG and several IFNG-inducible genes (GBP1, CXCL9, CXCL10, CXCL11, STAT1, IFIT5), and of various antiviral molecules, either IFN-induced (PLSCR1, IFIH1) or involved in the innate antiviral response such as IFIH1 (MDA5), TLR1, TLR8, OAS1, and IVNS1ABP.
EBV+ cHL samples were also associated with higher expression of genes related either to activated T cells such as SLAMF7, SLAMF8, TNFSF10 (TRAIL), and CD38, or to macrophages such as CXCL9, CXCL10, STAT1, CD14, CD68, cathepsins (CTSD/B/S), CASP1, C3AR1, CCL5 (RANTES), FCGR1A (CD64), CD163, TNFSF13 (APRIL), CCR1, and complement factors.
Using GSEA based on previously published T cell–related gene sets, we found that the gene signature of EBV+ cHL tissues was significantly enriched in genes up-regulated in Th1 T cells (GS9 and GS11, P < .001; GS10, P = .063). In contrast, we found no significant association between our EBV signature and the different gene sets overexpressed in Th2 (GS12 to GS14) T-cell subsets.
The differential expression of 2 selected genes of the signature, IFIH1 (MDA5) and IFNG, was validated using RQ-PCR. The levels of mRNA measured by RQ-PCR confirmed those observed with the DNA microarrays, that is, a higher level of expression in the 4 EBV+ samples compared with the 3 EBV− samples for IFIH1 (MDA5) (fold change = 5.1; P = .12, Student t test) and IFNG (fold-change = 2.6; P = .008, Student t test).
Nodular sclerosis and mixed cellularity cHL samples have distinct molecular features
We next compared the gene expression profiles of cHL samples with respect to their histologic type. A total of 76 probe sets corresponding to 67 unique genes/ESTs was differentially expressed between the 42 NS cHL samples and the 17 MC cHL samples. Sixty-four probe sets, representing 55 unique genes, were overexpressed in NS samples, and 12 probe sets, representing 12 unique genes, were overexpressed in MC samples (Table S4). The resulting LDA-based classification of samples is shown in Figure S1.
Among the 55 genes overexpressed in NS cHL, ontology analysis revealed an overrepresentation of genes related to extracellular matrix (CALD1, COL6A3, ITGB1, ITGAV), and to nonspecific cellular functions (PDGFRA, THY 1, RHOC). Conversely, the functional categories of the genes overexpressed in MC cHL were reflective of lymphocyte activation (TLR8, CD38, SLAMF7).
The stroma of H/TCRBCL is highly enriched in PD-1+-reactive T cells that do not express usual Tfh or Treg markers
We chose to include H/TCRBCL samples as controls because they represent the only lymphoma type that harbors an equivalent amount of reactive T cells and macrophages as found in cHL. Thus the differences evidenced by gene expression profiling are expected to be due mainly to stromal components, rather than to the nature of neoplastic cells. We searched for stromal markers that could reliably differentiate cHL and H/TCRBCL samples.
Global clustering (Figure 1A) suggested strong differences between the 2 entities. We applied supervised analysis to search for a gene expression signature that would discriminate between the 5 H/TCRBCL and the 63 cHL samples. With a 0.01% risk, we identified 666 probe sets corresponding to 614 unique genes/ESTs as differentially expressed (Table S5). The resulting LDA-based classification of samples is shown in Figure S2.
Differentially expressed probe sets included 310 probe sets, corresponding to 276 unique genes overexpressed in H/TCRBCL. These overexpressed genes belonged to miscellaneous functional categories including genes related to macrophages (STAT1, LAMP1), Th1 response (IFNG, CCR5), and MUC1, coding for a protein closely related to the epithelial membrane antigen. One of the most significantly overrepresented genes in H/TCRBCL was PDCD1/PD-1, which codes for a lymphocyte inhibitory receptor. The fact that PD-1 was recently reported by others as well as ourselves to be expressed at high levels by T-follicular helper (Tfh) cells38,42 led us to verify the RNA levels of other known Tfh markers, namely CXCL13 and ICOS43 : both were significantly lower in H/TCRBCL samples. In addition, we performed a GSEA analysis based on previously published Tfh-related gene set lists.30 We found no significant association between any of the different gene sets overexpressed in Tfh (GS1 to GS8) and the H/TCRBCL gene signature.
Conversely, compared with cHL, H/TCRBCL samples showed underexpression of PTEN, SOCS2, SOCS5, and genes related to the TGFB pathway (TGFBR3, the TGFB inducible genes KLF10 [TIEG] and TGFB1I1). Of note, TNFSFR8 (CD30) was also, as expected, underexpressed in H/TCRBCL.
To validate the H/TCRBCL gene signature, PD-1 and MUC1 protein expression were analyzed using IHC on the validation set of cHL samples and on a distinct set of 13 H/TCRBCL paraffin samples. As expected from DNA microarray data, a variable number of malignant B cells in 8 of 13 H/TCRBCL cases stained for MUC1, whereas all cHL cases were negative. The PD-1 antibody strongly stained (IHC score 2 or 3) the vast majority of reactive T lymphocytes in 13 of 13 H/TCRBCL cases (Figure 4G,H), whereas only 8 of 44 informative cHL cases displayed a 2 or 3 IHC score of PD-1 staining in reactive lymphocytes (Figure 4F). These data therefore confirm our mRNA data (P < .001).
We then determined at the protein level whether PD-1+ stromal T cells in H/TCRBCL could be derived from either Tfh or Treg cells, using their classical markers CXCL13 and FOXP3, respectively. Only a weak or absent CXCL13 IHC staining was evidenced in the 13 H/TCRBCLs analyzed. In these 13 cases, CXCL13+ reactive cells were much sparser than PD-1+ cells. FOXP3 immunodetection was virtually negative in 13 of 13 H/TCRBCL paraffin samples (< 5% of positive reactive lymphocytes). These results thus gave no clue as to a Tfh or Treg derivation of PD-1+ cells in H/TCRBCL.
Discussion
There have been few gene expression analyses of nonmicrodissected cHL tissues.23,24 In a preliminary study in which BCL11a was not analyzed, we observed that the B-cell marker CD22 and several genes regulating apoptosis were overexpressed in patients with favorable outcome, whereas genes involved in extracellular matrix remodeling had an adverse influence.23 Accordingly, the major finding of the present study is the favorable role of reactive B cells within cHL tissues. Also in agreement with our previous data is the influence of apoptotic and matrix remodeling genes that we describe herein. The present findings are also partly in line with a recent study suggesting that favorable outcome of cHL patients may be correlated with overexpression of genes specific for particular B-cell subpopulations, whereas high topo-IIα expression may be linked to poor survival.24 However, there is only limited overlap between the latter report and the present one, which might be explained by major methodologic differences. In fact, the present study analyzed patients with either localized or advanced disease and used Affymetrix oligonucleotide microarrays containing approximately 16 000 genes. In contrast, the Spanish study24 analyzed only advanced-stage patients and used spotted microarrays containing fewer than 10 000 genes in which BCL11A was not included.
We confirm herein that both low number of FOXP3+ cells and high number of TIA-1+ cytotoxic lymphocytes correlate with a poor outcome, as previously reported.19-22 We also confirm that cHL patients with high topo-IIα expression have inferior survival, and that the mere count of granzyme-B+ cells is devoid of predictive significance as reported elsewhere.16,21,24 These confirmations are of particular value, because we used an independent and previously unreported set of patients, different antibodies, and counting methods.
BCL11A is a transcription factor expressed in normal pDCs and B cells40,41 and in primary mediastinal B-cell lymphoma cells.44 We report herein that BCL11A expression has a high favorable prognostic value when localized in reactive pDCs and B cells, but not in RS cells. This led us to analyze separately the pDC and B-cell populations using antibodies against BDCA2 and CD20, respectively. The amount of CD20+ reactive cells was correlated with better outcome but was much less significant than BCL11A, whereas the number of BDCA2+ cells showed no significance. An interpretation of these findings could be that cooperation between pDCs and B cells within the tumor is required for an efficient antitumoral response. BCL11A+ pDCs may be implicated in the antitumor response, because it has been reported that EBV-stimulated pDCs can produce IFNA, promote activation of natural killer (NK) cells, and stimulate IFNG-producing CD3+ cells.45 This may not be contradictory with the lack of prognostic value of BDCA2 staining compared with BCL11A, because different pDC subsets may be recognized by either antibody.
Our major finding regarding the association of high intratumoral B-cell counts with better outcome is in accordance with the excellent prognosis of the particular variant of cHL referred to as “nodular, lymphocyte-rich cHL” (NLRHL), which contains important B-cell amounts.46 The histologic type of our B cell–rich cHL cases was either MC or NS, but we noticed that RS cells were often located in the outer rim of B-cell nodules, a feature reminiscent of the localization of RS cells in NLRHL.46 This raises the hypothesis that our B cell–rich cases could correspond to intermediate points on a putative spectrum spanning from NLRHL to NS or MC, depending on the B-cell content of the lesions.
H/TCRBCL and cHL lesions share morphologic similarities,6 and the differential diagnosis of H/TCRBCL versus cHL can be a pitfall for pathologists.47 We report herein that the majority of reactive T cells in H/TCRBCL express the PD-1 inhibitory receptor, whereas such PD-1+ T cells are sparser in cHL. This implies that PD1 immunodetection on paraffin sections can be helpful for the diagnosis of H/TCRBCL. Sustained PD1 expression in H/TCRBCL may be induced by IFNG, whose transcripts were also more abundant in H/TCRBCL than cHL tissues. Because it is known that macrophages produce high amounts of PDL-1,48 the natural PD-1 ligand, T cells in H/TCRBCL might be submitted to a continuous PD-1 inhibitory signal, and thus unable to react against tumor cells, which may explain the specific aggressive behavior of H/TCRBCL.47
To date, only a few differences have been found in the microenvironment of EBV+ versus EBV− RS cells, such as an increased expression of IP10/CXCL10,11 and CCL20.13 The present study demonstrates that EBV+ and EBV− cHL tissues can be clearly separated from each other by a robust gene signature involving innate immunity and antiviral responses in EBV+ tumors. In fact, the EBV+ cHL subset overexpressed antiviral genes such as IVNS1ABP (NS1BP), PLSCR1, and OAS, together with the pattern recognition receptor TLR8 and the MDA5 helicase, which are both involved in the recognition of viruses of various structures.49 Signaling through TLRs and helicases is known to converge toward induction of interferons, which are the principal cytokines mediating innate immunity against viral infection.49
Our gene profiling data also provide evidence of intratumoral Th1 activity in EBV+ cHL. The molecular profile of EBV+ tumors (simultaneous overexpression of IFNG, CXCL9, CXCL10, and CXCL11/ITAC) suggests that this Th1 reaction could be orchestrated by IFNG, which is capable of inducing expression of not only CXCL10, but also CXCL9/MIG and CXCL11/ITAC, both also known as CXCR3 ligands and potential chemoattractants of Th1 lymphocytes. Of note, CXCL9 and CXCL10 were also predominantly expressed in EBV+ cHL samples in a recent gene profiling analysis, but without any evidence of Th1 or antiviral reaction.13 This discrepancy may be because the latter study was focused on cHL samples of the NS type only.13 It is noteworthy that our patients with EBV+ cHL did not have a better outcome, thereby suggesting that the described intratumoral immune reaction is inadequate to eliminate tumor cells. Nonetheless, our data raise the possibility that it could be further stimulated to design future therapies.
In conclusion, the present report suggests that B cells have an important influence on cHL outcome and that EBV favors a Th1 reaction in the cHL microenvironment. The latter point could be considered as a basis for targeted immunotherapies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Special thanks to Coralie Attias for technical assistance and Karen Leroy and the Ligue Nationale Contre le Cancer (Programme Carte d'Identité des Tumeurs) for their help in the analysis of H/TCRBCL samples. We are grateful to Karen Pulford for the gift of the anti-BCL11A antibody.
We thank all the members of the GELA network who provided tumor samples or clinical data that could not be included in the study due to technical concerns, especially Gilles Salles, Françoise Berger, Philippe Gaulard, Josette Briére, Anne Janin, Pauline Brice, Pierre Brousset, Jean-François Michiels, Catherine Chassagne, Catherine Seban, Gérard Sebahoun, Frédérique Retornaz, Celia Salanouba, Gérard Michel, and Hervé Chambost.
This work was supported by GELA and by grant 3684XA0431F from the Association de la Recherche contre le Cancer (Villejuif, France) and by the Programme Hospitalier de Recherche Clinique (PHRC 2004).
Authorship
Contribution: B.C., B.E., and L.X. analyzed TMA results; F.B. and P.F. performed and analyzed cDNA array experiments; A.S., J.M.P., M.C.C., F.M., O.C., T.P., T.M., A.V., P.F., and R.B. collected tissue samples and clinical data; D.B. supervised statistical analyses; D.O. supervised PCR experiments and helped to discuss the results; L.X. and F.B. wrote the paper; and L.X. designed the research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Luc Xerri, Department of Bio-Pathology, Institut Paoli-Calmettes, 232 Boulevard de Sainte Marguerite, BP 156, 13273 Marseille Cedex 9, France; e-mail: xerril@marseille.fnclcc.fr.
References
Author notes
*B.C. and F.B. contributed equally to this work.
†D.B. and D.O. contributed equally to this work.