Abstract
The thymus provides a microenvironment that induces the differentiation of T-progenitor cells into functional T cells and that establishes a diverse yet self-tolerant T-cell repertoire. However, the mechanisms that lead to the development of the thymus are incompletely understood. We report herein the results of screening for genes that are expressed in the third pharyngeal pouch, which contains thymic primordium. Polymerase chain reaction (PCR)–based cDNA subtraction screening for genes expressed in microdissected tissues of the third pharyngeal pouch rather than the second pharyngeal arch yielded one transcription factor, MafB, which was predominantly expressed in CD45−IA−PDGFRα+ mesenchymal cells and was detectable even in the third pharyngeal pouch of FoxN1-deficient nude mice. Interestingly, the number of CD45+ cells that initially accumulated in the embryonic thymus was significantly decreased in MafB-deficient mice. Alterations of gene expression in the embryonic thymi of MafB-deficient mice included the reduced expression of Wnt3 and BMP4 in mesenchymal cells and of CCL21 and CCL25 in epithelial cells. These results suggest that MafB expressed in third pharyngeal pouch mesenchymal cells critically regulates lymphocyte accumulation in the embryonic thymus.
Introduction
A functionally competent T-cell pool with a diverse repertoire of T-cell antigen receptors (TCRs) is essential in mounting immune responses to invading pathogens.1,2 Most peripheral T cells bearing αβ TCRs are generated in the thymus.2,3 The migration of hematopoietic stem cell–derived T-lymphoid progenitor cells into the thymus and the subsequent interactions with thymic stromal microenvironments are essential for T-cell development in the thymus.4,5 The entry of hematopoietic cells into thymus primordium is initiated during embryogenesis as early as embryonic day 11.5 (E11.5) in mice.6
The initial formation of thymic primordium occurs before the migration of hematopoietic cells and involves interactions between epithelial cells of the third pharyngeal pouch (3PP) endoderm and mesenchymal cells derived from neural crest at earlier stages (E9.5 to E11.5) of embryogenesis.7,8 This initial thymus development is governed by several transcription factors, including Tbx1, Hoxa3, Pax1, Pax9, Eya1, and Six1.9-18 Tbx1 is expressed in pharyngeal endoderm9,10 and is required for pharyngeal segmentation; its deficiency causes various pharyngeal defects, including impaired generation of the thymus and the parathyroid glands.11,12 Hoxa3 is expressed in the 3PP endoderm and neural crest mesenchyme;13,14 the lack of Hoxa3 reduces the expression of Pax1 and Pax9, which in turn causes defective formation of the thymus and the parathyroid glands.15-17 Eya1 and Six1 are expressed in pouch endoderm and neural crest mesenchyme of the third pharyngeal clefts, and are required for the development of the thymus and the parathyroid glands.18 Thus, transcriptional regulations of epithelial-mesenchymal interactions during this prelymphocyte stage of thymus development are mostly shared between epithelial and mesenchymal cells and between thymus and parathyroid development.
Subsequently, at E11.5, FoxN1 is detectable in the ventral aspect of the 3PP.19 FoxN1 is specifically expressed in epithelial cells (but not in mesenchymal cells) of the thymus but not the parathyroid glands.19 The formation of thymic primordium before the entry of T-lymphoid progenitor cells does not seem to require FoxN1, but the subsequent differentiation of thymic primordium into functional thymus is dependent on FoxN1.20 Thus, FoxN1-dependent development of thymic epithelial cells is implicated in the interactions between thymic epithelial cells and developing lymphoid cells.20-22 Indeed, FoxN1 is required for thymic epithelial cells to optimally produce CCL25, a chemokine involved in attracting T-lymphoid progenitor cells to the thymus,23,24 and to produce DLL1 and DLL4, the Notch ligands involved in supporting T-cell development in the thymus.25 However, FoxN1-dependent epithelial-lymphocyte interactions are not sufficient for thymus development. Rather, the contribution of mesenchymal cells appears crucial for the optimal generation of the thymus even at this stage. Several investigators have reported that neural crest–derived mesenchymal cells critically regulate the growth and development of thymic epithelial cells26 by secreting molecules, including Wnts, bone morphogenetic proteins (BMPs), and fibroblast growth factors (FGFs).27-30 However, the transcriptional mechanisms that govern these epithelial-mesenchymal interactions at this lymphocyte accumulation stage have remained elusive.
By screening for genes that are expressed in E11.5 3PPs, we have identified that the transcription factor MafB is strongly expressed in thymic mesenchymal cells rather than thymic epithelial cells or thymocytes. We show that MafB-deficient mice generate thymic primordium containing FoxN1-expressing epithelial cells by E11.5. However, subsequent generation of the thymus with developing thymocytes is significantly impaired in MafB-deficient mice. The E11.5 3PP of MafB-deficient mice exhibits reduced expression of several mesenchymal molecules, including Wnt3 and BMP4, and several epithelial molecules, including CCL21 and CCL25, suggesting that MafB expressed in thymic mesenchymal cells critically regulates embryonic thymus development at the lymphocyte accumulation stage.
Methods
Mice
All mice were maintained under specific pathogen–free conditions in accordance with intramural guidelines. C57BL/6 (B6), BALB/c-nu/nu, and BALB/c-nu/+ mice were obtained from SLC (Shizuoka, Japan). MafB-deficient mice31 were described previously. The day when a vaginal plug was first observed was designated as gestation day 0.5 (E0.5). All experiments were carried out under the approval of the Institutional Animal Care and Use Committee of the University of Tokushima.
Laser-capture microdissection
E11.5 embryos were embedded in OCT compound (Sakura Finetek, Tokyo, Japan), sliced into 5-μm–thick sections, and attached to glass slides for staining with Histogene reagent (Arcturus Engineering, Mountain View, CA). Sections were placed on a thin-laser pressure-catapulting membrane (Arcturus). Epithelial linings of 3PPs, second pharyngeal arches (2PAs), and several other tissues were dissected with a PixCell II laser capture microdissection system (Arcturus).
cDNA amplification, subtraction, and cDNA library construction
3PP and 2PA tissues were microdissected from 34 sections of E11.5 embryos. RNA was extracted from the microdissected tissues using CapSure HS LCM Caps and PicoPure RNA Isolation Kit (Arcturus). DNaseI-treated RNA was polymerase chain reaction (PCR)–amplified using the Super SMART PCR cDNA Synthesis Kit (Clontech, Mountain View, CA). Amplified cDNA was digested with RsaI and then subjected to the subtraction using the PCR-Select cDNA Subtraction Kit (Clontech) for genes expressed in 3PPs rather than 2PAs. Subtracted cDNA was cloned in TA cloning vector. A total of 1000 randomly picked clones were sequenced.
Isolation of thymic stromal cells
Thymic stromal cells were prepared from E14.5 embryonic thymi and 6-week-old adult thymi by digestion with collagenase, dispase, and DNaseI, as described.32 For isolation of fetal thymic stromal cells, cells were stained with allophycocyanin-conjugated anti-CD45 antibody, FITC-conjugated anti–I-A antibody, and biotinylated anti-PDGFRα antibody (eBioscience, San Diego, CA) followed by phycoerythrin-conjugated streptavidin. Cells were isolated with FACSVantage cell sorter (BD Biosciences, San Jose, CA) as described.33 For isolation of adult thymic stromal cells, CD45− cells enriched with a magnetic cell sorter (Miltenyi Biotec, Auburn, CA) were stained with allophycocyanin-conjugated anti-CD45 antibody, FITC-conjugated anti–I-A antibody, and fibroblast-specific MTS15 antibody,34 followed by biotinylated goat anti–rat antibody and phycoerythrin-conjugated streptavidin. Propidium iodide was used to exclude dead cells. Sorted cells with at least 80% purity were used for further analysis.
RNA amplification for quantitative RT-PCR analysis
RNA was extracted from microdissected E11.5 embryonic tissues using the PicoPure RNA Isolation Kit (Arcturus). DNaseI-treated RNA was amplified by the Message Amp II aRNA Kit (Ambion, Austin, TX), which consisted of reverse transcription (RT) with an oligo-dT primer containing the T7 promoter sequence and in vitro transcription of resulting cDNA with T7 RNA polymerase to amplify RNA copies. RNA purity and quantification was determined with a Nanodrop spectrophotometer (Nanodrop Technologies).
Quantitative RT-PCR analysis
Amplified RNA or total cellular RNA extracted using Isogen (Wako Pure Chemical Industries, Osaka, Japan) was reverse-transcribed with oligo-dT primer and Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA). Real-time PCR was performed with SYBER Premix Ex Taq (Takara, Otsu, Japan) and ABI Prism 7900HT Sequence Detection System with SDS 2.0 software (Applied Biosystems, Foster City, CA). Amplified signals were confirmed to be single bands over gel electrophoresis. Gene expression was normalized to GAPDH mRNA using the Δ cycle threshold (Ct) method.35 Primers for PCR are listed in Table 1.
PCR primers
| Gene . | Primer . | Primer . | PCR product size, bp . |
|---|---|---|---|
| GAPDH | 5′-CCGGTGCTGAGTATGTCGTG-3′ | 5′-CAGTCTTCTGGGTGGCAGTG-3′ | 349 |
| CCL21 | 5′-GGGGAACCTCTAAGTCTGGAA-3′ | 5′-CTCATTCCCTGGGAGACACT-3′ | 341 |
| PTH | 5′-GCTGGCAGTCTGTCTTCTTACCC-3′ | 5′-TGTCAGTGCCCTGCACTGTC-3′ | 343 |
| Casr | 5′-GGTATACAGCGCACACGTGA-3′ | 5′-CCTTTGCCTCTCCTGCACTT-3′ | 282 |
| Spink8 | 5′-AGTTGCAGTCCTTGTTCTGG-3′ | 5′-CTGTCTCCCAAACTGGGTATC-3′ | 331 |
| Pla2g7 | 5′-CAGACTCCAAAGGACATCGCAA-3′ | 5′-GTCAAAGGGTGACCCAGGAA-3′ | 303 |
| Elovl2 | 5′-CTACTACGGCCTGTCTGTG-3′ | 5′-GGGCTTTGGGGAAACCATTC-3′ | 281 |
| MafB | 5′-GTTATAGGGGAGGTCTAGGTGT-3′ | 5′-AAGCTCGTTTCCGATGCAG-3′ | 292 |
| Irs4 | 5′-AGACAGGGTACAGAAGGTAGAG-3′ | 5′-GCAACTCAAAGCAGGTCTGTGAAG-3′ | 407 |
| IL7 | 5′-CTGGTGAACTGCACAAGTAAGG-3′ | 5′-AGCTGCATTTCTGTGTCCATC-3′ | 287 |
| Nedd4 | 5′-TTGAGGAACAGCCTACACTTCC-3′ | 5′-TCCAGTGATTGCCACGTTCT-3′ | 329 |
| Pax1 | 5′-TGGAGCTAGCACAGCTGGGTATC-3′ | 5′-GCCCCTGTTTGCTCCATAAACGTC-3′ | 440 |
| Gcm2 | 5′-GCCAGCCATCTGTGACAAGG-3′ | 5′-GCCCTGGAATAGGGAAGCTG-3′ | 383 |
| Angpt2 | 5′-AATGTTCCGTGGGAGTTCAG-3′ | 5′-GTACAGTCTCCGCATTCACC-3′ | 388 |
| Eeflg | 5′-AGCCCCAGTTCAGGGCTATC-3′ | 5′-AGCGGTACTCGGCATACCAC-3′ | 394 |
| 2610528KiiRik | 5′-GAGCACCAACAGGCTACGAC-3′ | 5′-ATGTGCCTGGGAAGGTTTGG-3′ | 500 |
| Gpsn2 | 5′-CCCATACCCCACCAAGAACC-3′ | 5′-GGTAGTCGCGGAACTCCTTC-3′ | 206 |
| Hnrpf | 5′-GGGCTGCCCTACAAAGCAAC-3′ | 5′-CCCCGTAACAGCCACTCACT-3′ | 325 |
| PD-L1 | 5′-GAGAGCCTCGCGTCCAAAG-3′ | 5′-GTGGTTTTGCCCTGGCTGTGATCT-3′ | 409 |
| FGF7 | 5′-CTCTACAGGTCATGCTTCCACC-3′ | 5′-ACAGAACAGTCTTCTCACCCT-3′ | 174 |
| FGF10 | 5′-CAGCGGGACCAAGAATGAAG-3′ | 5′-TGACGGCAACAACTCCGATTT-3′ | 77 |
| IGF1 | 5′-CAGGCTATGGCTCCAGCATTCGG-3′ | 5′-CAGATCACAGCTCCGGAAGC-3′ | 77 |
| IGF2 | 5′-GAGCTTGTTGACACGCTTCAGTTTGTC-3′ | 5′-GTTTGGCCTCTCTGAACTCTTTGAG-3′ | 357 |
| IGF1R | 5′-GACATCCGCAACGACTATCAG-3′ | 5′-GTAGTTATTGGACACCGCATC-3′ | 393 |
| Wnt3 | 5′-ACCTGGAGAAGGCTGGAAGT-3′ | 5′-CTTGTCCTTGAGGAAGTCGC-3′ | 280 |
| Wnt4 | 5′-CTCAAAGGCCTGATCCAGAG-3′ | 5′-TCACAGCCACACTTCTCCAG-3′ | 293 |
| Wnt7a | 5′-GGCTTCGCCAAGGTCTTCG-3′ | 5′-CATGAGGTCACAGCCACTGG-3′ | 440 |
| Wnt7b | 5′-TGCCCGTGAGATCAAAAAG-3′ | 5′-CTGCGTTGTACTTCTCCTTG-3′ | 200 |
| Wnt10a | 5′-CCTGGAGACTCGGAACAAAG-3′ | 5′-AACCGCAAGCCTTCAGTTTA-3′ | 157 |
| Wnt10b | 5′-GATACCCACAACCGCAACTC-3′ | 5′-GGCTCACCTTCATTTACACACA-3′ | 323 |
| Wnt11 | 5′-GCTCCATCCGCACCTGTT-3′ | 5′-CGCTCCACCACTCTGTCC-3′ | 331 |
| FGFR2IIIb | 5′-AACGGTCACCACACCGGC-3′ | 5′-AGGCAGACTGGTTGGCCTG-3′ | 311 |
| BMP2 | 5′-TGTGACCAGACTATTGGACACC-3′ | 5′-AGTTCAGGTGGTCAGCAAGG-3′ | 460 |
| BMP4 | 5′-CGAGGCGACACTTCTACAG-3′ | 5′-TGGGGGCTTCATAACCT-3′ | 394 |
| FoxN1 | 5′-CTCGTCGTTTGTGCCTGAC-3′ | 5′-TGCCTCTTGTAGGGGTGGAAA-3′ | 243 |
| DLL1 | 5′-GTTAGCATCATTGGGGCTACCCAG-3′ | 5′-CGCCTCTGCTAACTCTGAGAGAACC-3′ | 519 |
| DLL4 | 5′-CTGTCCTTATGGCTTTGTGG-3′ | 5′-GCTCCTTCTTCTGGTTTGTG-3′ | 260 |
| CCL25 | 5′-GTTTTGTTGGGGCCTGGATG-3′ | 5′-ACTCCTCACGCTTGTACTGTTG-3′ | 345 |
| PDGFRa | 5′-TCGTCCTTTTCTCTGAGATGCGTT-3′ | 5′-CCCACGCTGAAGGTTCCGTT-3′ | 428 |
| RARb | 5′-ACCGGCATACTGCTCAATCC-3′ | 5′-ACTGGCATCGGTTCCTAGTG-3′ | 264 |
| LCK | 5′-ACGGTGGCTTCTACATCTCC-3 | 5′-GGCCGTGTACTCATTGTCCT-3 | 626 |
| Gene . | Primer . | Primer . | PCR product size, bp . |
|---|---|---|---|
| GAPDH | 5′-CCGGTGCTGAGTATGTCGTG-3′ | 5′-CAGTCTTCTGGGTGGCAGTG-3′ | 349 |
| CCL21 | 5′-GGGGAACCTCTAAGTCTGGAA-3′ | 5′-CTCATTCCCTGGGAGACACT-3′ | 341 |
| PTH | 5′-GCTGGCAGTCTGTCTTCTTACCC-3′ | 5′-TGTCAGTGCCCTGCACTGTC-3′ | 343 |
| Casr | 5′-GGTATACAGCGCACACGTGA-3′ | 5′-CCTTTGCCTCTCCTGCACTT-3′ | 282 |
| Spink8 | 5′-AGTTGCAGTCCTTGTTCTGG-3′ | 5′-CTGTCTCCCAAACTGGGTATC-3′ | 331 |
| Pla2g7 | 5′-CAGACTCCAAAGGACATCGCAA-3′ | 5′-GTCAAAGGGTGACCCAGGAA-3′ | 303 |
| Elovl2 | 5′-CTACTACGGCCTGTCTGTG-3′ | 5′-GGGCTTTGGGGAAACCATTC-3′ | 281 |
| MafB | 5′-GTTATAGGGGAGGTCTAGGTGT-3′ | 5′-AAGCTCGTTTCCGATGCAG-3′ | 292 |
| Irs4 | 5′-AGACAGGGTACAGAAGGTAGAG-3′ | 5′-GCAACTCAAAGCAGGTCTGTGAAG-3′ | 407 |
| IL7 | 5′-CTGGTGAACTGCACAAGTAAGG-3′ | 5′-AGCTGCATTTCTGTGTCCATC-3′ | 287 |
| Nedd4 | 5′-TTGAGGAACAGCCTACACTTCC-3′ | 5′-TCCAGTGATTGCCACGTTCT-3′ | 329 |
| Pax1 | 5′-TGGAGCTAGCACAGCTGGGTATC-3′ | 5′-GCCCCTGTTTGCTCCATAAACGTC-3′ | 440 |
| Gcm2 | 5′-GCCAGCCATCTGTGACAAGG-3′ | 5′-GCCCTGGAATAGGGAAGCTG-3′ | 383 |
| Angpt2 | 5′-AATGTTCCGTGGGAGTTCAG-3′ | 5′-GTACAGTCTCCGCATTCACC-3′ | 388 |
| Eeflg | 5′-AGCCCCAGTTCAGGGCTATC-3′ | 5′-AGCGGTACTCGGCATACCAC-3′ | 394 |
| 2610528KiiRik | 5′-GAGCACCAACAGGCTACGAC-3′ | 5′-ATGTGCCTGGGAAGGTTTGG-3′ | 500 |
| Gpsn2 | 5′-CCCATACCCCACCAAGAACC-3′ | 5′-GGTAGTCGCGGAACTCCTTC-3′ | 206 |
| Hnrpf | 5′-GGGCTGCCCTACAAAGCAAC-3′ | 5′-CCCCGTAACAGCCACTCACT-3′ | 325 |
| PD-L1 | 5′-GAGAGCCTCGCGTCCAAAG-3′ | 5′-GTGGTTTTGCCCTGGCTGTGATCT-3′ | 409 |
| FGF7 | 5′-CTCTACAGGTCATGCTTCCACC-3′ | 5′-ACAGAACAGTCTTCTCACCCT-3′ | 174 |
| FGF10 | 5′-CAGCGGGACCAAGAATGAAG-3′ | 5′-TGACGGCAACAACTCCGATTT-3′ | 77 |
| IGF1 | 5′-CAGGCTATGGCTCCAGCATTCGG-3′ | 5′-CAGATCACAGCTCCGGAAGC-3′ | 77 |
| IGF2 | 5′-GAGCTTGTTGACACGCTTCAGTTTGTC-3′ | 5′-GTTTGGCCTCTCTGAACTCTTTGAG-3′ | 357 |
| IGF1R | 5′-GACATCCGCAACGACTATCAG-3′ | 5′-GTAGTTATTGGACACCGCATC-3′ | 393 |
| Wnt3 | 5′-ACCTGGAGAAGGCTGGAAGT-3′ | 5′-CTTGTCCTTGAGGAAGTCGC-3′ | 280 |
| Wnt4 | 5′-CTCAAAGGCCTGATCCAGAG-3′ | 5′-TCACAGCCACACTTCTCCAG-3′ | 293 |
| Wnt7a | 5′-GGCTTCGCCAAGGTCTTCG-3′ | 5′-CATGAGGTCACAGCCACTGG-3′ | 440 |
| Wnt7b | 5′-TGCCCGTGAGATCAAAAAG-3′ | 5′-CTGCGTTGTACTTCTCCTTG-3′ | 200 |
| Wnt10a | 5′-CCTGGAGACTCGGAACAAAG-3′ | 5′-AACCGCAAGCCTTCAGTTTA-3′ | 157 |
| Wnt10b | 5′-GATACCCACAACCGCAACTC-3′ | 5′-GGCTCACCTTCATTTACACACA-3′ | 323 |
| Wnt11 | 5′-GCTCCATCCGCACCTGTT-3′ | 5′-CGCTCCACCACTCTGTCC-3′ | 331 |
| FGFR2IIIb | 5′-AACGGTCACCACACCGGC-3′ | 5′-AGGCAGACTGGTTGGCCTG-3′ | 311 |
| BMP2 | 5′-TGTGACCAGACTATTGGACACC-3′ | 5′-AGTTCAGGTGGTCAGCAAGG-3′ | 460 |
| BMP4 | 5′-CGAGGCGACACTTCTACAG-3′ | 5′-TGGGGGCTTCATAACCT-3′ | 394 |
| FoxN1 | 5′-CTCGTCGTTTGTGCCTGAC-3′ | 5′-TGCCTCTTGTAGGGGTGGAAA-3′ | 243 |
| DLL1 | 5′-GTTAGCATCATTGGGGCTACCCAG-3′ | 5′-CGCCTCTGCTAACTCTGAGAGAACC-3′ | 519 |
| DLL4 | 5′-CTGTCCTTATGGCTTTGTGG-3′ | 5′-GCTCCTTCTTCTGGTTTGTG-3′ | 260 |
| CCL25 | 5′-GTTTTGTTGGGGCCTGGATG-3′ | 5′-ACTCCTCACGCTTGTACTGTTG-3′ | 345 |
| PDGFRa | 5′-TCGTCCTTTTCTCTGAGATGCGTT-3′ | 5′-CCCACGCTGAAGGTTCCGTT-3′ | 428 |
| RARb | 5′-ACCGGCATACTGCTCAATCC-3′ | 5′-ACTGGCATCGGTTCCTAGTG-3′ | 264 |
| LCK | 5′-ACGGTGGCTTCTACATCTCC-3 | 5′-GGCCGTGTACTCATTGTCCT-3 | 626 |
Multicolor confocal microscopy analysis
Fresh tissues were embedded in OCT compound (Sakura Finetek). Frozen sections measuring 5-μm thick were fixed with either acetone or paraformaldehyde and stained with the following antibodies: anti–mouse FoxN1 antibody,24 ER-TR5 monoclonal antibody specific for medullary epithelial cells,36 biotinylated monoclonal antibodies specific for mouse CCL21 and CCL25 (R&D Systems, Minneapolis, MN), rabbit anti–mouse AIRE antibody (M-300; Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-pancytokeratin polyclonal antibody (Dako Cytomation, Carpinteria, CA), Alexa 488–conjugated anti–rabbit IgG antibody, and Alexa 633–conjugated anti–mouse IgG antibody. To analyze green fluorescent protein (GFP) expression derived from the MafB− allele, in which the coding sequence of MafB was replaced with GFP,31 of MafB+/− heterozygous mice, E15.5 thymus was fixed in 4% paraformaldehyde, sectioned, and stained with anti-pancytokeratin antibody. Stained sections were mounted with a fluorescence mounting medium (Dako). Images were acquired with a TCS SP2 laser scanning microscope (Leica, Mannheim, Germany) equipped with argon and helium-neon lasers (excitation at 488, 546, and 633 nm), 20× 1.25-0.75 NA and 40× 1.25-0.75 CS oil objectives, and Leica confocal software version 2.0.
Bioinformatics
Sequences were analyzed using the public databases of the National Center for Biotechnology Information (NCBI),37 JAX–Mouse Genome Informatics (MGI),38 and European Molecular Biology Laboratory (EMBL)–InterPro.39 Domains and motifs were identified using the NCBI–Conserved Domain Database (CDD)40 and the EMBL-SMART tool.41
Statistics
Statistical comparison was performed by the Student t test using Excel software (Microsoft, Redmond, WA).
Results
Subtraction screening for genes expressed in the 3PP
To identify genes that are selectively expressed in the 3PP that contains thymic primordium, tissues of the 3PP and the 2PA were isolated from E11.5 mouse embryos by laser capture microdissection (Figure 1A,B). Total RNA isolated from these tissues was reverse-transcribed and amplified. The 3PP-derived cDNA was hybridized with an excess amount of 2PA-derived cDNA, and subtracted cDNA was used for the preparation of a plasmid library (Figure 1C). The representative size of library inserts was 0.5 to 3 kb. Analysis of 1000 randomly picked clones yielded sequences of 678 inserts. A total of 217 genes were found in these inserts: 17 genes appeared no less than 3 times (Table 2; group 1); 50 genes appeared twice (Table 2; group 2); and 150 genes appeared only once (Table 2; group 3). Evaluation of gene expression in the 3PP and the 2PA by quantitative RT-PCR confirmed that 12 of the 17 group 1 genes were expressed at higher levels in the 3PP than in the 2PA, whereas none of the 8 genes randomly chosen from groups 2 and 3 showed any apparent differences in expression levels between the 2 tissues (Table 2). Table 3lists the 12 genes that were expressed at higher levels in the 3PP than in the 2PA. Among these genes, 6 encoding Pax1, Gcm2, calcium-sensing receptor (Casr), parathyroid hormone (PTH), CCL21, and IL7 are known for their roles in the development and function of the thymus and the parathyroid glands,15,16,19,23,42-46 suggesting that this library screening effectively profiled genes that are involved in the development and function of the 3PP and its descendant organs. The rest of the genes were MafB, Irs4, Pla2g7, Nedd4, Agpt2, and Spink8, which have not been previously identified for their roles in the development or function of the thymus or the parathyroid glands (Table 3). Therefore, we further analyzed their expression and function.
Subtraction screening for genes expressed in the 3PP. (A) Sagittal sections of E11.5 C57BL/6 mouse embryos were 2-color stained with anti-CD45 (green) and anti-keratin (red) antibodies. Regions of the 3PP and the 2PA are marked with blue lines. (B) Regions of 3PP (top panels, marked with dashed lines) and 2PA (bottom panels, marked with dashed lines) in histogene-stained sagittal sections of E11.5 C57BL/6 mouse embryos were microdissected for RNA preparation. Left and right panels show the sections before and after microdissection, respectively. (C) Schematic diagrams of the screening strategy for genes expressed in E11.5 3PPs.
Subtraction screening for genes expressed in the 3PP. (A) Sagittal sections of E11.5 C57BL/6 mouse embryos were 2-color stained with anti-CD45 (green) and anti-keratin (red) antibodies. Regions of the 3PP and the 2PA are marked with blue lines. (B) Regions of 3PP (top panels, marked with dashed lines) and 2PA (bottom panels, marked with dashed lines) in histogene-stained sagittal sections of E11.5 C57BL/6 mouse embryos were microdissected for RNA preparation. Left and right panels show the sections before and after microdissection, respectively. (C) Schematic diagrams of the screening strategy for genes expressed in E11.5 3PPs.
Screening for genes expressed in E11.5 3PPs
| Gene from subtraction screening . | Verification of expression specificity . | ||||
|---|---|---|---|---|---|
| Group/No. . | Name . | Accession no. . | Frequency . | 3PP/2PA (SE) . | . |
| 1 | |||||
| 1 | CCL21 | NM_011124 | 254 | 407.7 (86) | * |
| 2 | PTH | NM_020623 | 104 | 2.1 (1.1) | * |
| 3 | Casr | NM_013803 | 10 | 19 088.5 (1963) | * |
| 4 | Spink8 | NM_183136 | 8 | 53.9 (19) | * |
| 5 | Pla2g7 | NM_013737 | 7 | 5.3 (3.8) | * |
| 6 | Elovl2 | NM_019423 | 6 | 0.5 (0.3) | |
| 7 | MafB | NM_010658 | 6 | 1 842.7 (542) | * |
| 8 | Irs4 | NM_010572 | 5 | 4 166.0 (958) | * |
| 9 | IL7 | NM_008371 | 4 | 658.7 (32) | * |
| 10 | Nedd4 | NM_010890 | 3 | 210.3 (94) | * |
| 11 | Pax1 | NM_008780 | 3 | 2.5 (2.1) | * |
| 12 | Gcm2 | NM_008104 | 3 | 8.2 (4.6) | * |
| 13 | Angpt2 | NM_007426 | 3 | 3.2 (0.7) | * |
| 14 | Eeflg | NM_026007 | 3 | 0.1 (0.1) | |
| 15 | 2610528KiiRik | NM_175184 | 3 | 0.8 (0.1) | |
| 16 | Gpsn2 | NM_134118 | 3 | 0.3 (0.1) | |
| 17 | Hnrnpf | NM_133834 | 3 | 0.5 (0.1) | |
| 17 genes | 12 genes | ||||
| 2 | |||||
| 1 | Dtprp | NM_010088 | 2 | 0.1 (0.0) | |
| 2 | Pbx1 | NM_008783 | 2 | 1.6 (0.7) | |
| 3 | Atp5a1 | NM_007505 | 2 | 0.4 (0.1) | |
| 4 | Zfp9 | NM_011763 | 2 | 1.1 (0.5) | |
| 50 genes | |||||
| 3 | |||||
| 1 | Ndufv1 | NM_133666 | 1 | 0.1 (0.0) | |
| 2 | Neurod | NM_010894 | 1 | 0.3 (0.1) | |
| 3 | Cks2 | NM_025415 | 1 | 0.3 (0.1) | |
| 4 | Ptpn18 | NM_011206 | 1 | 0.1 (0.0) | |
| 150 genes | |||||
| Total | 217 genes | 12 genes | |||
| Gene from subtraction screening . | Verification of expression specificity . | ||||
|---|---|---|---|---|---|
| Group/No. . | Name . | Accession no. . | Frequency . | 3PP/2PA (SE) . | . |
| 1 | |||||
| 1 | CCL21 | NM_011124 | 254 | 407.7 (86) | * |
| 2 | PTH | NM_020623 | 104 | 2.1 (1.1) | * |
| 3 | Casr | NM_013803 | 10 | 19 088.5 (1963) | * |
| 4 | Spink8 | NM_183136 | 8 | 53.9 (19) | * |
| 5 | Pla2g7 | NM_013737 | 7 | 5.3 (3.8) | * |
| 6 | Elovl2 | NM_019423 | 6 | 0.5 (0.3) | |
| 7 | MafB | NM_010658 | 6 | 1 842.7 (542) | * |
| 8 | Irs4 | NM_010572 | 5 | 4 166.0 (958) | * |
| 9 | IL7 | NM_008371 | 4 | 658.7 (32) | * |
| 10 | Nedd4 | NM_010890 | 3 | 210.3 (94) | * |
| 11 | Pax1 | NM_008780 | 3 | 2.5 (2.1) | * |
| 12 | Gcm2 | NM_008104 | 3 | 8.2 (4.6) | * |
| 13 | Angpt2 | NM_007426 | 3 | 3.2 (0.7) | * |
| 14 | Eeflg | NM_026007 | 3 | 0.1 (0.1) | |
| 15 | 2610528KiiRik | NM_175184 | 3 | 0.8 (0.1) | |
| 16 | Gpsn2 | NM_134118 | 3 | 0.3 (0.1) | |
| 17 | Hnrnpf | NM_133834 | 3 | 0.5 (0.1) | |
| 17 genes | 12 genes | ||||
| 2 | |||||
| 1 | Dtprp | NM_010088 | 2 | 0.1 (0.0) | |
| 2 | Pbx1 | NM_008783 | 2 | 1.6 (0.7) | |
| 3 | Atp5a1 | NM_007505 | 2 | 0.4 (0.1) | |
| 4 | Zfp9 | NM_011763 | 2 | 1.1 (0.5) | |
| 50 genes | |||||
| 3 | |||||
| 1 | Ndufv1 | NM_133666 | 1 | 0.1 (0.0) | |
| 2 | Neurod | NM_010894 | 1 | 0.3 (0.1) | |
| 3 | Cks2 | NM_025415 | 1 | 0.3 (0.1) | |
| 4 | Ptpn18 | NM_011206 | 1 | 0.1 (0.0) | |
| 150 genes | |||||
| Total | 217 genes | 12 genes | |||
Subtraction screening was carried out as described in ″Methods.″ Of the 217 genes identified in the screening, 17 that appeared more than twice are categorized in group 1. Groups 2 and 3 consist of genes that appeared twice (50 genes) and once (150 genes), respectively. Ratios of expression levels in the 3PP to expression levels in the 2PA were verified by quantitative RT-PCR, and the values indicate averages and standard errors (SEs) of 3 independent measurements.
Twelve of the 17 group 1 genes were confirmed to be expressed at higher levels in 3PP than in 2PA. None of 4 randomly selected genes from group 2 and 4 randomly selected genes from group 3 showed significantly higher expression in 3PP than in 2PA.
Summary of genes expressed in E11.5 3PPs
| Gene . | Synonym . | Category . | Pharyngeal functions . | References . |
|---|---|---|---|---|
| Pax1 | Paired box 1 | Transcription factor | Pharyngeal development | 15,16 |
| Gcm2 | Glial cells missing homolog 2 | Transcription factor | Parathyroid development | 19,42 |
| Casr | Calcium-sensing receptor | Receptor | Parathyroid function | 43 |
| PTH | Parathyroid hormone | Hormone | Parathyroid function | 44 |
| CCL21 | CC chemokine ligand 21 | Chemokine | Parathyroid/thymus function | 23,45 |
| IL7 | Interleukin 7 | Cytokine | Thymus function | 46 |
| MafB | Transcription factor | Unknown | ||
| Irs4 | Insulin receptor substrate 4 | Receptor signaling | Unknown | |
| Pla2g7 | PAF acetylhydrolase | Phospholipase | Unknown | |
| Nedd4 | Ubiquitin ligase | Unknown | ||
| Agpt2 | Angiopoietin 2 | Angiogenesis | Unknown | |
| Spink8 | Tof2 | Peptidase inhibitor | Unknown |
| Gene . | Synonym . | Category . | Pharyngeal functions . | References . |
|---|---|---|---|---|
| Pax1 | Paired box 1 | Transcription factor | Pharyngeal development | 15,16 |
| Gcm2 | Glial cells missing homolog 2 | Transcription factor | Parathyroid development | 19,42 |
| Casr | Calcium-sensing receptor | Receptor | Parathyroid function | 43 |
| PTH | Parathyroid hormone | Hormone | Parathyroid function | 44 |
| CCL21 | CC chemokine ligand 21 | Chemokine | Parathyroid/thymus function | 23,45 |
| IL7 | Interleukin 7 | Cytokine | Thymus function | 46 |
| MafB | Transcription factor | Unknown | ||
| Irs4 | Insulin receptor substrate 4 | Receptor signaling | Unknown | |
| Pla2g7 | PAF acetylhydrolase | Phospholipase | Unknown | |
| Nedd4 | Ubiquitin ligase | Unknown | ||
| Agpt2 | Angiopoietin 2 | Angiogenesis | Unknown | |
| Spink8 | Tof2 | Peptidase inhibitor | Unknown |
The 12 genes selectively expressed in E11.5 3PPs are divided into two groups. The 6 genes in the top group are known for their roles in the development or function of parathyroid glands or thymus. The involvement in pharyngeal region of the 6 genes in the bottom group has not been identified.
Expression profiles of 3PP genes
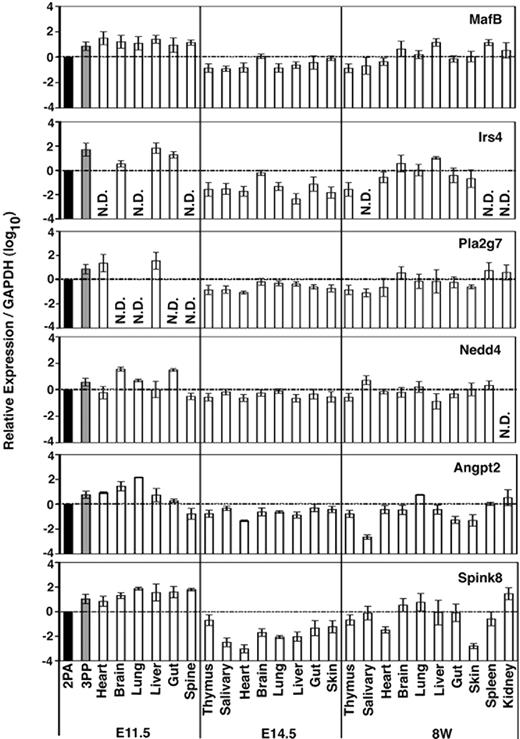
We next examined the expression of these functionally unclear 3PP genes (MafB, Irs4, Pla2g7, Nedd4, Agpt2, and Spink8) in different tissues isolated from different ontogenies of mice. Total RNA isolated from indicated tissues of E11.5, E14.5, and 8-week-old mice was reverse-transcribed (Figure 2). Quantitative RT-PCR confirmed that all of these 6 genes were expressed at higher levels in the 3PP than in the 2PA at E11.5 (Figure 2), in good agreement with the results of initial screening (Table 2). However, none of them was expressed strictly in the 3PP or specifically in the thymus; rather, all 6 genes were expressed in various organs throughout the ontogeny (Figure 2). It may be interesting to note that Irs4 and Pla2g7 were undetectable in several organs of E11.5 mice, and Irs4 was undetectable in several organs of 8-week-old mice (Figure 2), suggesting that Irs4 and Pla2g7 show higher tissue expression specificity than the other 4 genes.
Expression profiles of 3PP genes. Quantitative RT-PCR analysis of total cellular RNA from indicated tissues of E11.5, E14.5, and 8-week-old (8W) C57BL/6 mice. mRNA levels of indicated genes were normalized to GAPDH mRNA levels, and are indicated as the ratio to the amount expressed in E11.5 2PAs (■). Shown are geometric means (bars) and standard errors (lines) of 3 independent measurements.
Expression profiles of 3PP genes. Quantitative RT-PCR analysis of total cellular RNA from indicated tissues of E11.5, E14.5, and 8-week-old (8W) C57BL/6 mice. mRNA levels of indicated genes were normalized to GAPDH mRNA levels, and are indicated as the ratio to the amount expressed in E11.5 2PAs (■). Shown are geometric means (bars) and standard errors (lines) of 3 independent measurements.
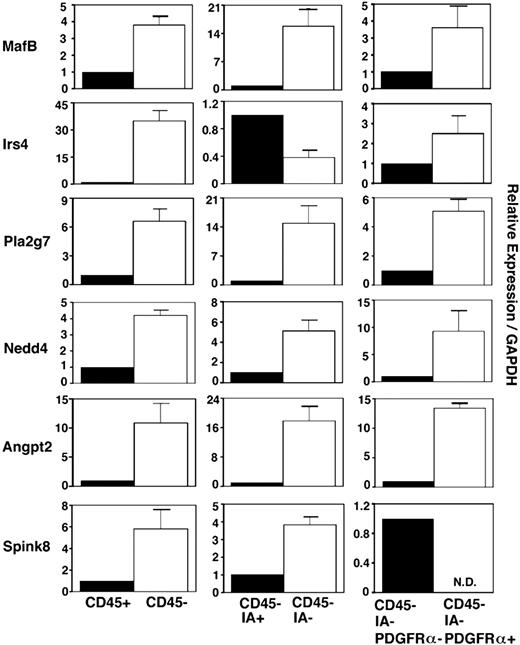
We also examined the expression of these 6 genes in various cell subsets in embryonic thymus. Freshly isolated E14.5 fetal thymus lobes were digested with collagenase, dispase, and DNase, and single-cell suspensions were multicolor-stained for flow cytometry sorting of CD45+ leukocytes and CD45− stromal cells. CD45− stromal cells were additionally fractionated into IA+ thymic epithelial cells and IA− nonthymic epithelial cells. CD45−IA− nonthymic epithelial cells were further fractionated into PDGFRα+ mesenchymal cells and PDGFRα− nonmesenchymal cells47 (Figure 3). Quantitative RT-PCR analysis of the thymic epithelial cell–specific gene FoxN1, T-lymphoid cell–specific gene Lck, and mesenchymal cell–specific gene PDGFRα verified the successful fractionation of sorted cell populations (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). In this analysis of sorted cell populations, all 6 genes were more strongly expressed in CD45− stromal cells than CD45+ leukocytes (Figure 3). Of the 6 genes, 5 (MafB, Pla2g7, Nedd4, Agpt2, and Spink8) were more strongly expressed in CD45−IA− nonthymic epithelial cells than CD45−IA+ thymic epithelial cells, whereas Irs4 was more strongly expressed in thymic epithelial cells than nonepithelial thymic stromal cells. MafB, Irs4, Pla2g7, Nedd4, and Agpt2 were more strongly expressed in CD45−IA−PDGFRα+ thymic mesenchymal cells than PDGFRα− nonmesenchymal thymic stromal cells, whereas Spink8 was more strongly expressed in nonmesenchymal, nonepithelial thymic stromal cells than thymic mesenchymal cells (Figure 3). These results indicate that among the 6 genes that are expressed in the 3PP but whose functions are unclear, MafB, Pla2g7, Nedd4, and Agpt2 are predominantly expressed in mesenchymal cells in the embryonic thymus, whereas Irs4 and Spink8 are predominantly expressed in thymic epithelial cells and nonepithelial, nonmesenchymal stromal cells, respectively, in the embryonic thymus.
Expression of 3PP genes in fetal thymus cells. Quantitative PCR analysis of total cellular RNA from indicated cell fractions purified from E14.5 C57BL/6 mice. mRNA levels of indicated genes were normalized to GAPDH mRNA levels and are indicated as the ratio to the amount expressed in CD45+ leukocytes (left panels), CD45−IA+ epithelial cells (middle panels), or CD45−IA−PDGFRα− nonepithelial, nonmesenchymal stromal cells (right panels). Means and standard errors of the results obtained from 3 independent measurements are shown.
Expression of 3PP genes in fetal thymus cells. Quantitative PCR analysis of total cellular RNA from indicated cell fractions purified from E14.5 C57BL/6 mice. mRNA levels of indicated genes were normalized to GAPDH mRNA levels and are indicated as the ratio to the amount expressed in CD45+ leukocytes (left panels), CD45−IA+ epithelial cells (middle panels), or CD45−IA−PDGFRα− nonepithelial, nonmesenchymal stromal cells (right panels). Means and standard errors of the results obtained from 3 independent measurements are shown.
MafB is expressed in thymic mesenchymal cells
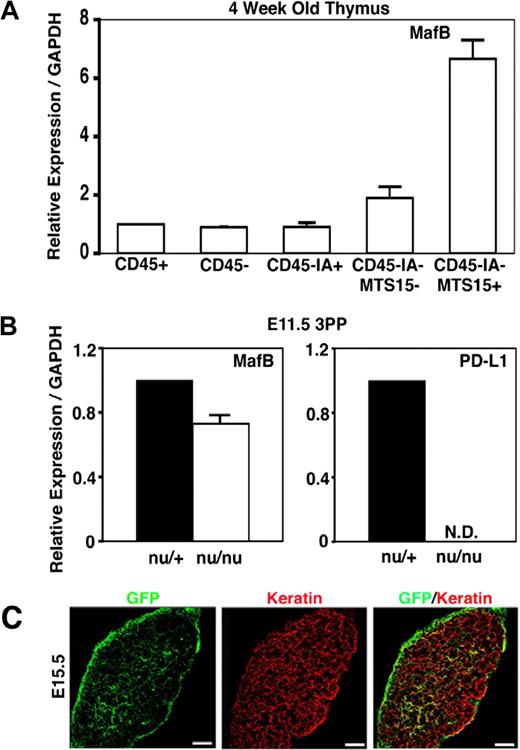
Because MafB is the only transcription factor in the 6 3PP genes whose roles are unclear in the development and function of 3PP-derived organs, we further analyzed the expression and function of MafB in thymus development. To do so, we initially examined MafB expression in cell subpopulations isolated from embryonic and adult thymi. As described above, MafB in E14.5 embryonic thymus was predominantly expressed in mesenchymal cells (Figure 3). To examine the expression of MafB in postnatal thymus, adult thymus lobes were digested with collagenase, dispase, and DNase, and the single-cell suspension was fractionated into CD45+ leukocytes and CD45− stromal cells. CD45− stromal cells were further fractionated into CD45−IA+ thymic epithelial cells, CD45−IA+MTS15+ thymic mesenchymal cells, and CD45−IA+MTS15− nonepithelial, nonmesenchymal thymic stromal cells. Quantitative RT-PCR analysis of MafB mRNA levels showed that MafB was most strongly expressed in CD45−IA+MTS15+ thymic mesenchymal cells in the adult thymus (Figure 4A). These results indicate that MafB in the thymus is most strongly expressed in mesenchymal cells in either the embryonic or postnatal period.
Expression of MafB in adult thymus cells and nude 3PPs. (A) Quantitative PCR analysis of MafB transcript levels in indicated cell fractions isolated from adult C57BL/6 mice. MafB mRNA levels were normalized to GAPDH mRNA levels and are indicated as the ratio to the amount expressed in CD45+ leukocytes. Means and standard errors of the results obtained from 4 independent measurements are shown. (B) Quantitative PCR analysis of MafB and PD-L1 transcript levels in microdissected 3PPs from E11.5 nu/+ mice or E11.5 nu/nu mice. mRNA levels were normalized to GAPDH mRNA levels and are indicated as the ratio to the amount expressed in nu/+ 3PPs. Means and standard errors of the results obtained from 3 independent measurements are shown. (C) In situ expression analysis of MafB in developing thymus by monitoring GFP expression (green) derived from the MafB− allele, in which the coding sequence of MafB was replaced with GFP, of MafB+/− heterozygous mice.31 Paraformaldehyde-fixed E15.5 thymus sections were stained with anti-pancytokeratin antibody (red). Scale bars indicate 100 μm. Representative results of 10 different sections are shown.
Expression of MafB in adult thymus cells and nude 3PPs. (A) Quantitative PCR analysis of MafB transcript levels in indicated cell fractions isolated from adult C57BL/6 mice. MafB mRNA levels were normalized to GAPDH mRNA levels and are indicated as the ratio to the amount expressed in CD45+ leukocytes. Means and standard errors of the results obtained from 4 independent measurements are shown. (B) Quantitative PCR analysis of MafB and PD-L1 transcript levels in microdissected 3PPs from E11.5 nu/+ mice or E11.5 nu/nu mice. mRNA levels were normalized to GAPDH mRNA levels and are indicated as the ratio to the amount expressed in nu/+ 3PPs. Means and standard errors of the results obtained from 3 independent measurements are shown. (C) In situ expression analysis of MafB in developing thymus by monitoring GFP expression (green) derived from the MafB− allele, in which the coding sequence of MafB was replaced with GFP, of MafB+/− heterozygous mice.31 Paraformaldehyde-fixed E15.5 thymus sections were stained with anti-pancytokeratin antibody (red). Scale bars indicate 100 μm. Representative results of 10 different sections are shown.
MafB expression in microdissected tissues from E11.5 3PPs of FoxN1-deficient nude mice was comparable with that of control heterozygotes, whereas the expression of PD-L1, which was previously shown to be dependent on thymic epithelial cell–specific transcription factor FoxN1,48 was undetectable in E11.5 3PPs of nude mice (Figure 4B). These results indicate that MafB expression in E11.5 3PPs is independent of FoxN1.
In situ expression analysis of MafB in the developing thymus by monitoring GFP expression derived from the MafB− allele, in which the coding sequence of MafB was replaced with GFP,31 of MafB+/− heterozygous mice showed that MafB expression in embryonic thymus was most prominently detected at the capsules and also detectable within the thymus (Figure 4C). This distribution of MafB-expressing cells agrees with the reported distribution of neural crest–derived mesenchymal cells.49-51
Thymus development in MafB-deficient mice
MafB is essential for central respiratory control, possibly involving the specification of rhythmogenic neurons. MafB-deficient mice die from central apnea at birth and are defective for respiratory rhythmogenesis.31,52 MafB-deficient newborn mice also display renal dysgenesis.31 We thus examined thymus development in MafB-deficient mice during embryogenesis before death at birth. We found that thymus containing keratin-expressing thymic epithelial cells was formed in MafB-deficient mice at E12.5 (Figure 5A). However, the number of CD45+ lymphoid cells that accumulated within E12.5 thymus of MafB-deficient mice was significantly reduced and approximately half of that of wild-type mice (Figure 5A, D). The number of CD45+ cells in the E12.5 thymus of MafB+/− heterozygous mice was less severely reduced and approximately 75% of that of wild-type mice (Figure 5A,D), suggesting that haploid dosage of the MafB gene affects the number of E12.5 thymocytes. Similarly, the numbers of CD45+ cells in E13.5 thymus, which were 4-fold larger than those in E12.5 thymus, were reduced in MafB−/− mice (approximately 50% of wild-type mice) and MafB+/− mice (approximately 75% of wild-type mice; Figure 5B,E). On the other hand, the numbers of thymic epithelial cells identified by the coexpression of FoxN1 and keratin were comparable and not significantly different among MafB−/−, MafB+/−, and wild-type mice (Figure 5C,F). These results indicate that the number of thymocytes that initially accumulate in the fetal thymus is significantly reduced in MafB-deficient mice, and that MafB affects the optimal cellularity of thymocytes but not thymic epithelial cells.
Early development of embryonic thymus in MafB-deficient mice. (A-C) Sagittal sections of indicated mice at E12.5 (A) or E13.5 (B) were 2-color–stained for CD45 (green) and keratin (red). Sagittal E13.5 sections of frozen embryos from indicated mice were also 2-color–stained for FoxN1 (red) and keratin (blue; C). Scale bars indicate 50 μm in panel A and 150 μm in panels B and C. (D-F) Means and standard errors (n = 10 for panel D, n = 4 for panel E, and n = 2 for panel F) of the numbers of CD45+ leukocytes (D,E) or FoxN1+ cells (F) in keratin+ thymic areas per section of E12.5 (D) and E13.5 (E,F) embryos are shown. ***P < .001; **P < .01; N.S., not significant.
Early development of embryonic thymus in MafB-deficient mice. (A-C) Sagittal sections of indicated mice at E12.5 (A) or E13.5 (B) were 2-color–stained for CD45 (green) and keratin (red). Sagittal E13.5 sections of frozen embryos from indicated mice were also 2-color–stained for FoxN1 (red) and keratin (blue; C). Scale bars indicate 50 μm in panel A and 150 μm in panels B and C. (D-F) Means and standard errors (n = 10 for panel D, n = 4 for panel E, and n = 2 for panel F) of the numbers of CD45+ leukocytes (D,E) or FoxN1+ cells (F) in keratin+ thymic areas per section of E12.5 (D) and E13.5 (E,F) embryos are shown. ***P < .001; **P < .01; N.S., not significant.
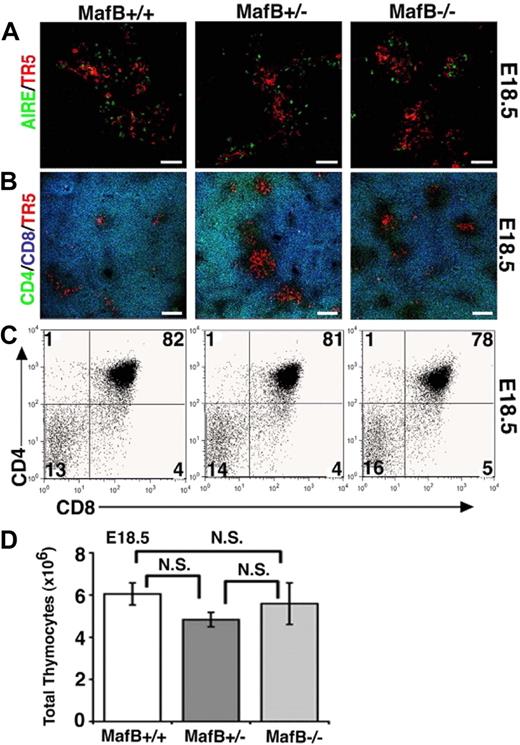
Later in development at E18.5, the thymi of MafB-deficient mice contained ER-TR5–expressing medullary epithelial cells that included AIRE+ cells (Figure 6A) and generated unimpaired CD4/CD8 profiles of thymocytes that included CD4+CD8+ thymocytes (Figure 6B,C). The number of thymocytes was not significantly impaired in E18.5 MafB-deficient mice (Figure 6D). These results indicate that unlike initial thymus development at E12.5 and E13.5, thymus development at E18.5 is restored to normalcy in MafB-deficient mice, and therefore suggest that MafB is involved in embryonic thymus development only during initial ontogeny.
Late development of embryonic thymus in MafB-deficient mice. (A,B) Frozen sections of embryonic thymus isolated from indicated mice at E18.5 were multicolor-stained either for AIRE (green) and ER-TR5 (a marker for medullary thymic epithelial cells; red) or for CD4 (green), CD8 (blue), and ER-TR5 (red). Representative results of 4 independent experiments are shown. Scale bars indicate 50 μm in panel A and 100 μm in panel B. (C) Two-color flow cytometry analysis of CD4 and CD8 in E18.5 thymocytes from indicated mice. Numbers indicate percentage of cells in quadrant. Representative results of 4 independent experiments are shown. (D) Means and standard errors (n = 3-4) of the numbers of thymocytes from indicated mice at E18.5 are shown. N.S. indicates not significant.
Late development of embryonic thymus in MafB-deficient mice. (A,B) Frozen sections of embryonic thymus isolated from indicated mice at E18.5 were multicolor-stained either for AIRE (green) and ER-TR5 (a marker for medullary thymic epithelial cells; red) or for CD4 (green), CD8 (blue), and ER-TR5 (red). Representative results of 4 independent experiments are shown. Scale bars indicate 50 μm in panel A and 100 μm in panel B. (C) Two-color flow cytometry analysis of CD4 and CD8 in E18.5 thymocytes from indicated mice. Numbers indicate percentage of cells in quadrant. Representative results of 4 independent experiments are shown. (D) Means and standard errors (n = 3-4) of the numbers of thymocytes from indicated mice at E18.5 are shown. N.S. indicates not significant.
Reduced chemokine expression in embryonic thymus of MafB-deficient mice
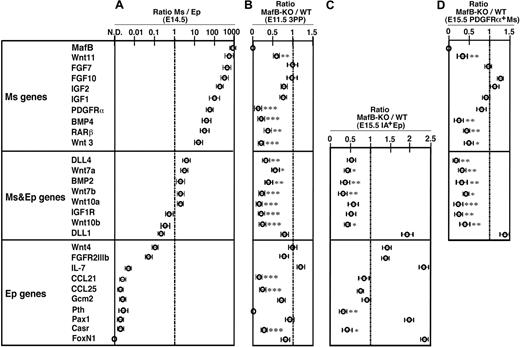
Finally, we examined the expression of genes that were previously shown to participate in thymus development6,8 (Figure 7) to understand how MafB regulates initial thymus development. Using quantitative RT-PCR, the genes were first examined for the ratios of expression between CD45−IA−PDGFRα+ thymic mesenchymal cells and CD45−IA+ thymic epithelial cells isolated from the embryonic thymus (Figure 7A). Wnt11, FGF7, and PDGFRα genes were more than 10 times more strongly detectable in mesenchymal cells than in epithelial cells (Figure 7A mesenchymal genes). In contrast, the expression levels of DLL4, Wnt7a, and BMP2 genes showed a less than 10-fold difference between mesenchymal and epithelial cells (Figure 7A mesenchymal and epithelial genes). Wnt4, FGFR2IIIb, Pax1, and FoxN1 genes were more than 10 times more strongly detectable in epithelial cells than in mesenchymal cells (Figure 7A epithelial genes). We then isolated cellular RNA from microdissected tissues of E11.5 3PPs of MafB-deficient mice as well as wild-type mice (Figure 7B). Quantitative RT-PCR analysis of mesenchymal and/or epithelial genes in the 3PPs of MafB-deficient mice showed significantly reduced expression of mesenchymal genes, such as Wnt11, PDGFRα, BMP4, RARβ, and Wnt3; mesenchymal and epithelial genes, such as DLL4, Wnt7a, BMP2, Wnt7b, Wnt10a, IGF1R, and Wnt10b; and epithelial genes, such as CCL21 and CCL25 (Figure 7B). Other genes in the 3PPs of MafB-deficient mice, including mesenchymal FGF7 and FGF10 and epithelial IL7, Pax1, Gcm2, and FoxN1, showed no expression reduction (Figure 7B). These results indicate that MafB, which in the thymus is most strongly expressed in mesenchymal cells, regulates the expression of several epithelial genes, including CCL21 and CCL25, in addition to several mesenchymal genes, including Wnt3 and BMP4, in E11.5 3PPs.
Gene expression profiles of thymic stromal cells in MafB-deficient mice. (A) Quantitative RT-PCR analysis of indicated genes in CD45−IA−PDGFRα+ thymic mesenchymal cells (Ms) and CD45−IA+ thymic epithelial cells (Ep) isolated from E14.5 C57BL/6 embryonic thymus. mRNA levels were normalized to GAPDH mRNA levels and are indicated as the ratios of the levels in Ms cells to the levels in Ep cells. Shown are geometric means (bars) and standard errors (lines) of 3 independent measurements. The genes are aligned according to the values of the ratios. In this list, genes that are more than 10 times more strongly detectable in Ms cells than in Ep cells are categorized as Ms genes. Genes that are not more than 10 times different in Ms cells and Ep cells are categorized as Ms and Ep genes. Genes that are more than 10 times more strongly detectable in Ep cells than in Ms cells are categorized as Ep genes. (B) Quantitative RT-PCR analysis of indicated genes in microdissected 3PP tissues from either E11.5 MafB-deficient (MafB-KO) or wild-type mice (WT). mRNA levels were normalized to GAPDH mRNA levels and are indicated as the ratios of the levels in MafB-KO 3PP to the levels in WT 3PP. Data are means and standard errors of 5 separate measurements. (C,D) Quantitative RT-PCR analysis of indicated genes in CD45−IA+ thymic epithelial cells (C) and CD45−PDGFRα+ thymic mesenchymal cells (D) isolated from E15.5 MafB-deficient (MafB-KO) or wild-type mice (WT). mRNA levels were normalized to GAPDH mRNA levels and are indicated as the ratios of the levels in MafB-KO samples to those in WT samples. Data are means and standard errors of 4 separate measurements. ***P < .001; **P < .01; *P < .05.
Gene expression profiles of thymic stromal cells in MafB-deficient mice. (A) Quantitative RT-PCR analysis of indicated genes in CD45−IA−PDGFRα+ thymic mesenchymal cells (Ms) and CD45−IA+ thymic epithelial cells (Ep) isolated from E14.5 C57BL/6 embryonic thymus. mRNA levels were normalized to GAPDH mRNA levels and are indicated as the ratios of the levels in Ms cells to the levels in Ep cells. Shown are geometric means (bars) and standard errors (lines) of 3 independent measurements. The genes are aligned according to the values of the ratios. In this list, genes that are more than 10 times more strongly detectable in Ms cells than in Ep cells are categorized as Ms genes. Genes that are not more than 10 times different in Ms cells and Ep cells are categorized as Ms and Ep genes. Genes that are more than 10 times more strongly detectable in Ep cells than in Ms cells are categorized as Ep genes. (B) Quantitative RT-PCR analysis of indicated genes in microdissected 3PP tissues from either E11.5 MafB-deficient (MafB-KO) or wild-type mice (WT). mRNA levels were normalized to GAPDH mRNA levels and are indicated as the ratios of the levels in MafB-KO 3PP to the levels in WT 3PP. Data are means and standard errors of 5 separate measurements. (C,D) Quantitative RT-PCR analysis of indicated genes in CD45−IA+ thymic epithelial cells (C) and CD45−PDGFRα+ thymic mesenchymal cells (D) isolated from E15.5 MafB-deficient (MafB-KO) or wild-type mice (WT). mRNA levels were normalized to GAPDH mRNA levels and are indicated as the ratios of the levels in MafB-KO samples to those in WT samples. Data are means and standard errors of 4 separate measurements. ***P < .001; **P < .01; *P < .05.
It was previously shown that FoxN1 essentially promotes the expression of CCL25, DLL1, and DLL4,19-25 whereas Gcm2 is essential for the expression of CCL21, PTH, and Casr.19,23,42-44 Our results demonstrate that the expression of some FoxN1-dependent genes, including CCL25 and DLL4, and some Gcm2-dependent genes, including CCL21, PTH, and Casr, was reduced in the 3PPs of MafB-deficient mice, whereas both FoxN1 and Gcm2 were normally detectable in the absence of MafB (Figure 7B). These results suggest that FoxN1 and Gcm2 expressed in the 3PP are not sufficient for the expression of CCL21 and CCL25 as well as several other epithelial genes, and that mesenchymal MafB, in addition to epithelial FoxN1 and Gcm2, critically regulates the development of the thymus and the parathyroid glands.
We further found that the expression of some parathyroid epithelial genes, such as PTH and Casr, was significantly reduced in IA+ thymic epithelial cells of MafB-deficient mice even at E15.5 (Figure 7C), suggesting that MafB affects the maturation of the epithelial compartment in the common primordia for the thymus and the parathyroid glands. Interestingly, the numbers of PDGFRα+ mesenchymal cells, IA+ epithelial cells, and PDGFRα−IA− stromal cells were not significantly reduced in the thymus of E15.5 MafB-deficient embryos (Figure S2), whereas the expression of some mesenchymal genes, such as Wnt3 and BMP4, was significantly reduced in PDGFRα+ thymic mesenchymal cells of E15.5 MafB-deficient embryos (Figure 7D), suggesting that the reduced expression of such mesenchymal genes as Wnt3 and BMP4 reflects reduced gene expression per mesenchymal cell rather than fewer mesenchymal cells in the embryonic thymus.
We previously identified that the coordination between CCL21 and CCL25 is essential for chemotactic guidance of T-lymphoid progenitor cells to fetal thymic primordium before, but not after, vascularization of fetal thymus.23,45,53 It was therefore interesting to know whether or not CCL21 and CCL25 protein expression might be also decreased in the E11.5 3PPs of MafB-deficient mice. As shown in Figure 8, both CCL21 and CCL25 proteins in E11.5 3PPs were less detectable in MafB-deficient mice than in wild-type mice. These results indicate that the 3PP of MafB-deficient mice is defective in the expression of CCL21 and CCL25. The reduction of these 2 chemokines can explain why thymocyte cellularity is reduced only during initial development, rather than late development, of the embryonic thymus, because these 2 chemokines are required for lymphoid progenitor colonization before, but not after, vascularization of fetal thymus.
Expression of CCL21 and CCL25 proteins in E11.5 3PPs of MafB-deficient mice. Sagittal sections of frozen embryos from indicated mice were 2-color–stained either for CCL21 (pink) and keratin (blue) or for CCL25 (pink) and keratin (blue). Anterior-posterior (A-P) and dorsal-ventral (D-V) orientations of the sections are indicated. Scale bars indicate 100 μm. Two representative datasets (nos. 1 and 2) of 10 to 14 different sections are shown.
Expression of CCL21 and CCL25 proteins in E11.5 3PPs of MafB-deficient mice. Sagittal sections of frozen embryos from indicated mice were 2-color–stained either for CCL21 (pink) and keratin (blue) or for CCL25 (pink) and keratin (blue). Anterior-posterior (A-P) and dorsal-ventral (D-V) orientations of the sections are indicated. Scale bars indicate 100 μm. Two representative datasets (nos. 1 and 2) of 10 to 14 different sections are shown.
Discussion
Mesenchymal cells essentially contribute to thymus development before and after the entry of hematopoietic cells. In particular, their interactions with epithelial cells pivotally regulate the maturation of thymic epithelial cells via such mediators as Wnts, BMPs, and FGFs.26-30 However, the transcriptional mechanisms underlying the development of thymic mesenchymal cells were unclear. Our results show that the transcriptional factor MafB in the thymus is predominantly expressed in mesenchymal cells and is essential for optimum accumulation of thymocytes during embryonic thymus development. The expression of Wnt family genes, including Wnt3 and Wnt11, as well as BMP family genes, including BMP4, which were predominantly produced by thymic mesenchymal cells, was decreased in embryonic thymus of MafB-deficient mice. Coincident with the decrease in expression of these mesenchymal genes, the expression of several genes, including CCL21 and CCL25, which were predominantly produced by thymic epithelial cells, was reduced in the embryonic thymi of MafB-deficient mice. These results suggest that MafB expressed in mesenchymal cells of the embryonic thymus governs the production of several molecules, such as Wnt3, Wnt11, and BMP4, which in turn act on thymic epithelial cells to promote the production of CCL21 and CCL25, which attract lymphoid progenitor cells into the thymus. Thus, this study proposes a novel molecular pathway that mediates mesenchymal contribution to thymus development.
MafB is a basic leucine zipper transcription factor belonging to the large Maf family. It forms dimers with various basic leucine zipper proteins, thereby regulating a wide variety of genes.54 MafB is expressed in various tissues, including kidney, pancreas, lens, and hindbrain, as well as in hematopoietic cells.31,49,54,55 A deficiency in MafB results in death at birth from the defect of respiratory neurons in the hindbrain and subsequent central breathing failure.52 MafB-deficient mice also display renal dysgenesis.31 A missense mutation in MafB, called kreisler, causes segmentation abnormalities in the caudal hindbrain and defective inner ear development.55 The expression and function of MafB in mesenchymal cells of embryonic thymus revealed in the present study agree with the previous detection of MafB in rhombomere 6 of the hindbrain,55 which gives rise to neural crest cells that migrate to the third pharyngeal region.49,56 We also showed that MafB in the thymus is barely detectable in other cell types, including thymic epithelial cells, and that MafB deficiency does not result in a decrease in the number of FoxN1-expressing thymic epithelial cells. These results suggest that MafB regulates thymus development by specifically controlling mesenchymal functions that map downstream of and/or in parallel with FoxN1 expression in thymic epithelial cells.
Interestingly, a MafB ortholog in Drosophila melanogaster encoded by traffic jam (tj) is expressed in somatic gonadal cells that are in direct contact with germline cells, and in tj mutant gonads, somatic cells fail to intermingle and properly envelop germline cells, causing early blockade of germ cell differentiation.57 Thus, the possible role of MafB in mesenchymal cells and in cell-to-cell interactions for thymus development may be analogous to the role of tj in gonad morphogenesis in the fruit fly.
Regarding the mechanisms underlying thymus development regulation by MafB in mesenchymal cells, our results showed that the 3PPs of MafB-deficient mice express significantly reduced levels of Wnt3, Wnt11, and BMP4 genes among the genes that are expressed in thymic mesenchymal cells rather than thymic epithelial cells. Wnt7a, Wnt7b, Wnt10a, Wnt10b, and BMP2, which are expressed by both thymic mesenchymal cells and thymic epithelial cells, are also decreased in the 3PPs of MafB-deficient mice. Previous reports described that Wnts regulate thymocyte cellularity58,59 and can act on thymic epithelial cells27 and thymocytes.60 It was also reported that BMPs are expressed in thymic mesenchymal cells and act to regulate thymus development by controlling the epithelial-mesenchymal interaction that shapes thymic stroma.28,61-63 BMPs in the thymus may also directly act on immature thymocytes.64,65 On the other hand, FGF7 and FGF10, which are also expressed in thymic mesenchymal cells rather than thymic epithelial cells and regulate the development of thymic epithelial cells,26,29,66,67 are normally detectable in the 3PPs of MafB-deficient mice. Thus, we think it is possible that MafB in thymic mesenchymal cells regulates the expression of these Wnts and BMPs rather than FGFs, thereby regulating thymus development by acting on thymic epithelial cells and thymocytes. It is interesting to note that MafB does not affect FGF7 and FGF10 gene expression, in agreement with the finding that the number of FoxN1-expressing thymic epithelial cells is not reduced in MafB-deficient mice.
Genes that are severely affected in the 3PPs of MafB-deficient mice include CCL21 and CCL25. The expression of CCL21 and CCL25 proteins in thymic primordium is indeed reduced in MafB-deficient mice. CCL21 and CCL25 are expressed in Gcm2-dependent parathyroid epithelial cells and FoxN1-dependent thymic epithelial cells, respectively, rather than thymic mesenchymal cells and are involved in attracting T-lymphoid progenitor cells to the fetal thymus before vascularization.23,45,53 Thus, we think it is possible that MafB in 3PP mesenchymal cells regulates the expression of Wnts and BMPs, and these molecules, in turn, act on 3PP epithelial cells to produce CCL21 and CCL25. Later in embryonic development, thymocyte cellularity recovers to normalcy in MafB-deficient mice, in agreement with the possibility that the reduced thymocyte cellularity in early embryogenesis is due to impaired chemokine production by fetal thymus without MafB. These results indicate that unlike initial thymus development at E12.5 and E13.5, thymus development at E18.5 is restored to normalcy in MafB-deficient mice, therefore suggesting that MafB is involved in embryonic thymus development only during initial ontogeny. Indeed, the numbers of Vγ3-expressing TCR-γδ+ T cells, which represent the first wave of T cells in the ontogeny,23,68 in the thymus and the skin at E18.5 were not reduced in the embryos of MafB-deficient mice (Figure S3).
Itoi et al reported that in PDGFRα-deficient patch mutant mice, whose mesenchymal cells are primarily affected, epithelial cells in thymic primordium show a reduction in number and are defective in the expression of SCF, DLL4, and class II MHC molecules.69 Interestingly, thymic mesenchymal cells in patch mutant embryos are defective in producing FGF7 and FGF10, whereas the genes that exhibit reduced expression in thymic epithelial cells of patch mutant mice do not include CCL21 or CCL25.69 These results indicate that spectra of molecules affected in patch mutant mice and MafB-deficient mice are unequal either in thymic mesenchymal cells or in thymic epithelial cells. Thus, it is likely that epithelial-mesenchymal interactions during thymus development are mediated by multiple molecular pathways in parallel rather than a single molecular stimulus.
In this study, we used PCR-based cDNA subtraction screening for the genes expressed in microdissected tissues of the 3PP rather than the 2PA. Among the 12 genes extracted, 6 (Pax1, Gcm2, Casr, PTH, CCL21, and IL7) are known for their roles in the development and function of the thymus and the parathyroid glands,15,16,19,23,42-46 suggesting that our gene expression profiling has a good fidelity in extracting genes that are involved in the development and function of the 3PP and its derivative organs. However, it should be noted that our screening did not extract such genes as Tbx1, Hoxa3, and FoxN1, which are also specifically expressed in the 3PP and are also crucial for its development.8-15,19-21 These genes may be expressed in the 2PA and thus subtracted during the procedure (eg, Tbx1) or expressed in E11.5 3PPs at very low levels and thus undetectable in the present screening (eg, FoxN1).
In conclusion, the present study examining the gene expression profile of the mouse 3PP has identified the role of MafB in thymus development. The results have revealed a novel transcriptional regulation of thymus development by mesenchymal cells and offer a novel molecular pathway that mediates mesenchymal contribution to thymus development via Wnts, BMPs, and chemokines. Because it is shown that transcription factors belonging to the large Maf family, including MafB, MafA, and c-Maf, can exert partly overlapping roles in organ development and cellular functions,70,71 it is possible that MafA and c-Maf may also be involved in controlling thymus development. An improved understanding of the molecular mechanisms of thymus development should aid in the formulation of strategies to enhance thymus regeneration in the elderly or those following chemotherapy and/or radiotherapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Drs N. Iwanami, T. Nitta, and T. Ueno for critically reading the manuscript.
This study was supported by a Ministry of Education, Culture, Sports, Science, and Technology (MEXT; Tokyo, Japan) Grant-in-Aid for Scientific Research.
Authorship
Contribution: D.A.S., S. Tomita, and Y.T. designed the research and wrote the paper; D.A.S., S. Tomita, M.H., Y.I., Y.K., N.V.K., S.H., I.O., and S.N. performed research; D.A.S. and S. Tomita analyzed data; and T.A. and S. Takahashi contributed vital materials.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current address for Dr Tomita is Department of Pharmacology, Institute of Health Biosciences, University of Tokushima, Tokushima 770-8503, Japan.
Correspondence: Yousuke Takahama, Division of Experimental Immunology, Institute for Genome Research, University of Tokushima, 3-18-15 Kuramoto, Tokushima, Japan; e-mail: takahama@genome.tokushima-u.ac.jp.
References
Author notes
*D.A.S. and S. Tomita contributed equally to this work.