Abstract
Clinical trials with antiangiogenic agents have not been able to validate plasma or serum levels of angiogenesis regulators as reliable markers of cancer presence or therapeutic response. We recently reported that platelets contain numerous proteins that regulate angiogenesis. We now show that accumulation of angiogenesis regulators in platelets of animals bearing malignant tumors exceeds significantly their concentration in plasma or serum, as well as their levels in platelets from non–tumor-bearing animals. This process is selective, as platelets do not take up a proportional amount of other plasma proteins (eg, albumin), even though these may be present at higher concentrations. We also find that VEGF-enriched Matrigel pellets implanted subcutaneously into mice or the minute quantities of VEGF secreted by microscopic subcutaneous tumors (0.5-1 mm3) result in an elevation of VEGF levels in platelets, without any changes in its plasma levels. The profile of other angiogenesis regulatory proteins (eg, platelet-derived growth factor, basic fibroblast growth factor) sequestered by platelets also reflects the presence of tumors in vivo before they can be macroscopically evident. The ability of platelets to selectively take up angiogenesis regulators in cancer-bearing hosts may have implications for the diagnosis and management of many angiogenesis-related diseases and provide a guide for antiangiogenic therapies.
Introduction
Platelets play a major role in hemostasis, as well as in tissue repair, maintenance of endothelium, and vascular tone. They may also facilitate delivery of angiogenesis regulators and other growth factors to sites of pathologic angiogenesis.1,2 Correlative studies suggest that increasing platelet counts may be linked to tumor progression.3,4
We and others have reported previously that platelets contain several proteins that regulate angiogenesis.5-8 We have now discovered that the platelet concentrations of angiogenesis regulatory proteins, although relatively constant and stable under physiologic conditions, are modified by and reflect the presence of a tumor. In the presence of microscopic (< 1.0 mm) tumors in a mouse, circulating platelets sequester increased concentrations of angiogenesis regulatory proteins, without a corresponding elevation in their plasma levels. The uptake of angiogenesis regulatory proteins is selective, as platelets do not take up other plasma proteins. For example, although albumin is present in plasma at much higher concentrations than, for example, vascular endothelial growth factor (VEGF), albumin levels in platelets do not differ in the presence or absence of tumors.
In this study, we used a high-throughput surface-enhanced laser desorption/ionization–time-of-flight mass spectrometry (SELDI-ToF MS), which permitted a rapid analysis of a large number of samples in a highly efficient and reproducible manner.9,10 In this open-ended proteomic comparison of platelets from tumor-bearing and non–tumor-bearing animals, the majority of identified differentially expressed proteins were angiogenesis regulators. Our subsequent studies revealed that platelets can sequester selected proteins either in vitro or in vivo. The platelet angiogenesis proteome reflected the presence of dormant, microscopic-sized tumors in mice months before these tumors can be detectable by conventional methods, and before the angiogenesis regulatory proteins could be detected in plasma, suggesting that analysis of the “platelet angiogenesis proteome” may be used to detect tumor establishment or recurrence before a patient is symptomatic, providing an opportunity for early therapeutic intervention with nontoxic therapies.
Methods
In vitro uptake of angiogenesis regulators by freshly isolated platelets
Platelet-rich plasma (PRP) was isolated from the blood of healthy human volunteers by centrifugation of citrated whole blood at 180g for 20 minutes. The PRP was then transferred to a fresh polyethylene tube and incubated on a gentle rocker at room temperature for 1 hour with increasing concentrations of human recombinant endostatin (EntreMed, Rockville, MD) or basic fibroblast growth factor (bFGF; R&D Systems, Minneapolis, MN). Platelets were gently resuspended in Tyrode buffer containing 1 U/mL prostaglandin E2 and 1% Triton X-100 to remove the membrane fraction of platelets. After a second centrifugation at 800g, the pellets (platelets) were lysed for standard sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) analysis using 50 mM Tris HCl, 100 to 120 mM NaCl, 5 mM ethylenediaminetetraacetic acid, 1% Igepal, and Protease Inhibitor Tablet (complete TM mixture; Roche Diagnostics, Indianapolis, IN). Protein concentrations were equalized using the standard Bradford method (Bio-Rad, Hercules, CA) and an equivalent amount of protein standards or platelet protein lysate were mixed with sample buffer (Invitrogen, Carlsbad, CA) and loaded onto a 12% SDS-polyacrylamide gel (Invitrogen). After transfer to a polyvinylidene difluoride membrane (Millipore, Billerica, MA), the mixture was blocked with 7% milk and incubated with the following antibodies: antihuman endostatin (courtesy of Kashi Javaherian, Children's Hospital, Boston, MA) or antihuman bFGF (1:1000; Upstate Biotechnology, Charlottesville, VA). Positive signals were then detected using a Super Signal West Pico Chemiluminescence Kit (Pierce Chemical, Rockford, IL) and autoradiography.
To localize the proteins taken up by platelets, we used immunofluorescence microscopy. To this end, platelets in the form of PRP were incubated with 200 ng/mL to 2 μg/mL His-Tag endostatin (generous gift from Kashi Javaherian) for 1 hour at 37°C. Platelets were then fixed in solution with 4% paraformaldehyde in phosphate-buffered saline (PBS) at room temperature, plated onto coverslips in 24-well plate, permeabilized with 0.1% Triton X-100, and blocked for nonspecific binding sites by incubating for 1 hour in PBS containing 1% normal goat serum. The fixed platelets were then incubated overnight at 4°C with mouse anti His-Tag monoclonal primary antibody (Abcam, Cambridge, MA) diluted 1:500 in PBS, washed several times with PBS, and then incubated with Alexa 488 antimouse secondary antibody (Invitrogen) and diluted 1:500 for 1 hour at room temperature. Three controls (processed identically to treated specimens) were prepared for this experiment. The first one consisted of platelets not incubated with His-Tag endostatin. The second control consisted of substituting the primary antibody His-Tag by mouse IgG2a isotype control at the same dilution of His-Tag antibody. The third control consisted of processing the platelets omitting the primary antibody. Platelets were imaged using a Leica TCS SP2 AOBS confocal system (Leica, Wetzlar, Germany) fitted to a DM IRE2 inverted microscope with a 100× objective and 488-nm argon diode laser.
Analysis of sera obtained from VEGF- or bFGF-“loaded” platelets
A total of 1 mL PRP was isolated from fresh blood of human volunteers and incubated on a gentle rocker at room temperature for 1 hour with 600 ng/mL recombinant human VEGF or bFGF. After incubation, 1 unit of either thrombin or adenosine diphosphate (ADP) was added to 1 mL of each sample for 5 minutes, and clot and sera were separated by centrifugation. The serum was then analyzed for VEGF and bFGF by standard enzyme-linked immunosorbent assays (ELISAs; R&D Systems). In an alternative experiment geared to evaluate the completion of protein release, 1 mL PRP was incubated with 100 ng bFGF for 45 minutes on gentle rocker at room temperature and spun at 150g. The plasma containing excess of the protein was then removed, and platelets resuspended in 1 mL saline to which 1 unit of thrombin or 20 mM ADP was added. The sample was then spun again at 900g to pellet the platelets and the supernatant and platelet pellet analyzed using ELISA.
Localization of VEGF in activated platelets
Antihuman VEGF mouse monoclonal antibody was obtained from BD Biosciences (San Jose, CA) and used at 5 μg/mL. Rabbit anti-β1 tubulin antiserum (a kind gift from Nicholas Cowan, New York University Medical Center, New York, NY) was used at 1:1000 dilution. Alexa 488 antirabbit and Alexa 568 antimouse secondary antibodies with minimal cross-species reactivity were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA). Cells were analyzed on a Zeiss Axiovert 200 microscope (Carl Zeiss, Thornwood, NY) equipped with a 100× objective (NA 1.4), and a 100-W mercury lamp. Images were acquired with an Orca II cooled charged coupled device camera (Hamamatsu, Bridgewater, NJ). Electron shutters and image acquisition were under the control of Metamorph software.
Resting platelets were fixed for 20 minutes in suspension by the addition of 3.7% formaldehyde. The platelets were attached to polylysine-coated coverslips placed in wells of a 12-well microtiter plate and centrifuged at 250g for 5 minutes. For agonist-induced activation, platelets were sedimented onto coverslips in an identical fashion, and 1 U/mL thrombin was added for 5 minutes. Activated platelets were fixed for 20 minutes in 3.7% formaldehyde. Samples were permeabilized in Hank balanced solution containing 0.5% Triton X-100 and washed with PBS. Specimens were blocked overnight in PBS plus 1% bovine serum albumin, incubated in primary antibody for 2 to 3 hours at room temperature, washed, treated with appropriate secondary antibody for 1 hour, and again washed extensively in 1% PBS. Primary antibodies were used at 1 μg/mL in PBS plus 1% bovine serum albumin and secondary antibodies at a 1:500 dilution in the same buffer. Controls were processed identically except for omission of the primary antibody.
Platelet uptake of 125I-labeled VEGF in vivo
VEGF was iodinated using a previously established method.11 Briefly, Iodo Beads (Pierce Chemical) pre-equilibrated with 10 μL sodium phosphate buffer (0.2 M NaHPO4, pH 7.2) were incubated with 10 μg of carrier-free recombinant mouse VEGF (R&D Systems) and 1 mCi 125I. The sample was further diluted with 150 μL sodium phosphate buffer and passed through a 15-mL, pre-equilibrated NAD 5 column (GE Healthcare, Little Chalfont, United Kingdom) containing 0.2% gelatin in PBS. Fifteen fractions of 250 μL were then collected. Radioactivity in each fraction was quantified on a Gamma 5000 Beckman Iodine 125 (Beckman Coulter, Fullerton, CA), and the 2 fractions containing the greatest quantity of 125I-labeled VEGF (500 μL in total) were combined for use in the Matrigel assay on the day of the experiment. Briefly, the left flanks of C57/BL6 mice were shaved 1 day before Matrigel pellet implantation to avoid any interference by minor cutaneous inflammatory reaction. On the day of the experiment, 500 μL 125I-VEGF in buffer was mixed with 500 μL growth factor-free Matrigel (BD Biosciences), and 100 μL of this mixture was injected subcutaneously into the left flank of each mouse. Three days later, the mice were anesthetized using inhalational anesthesia (2% isofluorane in 1 L oxygen) and 1 mL whole blood was drawn into a citrated syringe (1% sodium citrate final concentration, 1/10 vol/vol) by direct cardiac puncture without opening the chest cavity.
The platelets were isolated in 2 centrifugation steps: the first at 180g to isolate PRP, followed by centrifugation at 400g to yield a platelet pellet and a platelet-poor plasma fraction. The radioactivity of each platelet sample was quantified on a gamma counter. The value was corrected for differences in tissue weight and expressed as counts per minute per gram of tissue.
Changes in platelet angiogenesis proteome in presence of tumor xenografts
We used previously established dormant and angiogenic xenografts of human liposarcoma (SW872).12-14 These were clones from microscopic, dormant, nonangiogenic tumors, or from rapidly growing, angiogenic tumors in immunodeficient mice. The angiogenic clone is already angiogenic at the time of implantation and expands rapidly. The nonangiogenic tumor subclone undergoes a switch to an angiogenic phenotype at approximately 133 days (median ± 2 weeks) and expands rapidly thereafter. The growth curves parallel the growth curve of angiogenic clones after the switch. Even though the tumor cell proliferation rates in vivo and in vitro are equivalent for both clones, the tumor cell apoptotic rates are higher in the nonangiogenic clone. Interestingly, the nonangiogenic clones secrete large amounts of VEGF and bFGF in tissue culture.12
All cell lines were cultured in Dulbecco modified Eagle medium (DMEM) containing 5% heat-inactivated fetal bovine serum (HyClone Laboratories, Logan, UT), 1% antibiotics (penicillin, streptomycin), and 0.29 mg/mL l-glutamine in a humidified 5% CO2 incubator at 37°C. For injections into mice, 80% to 90% confluent tumor cells were rinsed in PBS (Sigma-Aldrich, St Louis, MO), briefly trypsinized, and suspended in serum-free DMEM. The cells were washed twice in DMEM, and their final concentration was adjusted to 5 × 106 viable cells/200 μL.
Six-week-old male SCID mice from the Massachusetts General Hospital (Boston, MA) were injected subcutaneously in the flanks with 5 × 106 cells (in 0.2 mL) from a single clone. All experiments were conducted in compliance with Boston Children's Hospital guidelines using protocols approved by the Institutional Animal Care and Use Committee.
Platelet, plasma and tumor processing, and protein profiling
A total of 1 mL of blood was collected from anesthetized mice by direct cardiac puncture into citrated polyethylene tubes (1% sodium citrate final concentration, 1/10 vol/vol) and centrifuged immediately at 200g. The upper phase (∼ 500 μL), ie, the PRP, which still contained small platelets and platelet fragments, was then transferred into a fresh tube, and platelets were separated by a second centrifugation at 800g. The isolated platelet pellet and platelet poor plasma supernatant were analyzed separately using SELDI-ToF technology (Ciphergen, Fremont, CA) according to previously published methodology.15,16
The identities of some of the factors were confirmed by specific antibody immunoprecipitation. Platelet extracts were incubated with antibodies coupled to Reacti-gel (Pierce Chemical) in the presence of 1% PEG400 0.1% Tween-20, 0.15 M NaCl, 50 mM Tris-HCl, pH 7.5 (TTS). After extensive washing with PEG TTS and 1 M urea 0.1% 3-cholamidopropyl)dimethylamonio-1-propyl sulfonate, 50 mM Tris-HCl, pH 7.5, the immunoprecipitation complexes were analyzed on Normal Phase ProteinChip arrays.
Results
In vitro uptake of angiogenesis regulators by freshly isolated platelets
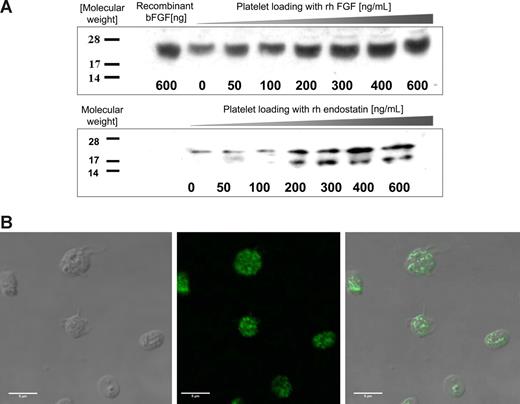
Platelets incubated with increasing concentrations of human recombinant endostatin or bFGF take up the protein in a dose-dependent manner (Figure 1A). Because the platelet surface expresses a high level of nonspecific protein binding sites, the platelet membrane fraction was removed by centrifugation with 1% Triton X-100 before protein lysis. A semiquantitative SDS-PAGE analysis reveals that the proteins are loaded into the cytoplasmic compartments.
Angiogenesis regulators are taken up by platelets in vitro. (A) PRP was incubated with increasing concentrations of recombinant human bFGF (rhbFGF) or recombinant human endostatin (rh endostatin) for an hour. The platelets were then isolated by sequential centrifugation, washed, treated with 1% Triton X to remove the membrane, and lysed for SDS-PAGE analysis. Standard Western blots using antihuman endostatin and antihuman bFGF antibodies reveal a dose-dependent increase in the respective proteins in the cytoplasmic fraction of fresh platelets. (B) To establish the localization of proteins taken up by platelets, the platelets were incubated with His-tag labeled endostatin, fixed using paraformaldehyde, and anti-His antibody was used to separate the platelet endogenous endostatin (not labeled) from the endostatin “loaded” into platelets (fluorescent label). (Left) DIC image of the platelets. (Middle) The fluorescent label of the His tag. (Right) The overlay of the 2 images. The pattern of the fluorescent signal indicates that endostatin is taken up into the granules of platelets rather than remaining membrane-associated.
Angiogenesis regulators are taken up by platelets in vitro. (A) PRP was incubated with increasing concentrations of recombinant human bFGF (rhbFGF) or recombinant human endostatin (rh endostatin) for an hour. The platelets were then isolated by sequential centrifugation, washed, treated with 1% Triton X to remove the membrane, and lysed for SDS-PAGE analysis. Standard Western blots using antihuman endostatin and antihuman bFGF antibodies reveal a dose-dependent increase in the respective proteins in the cytoplasmic fraction of fresh platelets. (B) To establish the localization of proteins taken up by platelets, the platelets were incubated with His-tag labeled endostatin, fixed using paraformaldehyde, and anti-His antibody was used to separate the platelet endogenous endostatin (not labeled) from the endostatin “loaded” into platelets (fluorescent label). (Left) DIC image of the platelets. (Middle) The fluorescent label of the His tag. (Right) The overlay of the 2 images. The pattern of the fluorescent signal indicates that endostatin is taken up into the granules of platelets rather than remaining membrane-associated.
To further confirm that these proteins were taken into the platelets, rather than remain associated with the membrane, we performed immunofluorescence studies. As seen in Figure 1B, platelets incubated with 200 ng/mL endostatin for 1 hour at 37°C take up the protein into the cytoplasmic compartment. A series of Z stack confocal images taken at different focal depths revealed that the nature of the fluorescent signal corresponded to a pattern expected of platelet granules (data not shown). The same set of experiments was performed with His-labeled VEGF164, and the same pattern of granular storage was observed (data not shown).
Analysis of sera obtained from VEGF- and bFGF-“loaded” platelets
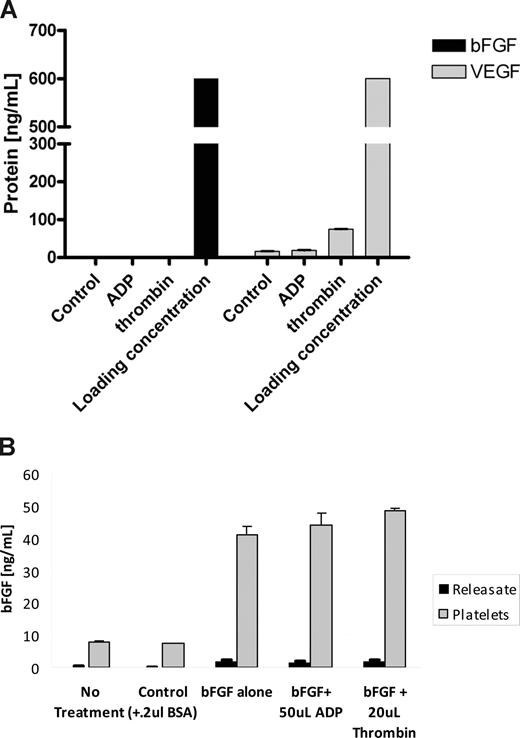
Based on the present understanding of platelet degranulation, we expected a release of angiogenesis regulators from platelets on their activation. Unexpectedly, the stimulation of fresh platelets loaded with 600 ng of either VEGF164 or bFGF with standard platelet activation agonists, such as ADP and thrombin, did not result in significant release of the angiogenesis regulatory proteins into the sera (Figure 2A), and the levels of the proteins did not exceed the levels released from the control (unstimulated) platelets. In the case of bFGF, essentially no protein was released with either agonist, and only a minor portion of VEGF was released with the more potent of the 2 agonists, thrombin. This was further confirmed by analyzing the platelet pellet for residual protein. To this end, PRP was incubated with bFGF, the plasma containing excess of the protein was removed, and platelets resuspended in 1 mL of saline to which 1 unit thrombin or 20 mM ADP was added. The analysis of both the platelet pellet as well as the “releasate” revealed that platelets retained the loaded protein (7-fold enhancement from releasate), and the amount of protein detected in plasma was negligible in comparison. The graph represents a triplicate analysis plus or minus SEM.
Angiogenesis regulators loaded into platelets are not released with agonists of platelet activation. (A) A total of 1 mL PRP was incubated with 600 ng/mL of either VEGF or bFGF for 30 minutes. This resulted in “loading” of these proteins into the α-granules of platelets similarly to Figure 1. After the incubation, the platelets were aggregated using a mild aggregation agonist (ADP), a more potent agonist (thrombin), or spun down without any stimulation (control). The resulting supernatants/sera were then analyzed for VEGF and bFGF using a commercially available ELISA. Neither of the platelet agonists was capable of releasing bFGF from platelets. In the case of VEGF, ADP-induced aggregation failed to release the VEGF and even the more potent aggregation with thrombin resulted in only a modest release of the loaded VEGF. (B) PRP was incubated with 100 ng bFGF/mL on gentle rocker at room temperature for 45 minutes, spun at 150g, and the plasma containing excess of the protein was removed. The platelets were then resuspended in 1 mL saline to which 20 mM ADP or 1 unit thrombin was added. The sample was then spun again at 900g to pellet the platelets, and the supernatant and platelet analyzed using bFGF ELISA. As evident, platelets retained the majority of the loaded protein, and protein was released with either agonist. Both experiments were repeated on 2 separate occasions, and the graph represents the mean of 5 samples per dose level plus or minus SEM. Loading concentration in panel A corresponds to 600 ng VEGF or bFGF per mL.
Angiogenesis regulators loaded into platelets are not released with agonists of platelet activation. (A) A total of 1 mL PRP was incubated with 600 ng/mL of either VEGF or bFGF for 30 minutes. This resulted in “loading” of these proteins into the α-granules of platelets similarly to Figure 1. After the incubation, the platelets were aggregated using a mild aggregation agonist (ADP), a more potent agonist (thrombin), or spun down without any stimulation (control). The resulting supernatants/sera were then analyzed for VEGF and bFGF using a commercially available ELISA. Neither of the platelet agonists was capable of releasing bFGF from platelets. In the case of VEGF, ADP-induced aggregation failed to release the VEGF and even the more potent aggregation with thrombin resulted in only a modest release of the loaded VEGF. (B) PRP was incubated with 100 ng bFGF/mL on gentle rocker at room temperature for 45 minutes, spun at 150g, and the plasma containing excess of the protein was removed. The platelets were then resuspended in 1 mL saline to which 20 mM ADP or 1 unit thrombin was added. The sample was then spun again at 900g to pellet the platelets, and the supernatant and platelet analyzed using bFGF ELISA. As evident, platelets retained the majority of the loaded protein, and protein was released with either agonist. Both experiments were repeated on 2 separate occasions, and the graph represents the mean of 5 samples per dose level plus or minus SEM. Loading concentration in panel A corresponds to 600 ng VEGF or bFGF per mL.
Localization of VEGF in activated platelets
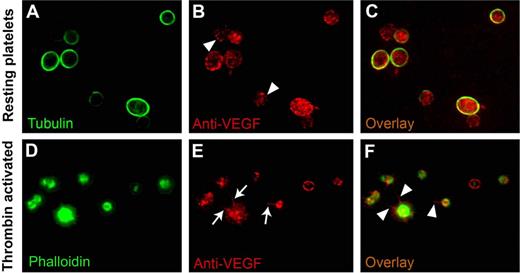
To establish the subcellular localization of VEGF on agonist activation, we used double-label immunofluorescence microscopy on fixed permeabilized platelets stained with antibodies for tubulin or phalloidin and VEGF. As expected, tubulin was concentrated in the marginal microtubule band in a resting platelet, and this structure defined the platelet periphery. In resting platelets, anti-VEGF antibodies consistently labeled punctate, vesicle-like structures distributed throughout the platelet cytoplasm (Figure 3B,C), and the sequential stacking of 4-μm slices of confocal microscope images supported a granular nature of the immunoreactive material.
VEGF localization in resting and activated platelets. Double-label immunofluorescence microscopy on fixed and permeabilized platelets was used to determine the intracellular localization of VEGF in the resting and activated states. In resting platelets, tubulin is concentrated, as expected, in the marginal microtubule band (A). In thrombin-activated platelets, phalloidin stain confirms the morphologic changes expected with activation (ie, the development of filopodia, lamellipodia, and pseudopodia) (D). The VEGF stain labels punctate, vesicle-like structures distributed throughout the platelet cytoplasm in both resting (arrowheads in panel B) and activated platelets (E), consistent with the lack of release of VEGF on activation, as noted in Figure 2. In the activated state, VEGF is redistributed to filopodia and lamellipodia (arrows in panel E and arrowheads in panel F) in contrast to its granular pattern of distribution in resting state (C).
VEGF localization in resting and activated platelets. Double-label immunofluorescence microscopy on fixed and permeabilized platelets was used to determine the intracellular localization of VEGF in the resting and activated states. In resting platelets, tubulin is concentrated, as expected, in the marginal microtubule band (A). In thrombin-activated platelets, phalloidin stain confirms the morphologic changes expected with activation (ie, the development of filopodia, lamellipodia, and pseudopodia) (D). The VEGF stain labels punctate, vesicle-like structures distributed throughout the platelet cytoplasm in both resting (arrowheads in panel B) and activated platelets (E), consistent with the lack of release of VEGF on activation, as noted in Figure 2. In the activated state, VEGF is redistributed to filopodia and lamellipodia (arrows in panel E and arrowheads in panel F) in contrast to its granular pattern of distribution in resting state (C).
Thrombin-induced activation of platelets was documented by the expected change in platelet shape and by the formation of lamellipodia and filopodia. In activated platelets, VEGF remained observable as punctate patterns, consistent with the notion that it remains associated with platelets, even after agonist-induced activation. More importantly, VEGF appeared to be preferentially redistributed along the filopodia and along the periphery of lamellipodia (Figure 3E,F).
Platelet uptake of 125I-labeled VEGF in vivo
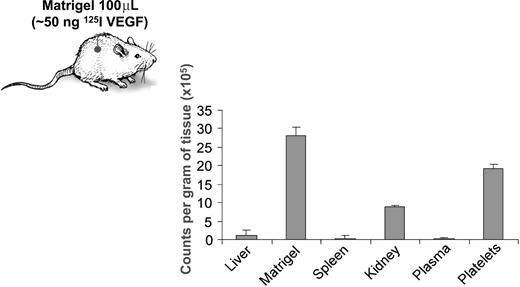
To confirm that the observed ability of platelets to take up proteins is not an in vitro artifact, we implanted Matrigel pellets containing 125I-labeled VEGF (50 ng labeled VEGF per 100 μL Matrigel) subcutaneously into mice. The analysis of the organ distribution of 125I-VEGF revealed that it accumulated in platelets at higher concentrations than in plasma or various organs (Figure 4).
Platelets selectively take up VEGF, without a corresponding increase of the protein in plasma. VEGF protein was labeled with radioactive iodine, and approximately 50 ng of 125I-labeled VEGF in 100 μL of Matrigel was implanted subcutaneously in the left flanks of C57/BL6 mice. Three days later, the mice were killed and 1 mL of citrated blood, as well as liver, kidney, spleen, and Matrigel pellet were collected. The radioactivity of each tissue sample was quantified on a gamma counter, the value corrected for differences in tissue weight, and expressed as counts per minute per gram of tissue. The experiment was repeated on 2 separate occasions with 5 mice per experiment, and the graph represents mean plus or minus SE of 1 of the experiments.
Platelets selectively take up VEGF, without a corresponding increase of the protein in plasma. VEGF protein was labeled with radioactive iodine, and approximately 50 ng of 125I-labeled VEGF in 100 μL of Matrigel was implanted subcutaneously in the left flanks of C57/BL6 mice. Three days later, the mice were killed and 1 mL of citrated blood, as well as liver, kidney, spleen, and Matrigel pellet were collected. The radioactivity of each tissue sample was quantified on a gamma counter, the value corrected for differences in tissue weight, and expressed as counts per minute per gram of tissue. The experiment was repeated on 2 separate occasions with 5 mice per experiment, and the graph represents mean plus or minus SE of 1 of the experiments.
Changes in the platelet angiogenesis proteome in presence of tumor xenografts
We used an Expression Difference Mapping system (Ciphergen) to characterize, in an open-ended fashion, the complete proteomic profile of platelets from tumor-bearing mice vs non–tumor-bearing controls (at 32 days after tumor implantation).
We used dormant subclones of human liposarcoma (SW872), which are known to secrete large amounts of VEGF and bFGF in tissue culture, but remain dormant for up to 133 days.12,14 We compared the platelet and plasma proteomes of 5 mice injected with either 200 μL serum free media (vehicle) or a cell suspension of 5 × 106 cells of the nonangiogenic or angiogenic clones of the liposarcoma cell line. The experiment was repeated twice for comparison of expression maps from separate analyses. Among the proteins identified in this analysis as differentially expressed in platelets of tumor-bearing mice vs tumor-free, sham-operated mice, angiogenesis regulators were prominently represented. Figure 5 depicts a representative analysis of a platelet angiogenesis proteome in gel view format, and Figure S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article) provides a semiquantitative analysis of the respective protein peaks. The levels of pro-angiogenic growth factors, such VEGF, bFGF, and platelet-derived growth factor (PDGF), tended to be higher in the platelets of tumor-bearing mice compared with tumor-free sham-operated mice (Figure 5). The levels of VEGF, bFGF, and PDGF in platelets from normal animals were lower and comparable with those in plasma. At 32 days after tumor implantation, the nonangiogenic dormant liposarcoma (∼ 1 mm3) tumors are approximately 100 times smaller than the angiogenic liposarcoma (∼ 1 cm3), yet platelets of the dormant tumor-bearing mice contained detectable levels of angiogenesis regulatory proteins. In contrast, there were negligible levels of these proteins in the plasma of either mouse. In mice with nonangiogenic dormant tumors, the pattern of elevated positive angiogenesis regulators tends to be accompanied by an elevation of the endogenous inhibitor endostatin (Figure 5). In contrast, platelets of mice bearing angiogenic tumors exhibit a decrease in negative angiogenesis regulators, such as endostatin (Figure 5). This suggests that, in the “angiogenic” tumors, the overall balance of angiogenesis regulators may be tipped toward a more pro-angiogenic phenotype in a manner reflected in the platelet proteome, but not in the plasma levels of the same proteins.
Platelet protein profiles of tumor-bearing mice correlate with tumor angiogenesis. SELDI-ToF MS expression difference maps from healthy mice (“Controls,” labeled gray), mice bearing nonangiogenic dormant tumor xenografts (“nonangiogenic,” labeled blue), and mice bearing angiogenic tumor xenografts of human liposarcoma (“angiogenic,” labeled red) are displayed in gel view. Differential expression patterns were detected for a range of peptides, and we present the most typical arrays: bFGF, PDGF, VEGF, endostatin, and albumin control. In each case, where differences in protein expressions were detected (eg, in the basic fraction [Q1 and Q2 fraction] of the platelet lysate, a difference was identified at 8200 Da), the band was further identified by purification and peptide mass fingerprinting and tandem MS (data not shown). (Abscissa) Relative MW computed from m/z value. (Ordinate) Identified peptide confirmed by immunodepletion and/or immunoprecipitation, and/or sequencing. The intensity of each of the bands correlates with relative expression level of the protein.
Platelet protein profiles of tumor-bearing mice correlate with tumor angiogenesis. SELDI-ToF MS expression difference maps from healthy mice (“Controls,” labeled gray), mice bearing nonangiogenic dormant tumor xenografts (“nonangiogenic,” labeled blue), and mice bearing angiogenic tumor xenografts of human liposarcoma (“angiogenic,” labeled red) are displayed in gel view. Differential expression patterns were detected for a range of peptides, and we present the most typical arrays: bFGF, PDGF, VEGF, endostatin, and albumin control. In each case, where differences in protein expressions were detected (eg, in the basic fraction [Q1 and Q2 fraction] of the platelet lysate, a difference was identified at 8200 Da), the band was further identified by purification and peptide mass fingerprinting and tandem MS (data not shown). (Abscissa) Relative MW computed from m/z value. (Ordinate) Identified peptide confirmed by immunodepletion and/or immunoprecipitation, and/or sequencing. The intensity of each of the bands correlates with relative expression level of the protein.
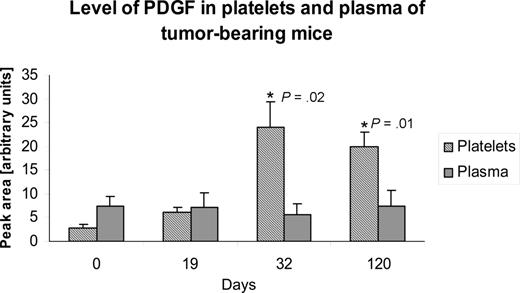
The elevation of PDGF in platelets, but not that of plasma, was sustained for up to 120 days after tumor implantation (Figure 6).
The tumor-associated change in platelet level of an angiogenesis regulator may be continuous. Platelet lysates from mice xenografted with human liposarcoma cells were longitudinally analyzed using SELDI-ToF at baseline, and up to 120 days after implantation of the tumor. The figure depicts an illustrative example of how the levels of an angiogenesis regulator (PDGF) are sustained in platelets for the duration of the experiment. Mice bearing dormant variant of human liposarcoma increase the platelet level of PDGF between 19 and 30 days after tumor implantation, and the level remains elevated for the duration of the experiment. The experiment was repeated on 2 separate occasions with 5 mice per experiment, and the graph represents mean plus or minus SEM.
The tumor-associated change in platelet level of an angiogenesis regulator may be continuous. Platelet lysates from mice xenografted with human liposarcoma cells were longitudinally analyzed using SELDI-ToF at baseline, and up to 120 days after implantation of the tumor. The figure depicts an illustrative example of how the levels of an angiogenesis regulator (PDGF) are sustained in platelets for the duration of the experiment. Mice bearing dormant variant of human liposarcoma increase the platelet level of PDGF between 19 and 30 days after tumor implantation, and the level remains elevated for the duration of the experiment. The experiment was repeated on 2 separate occasions with 5 mice per experiment, and the graph represents mean plus or minus SEM.
Discussion
Our results show that the levels of angiogenesis regulatory proteins in circulating platelets change in mice bearing human tumor xenografts compared with tumor-free, sham-operated controls. Using SELDI-ToF technology, we evaluated, in an open-ended fashion, the proteomic profiles of platelets from tumor-bearing and tumor-free, sham-operated mice. Among the proteins differentially expressed in platelets from these 2 groups, angiogenesis regulators were prominently represented. We have named this select group of angiogenic proteins the “platelet angiogenesis proteome” to emphasize the stability of the relative protein concentrations under physiologic conditions. Under normal conditions, the membership in this proteome appears to vary little. However, in a tumor-bearing mouse, the platelet content of angiogenesis regulators is altered and the changes persist, as suggested by the sustained elevation of PDGF in platelets, but not in plasma (Figure 6). The process appears selective for angiogenesis regulators, as we found that platelet levels of nonangiogenic proteins, such as albumin (Figures 5, S1), did not differ in presence or absence of tumors.
Circulating platelets can sequester angiogenesis regulatory proteins, even in the presence of a very small tumor mass, that is, cancers smaller than 1 mm3. Tumors of this size cannot be detected, at least at present, with conventional, clinically applicable methods. Our current study indicates that detection of these microscopic tumors cannot be accomplished using conventional measurements of angiogenesis regulatory proteins in plasma or serum because serum and plasma levels of these proteins are quite low and remain low even in the presence of much larger tumors. Although the presence of a microscopic tumor may be suspected on the basis of a single platelet protein marker, such as PF4,15 our current data indicate that platelets take up both positive and negative regulators of angiogenesis and that the balance of these 2 functionally opposing groups should be studied to obtain insight into the angiogenic potential of the tumor. This is in agreement with our finding of increased endostatin levels in the nonangiogenic clone compared with its angiogenic counterpart (Figure 5).
It appears that the platelet sequestration of angiogenesis regulatory proteins involves a process by which these proteins are internalized by circulating platelets, redistributed to different granular compartments within the platelets,17 and delivered to tissues as platelets adhere to abnormal vasculature. We have established previously that inhibitors and stimulators of angiogenesis are not necessarily in the same granular compartment in platelets and may not be released from platelets simultaneously.17 The platelet storage compartment, consisting of α-granules, dense granules, and lysosomes, is highly regulated. The exact manner of this regulation remains to be elucidated, but it is probable that the activation of platelets results in the redistribution of these proteins to the open canallicular system, which facilitates uptake and secretion of many proteins.17,18
Some platelet-specific proteins, such as PF4 and thrombomodulin, are synthesized by several cells, including megakaryocytes, and concentrate in platelets at 400-fold concentrations. As we had documented previously, it is the host (murine) PF4 that is up-regulated in platelets of human tumor xenograft bearing mice, suggesting that the synthesis of this angiogenesis suppressor is up-regulated in the host bone marrow megakaryocytes.15 Other proteins, such as factor V, thrombospondin, or P-selectin, are synthesized by other cells and taken up by platelets in the periphery. The most notable nonselective platelet protein is fibrinogen, which is synthesized by the hepatocytes and taken up into platelet α-granules.19-21
An important finding of our study was the relative absence of angiogenesis regulatory proteins in the plasma (Figure 5) or serum (Figure 2). These most common clinical analytes showed minimal or no differences in angiogenic proteins at baseline, or with tumor progression. This suggested that, contrary to commonly held beliefs, platelets may not undergo degranulation with an uncontrolled release of the growth regulators from α-granules into circulation, but rather that there may be a fairly strict spatial and temporal control of this release at the tumor or wound site. This was consistent with our in vitro analysis of the releasate of activated platelets, where minimal amounts of VEGF were released during agonist-induced platelet activation and bFGF was not released at all (Figure 2). The fact that a small amount of VEGF is released from platelets in response to thrombin is in agreement with previous studies,22 but these studies did not quantify the released vs retained proteins and could not therefore appreciate the incomplete degranulation. As we show in Figure 2B for bFGF, and in Figures 2A and 3 for VEGF, the proteins remain associated with platelets on activation. Using double-label immunofluorescence microscopy with antibodies against tubulin and VEGF for fixed and permeabilized resting platelets and phalloidin and VEGF for activated platelets, we showed that VEGF was distributed throughout the cytoplasm of resting and activated platelets and redistributed along the filopodia and the periphery of lamellipodia (Figure 3E,F). This tight association of angiogenesis-related proteins with platelets even after activation has been documented before23 but has not been clearly understood. It is probable that the redistribution of angiogenesis regulatory proteins to the filopodia and lamellipodia of platelets provides an avenue for a direct exchange of these proteins with the tissues. A publication by Ma et al24 demonstrated that the release of VEGF and endostatin is dependent on activation of specific proteinase-activated receptors (PARs). The authors show that the activation of PAR1 selectively releases a stimulator of angiogenesis (VEGF), whereas with the selective activation of PAR4 releases an inhibitor of angiogenesis (endostatin). Unfortunately, the tissue and platelet levels of these proteins were not reported, and the concentration of these 2 proteins in plasma was negligible.24,25
The potential for selective release of some platelet-associated angiogenesis proteins, but not others, may explain how platelets contribute to active angiogenesis early in wound healing and inhibit angiogenesis in the late stages of scar formation. Similarly, the dual ability of platelets to store angiogenesis regulators and to adhere to sites of abnormal endothelium also suggests an involvement of platelets in the amplification and/or dormancy of tumors. Variable concentrations of VEGF, bFGF, PF4, PDGF, endostatin, and other proteins important in angiogenesis can be taken up, internalized, and concentrated in platelets for a timely delivery to wounds or tumors. The redistribution of endogenous growth factors from cytoplasmic (granular) storage to the periphery of filopodia and lamellipodia of activated platelets (Figure 3E,F), as well as our previous report that pro- and antiangiogenic proteins are organized into separate platelet α-granular compartments and differentially released,17 provides an intact platelet-regulated mechanism that may be responsible for the regulation of angiogenesis in wounds and that the loss of this regulation may be the basis for angiogenesis in tumors. This would be consistent with the previously published notion that “tumors are wounds that do not heal.”26
Several other fundamental phenomena uncovered during these studies remain to be elucidated. For example, further studies will be needed to delineate the mechanism(s) by which platelets select, sequester, and retain only a relatively small number of proteins, excluding some much more abundant proteins or how these selected proteins bind in tissues.
The results reported here uncover new platelet biology with wide-ranging implications applicable to many angiogenesis-dependent physiologic states, such as development, wound repair, or reproduction, as well as disease conditions, such as cancer, atherosclerosis, diabetic ulcers, or inflammatory bowel disease. We have shown that the process of platelet uptake of angiogenesis regulators is highly specific and occurs well in advance of clinically detectable tumors. Platelet levels of angiogenesis-related proteins may therefore be superior to plasma and serum for the analysis of markers of angiogenesis in clinical trials in cancer. More importantly, however, the discovery that platelets actively sequester angiogenesis regulators and that these regulators can be differentially released from platelets would be expected to lead to many therapeutic applications in other angiogenesis-dependent conditions.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Ciphergen for their generosity in conducting many of the proteomic assays, sequencing the proteins free of charge, and sharing much of the scientific expertise. We also thank Drs Sean Downing and Constantine Mitsiades.
This work was supported by grants from the Breast Cancer Research Foundation (J.F.), the Department of Defense (DOD grant W81XWH-04-1-0316; J.F.), NASA (grant NNH04ZUU002N), and philanthropic funds (J.F.).
This work is being dedicated to Dr Judah Folkman's memory, with many thanks for his support, guidance, and vision.
National Institutes of Health
Authorship
Contribution: G.L.K. designed and performed the research, provided guidance for the group, analyzed the data, and wrote and revised the manuscript; T.-T.Y., V.P., and E.W. executed, analyzed, and evaluated the mass spectroscopy data and provided expertise with the SELDI-ToF technology; F.C. and L.K. performed research; D.C. analyzed and evaluated the mass spectroscopy data; J.E.I. performed research and contributed to evaluation of data; A.A.-S. executed and analyzed ELISA studies; E.B. performed animal experiments; N.A. designed and executed animal experiments; M.K. contributed to the analysis of the data; and J.F. provided mentorship for the team and expertise in preparation of the manuscript.
Conflict-of-interest disclosure: T.-T.Y., V.P., and E.W. were employees of Ciphergen during the execution of this study. The remaining authors declare no competing financial interests.
Correspondence: Giannoula Lakka Klement, Children's Hospital Boston, Karp Building 11.211, One Blackfan Circle, Boston, MA 02115; e-mail: giannoula_klement@dfci.harvard.edu and giannoula@gmail.com.

![Figure 5. Platelet protein profiles of tumor-bearing mice correlate with tumor angiogenesis. SELDI-ToF MS expression difference maps from healthy mice (“Controls,” labeled gray), mice bearing nonangiogenic dormant tumor xenografts (“nonangiogenic,” labeled blue), and mice bearing angiogenic tumor xenografts of human liposarcoma (“angiogenic,” labeled red) are displayed in gel view. Differential expression patterns were detected for a range of peptides, and we present the most typical arrays: bFGF, PDGF, VEGF, endostatin, and albumin control. In each case, where differences in protein expressions were detected (eg, in the basic fraction [Q1 and Q2 fraction] of the platelet lysate, a difference was identified at 8200 Da), the band was further identified by purification and peptide mass fingerprinting and tandem MS (data not shown). (Abscissa) Relative MW computed from m/z value. (Ordinate) Identified peptide confirmed by immunodepletion and/or immunoprecipitation, and/or sequencing. The intensity of each of the bands correlates with relative expression level of the protein.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/12/10.1182_blood-2008-06-159541/4/m_zh80070930900005.jpeg?Expires=1769181251&Signature=i3j2H58t9esS6HozOH-V0cfcHfg7Rc6WZIxuiFDCqukFxe41~y19iRi8wQnGp6do2ybJ~SPRN5H9X1LHMK40uXpc~6m4wXv0xYFsPciJGj4byM8hGONJjXwD7JuZ67uYO8n~-GsMFTbCgGVz2~WJMok33HlsdX2eOruTjY-1Uxe-0h1Ut3wRXaErXeO4tWsgzSxLj1cvegs-kLNa-HklqU4TFy~3vzkA6hOEO~ddZGFRCUNp3ZTjqg2skZuabnuARHwishRY8fqvsm5RMzvRf3Vb6Hw2WsBRbZvIJlAVIfkaKneJS0miSgC9Sf0~mAXGOsbDie79gZxqbNKoDuo9hA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal