Abstract
Nonrandom X-chromosome inactivation (XCI), also known as skewing, has been documented in the blood cells of a significant proportion of normal aging women by the use of methylation-based assays at the polymorphic human androgen receptor locus (HUMARA). Recent data obtained with a new transcription-based XCI determination method, termed suppressive polymerase chain reaction (PCR), has shed controversy over the validity of XCI ratio results obtained with HUMARA. To resolve this disparity, we analyzed XCI in polymorphonuclear leukocytes of a large cohort of women aged 43 to 100 years with the use of HUMARA (n = 100), a TaqMan single nucleotide polymorphism (SNP) assay (n = 90), and the suppressive polymerase chain reaction (PCR) assay (n = 67). The 3 methods yielded similar skewing incidences (42%, 38%, and 40%, respectively), and highly concordant XCI ratios. This confirms that the skewing of XCI ratio seen in blood cells of aging women is a bona fide and robust biologic phenomenon.
Introduction
Gene dosage compensation between XX females and XY males occurs by the random inactivation of one of parental X chromosomes in the female embryo.1 According to the Lyon hypothesis,1 the fraction of cells with inactivation of each X chromosome should be equivalent. A deviation from the 1:1 X-chromosome inactivation (XCI) ratio is referred to as skewing. We have previously shown that skewing of XCI increases significantly in the blood cells of female subjects from the neonatal period to old age.2 This observation has subsequently been confirmed by others3-6 and has important implications for understanding the physiology of aging hematopoiesis and the pathogenesis of age-related hematopoietic malignancies. The concordance of skewing documented in elderly monozygotic versus dizygotic twins7 supports a genetic component to the trait, and work by Abkowitz et al8 in a feline model suggests an age-dependent hemizygous cell-selection process.
A recent report9 argued against the skewing phenomenon in blood cells of aging women. In this study,9 the blood cells of 40 women older than 65 years of age were analyzed by both a methylation-based assay (ie, human androgen receptor locus [HUMARA]) and a novel real-time polymerase chain reaction (PCR) assay, termed suppressive PCR, in which allele-specific primers are used to identify coding single nucleotide polymorphisms (SNPs) in several X-linked genes, including the iduronate-2-sulphatase gene IDS. The use of HUMARA showed that 30% of women had skewed XCI in their blood cells, whereas the suppressive PCR assay did not show any significant skewing.9 These observations led the authors to conclude that the skewed XCI patterns observed with HUMARA were not an accurate reflection of the proportion of cells bearing a particular X chromosome in the active state but rather reflected age-related dysregulation of methylation patterns.9 Although this explanation is a plausible one for these data, it is at odds with previously published reports validating the HUMARA assay against transcription-based assays at the HUMARA or IDS loci10-12 and also with data showing concordance of skewing at the unlinked HUMARA and phosphoglycerate kinase (PGK) loci.2
To address the disparity suggested by the implementation of the suppressive PCR assay, we analyzed blood cells of aging women by using (1) HUMARA, (2) a TaqMan SNP assay at the IDS locus, and (3) the suppressive PCR assay.9
Methods
The study was approved by the Maisonneuve-Rosemont Hospital Ethics Committee, and informed consent was obtained in accordance with the Declaration of Helsinki. It included 100 women between 43 and 100 years of age (mean age, 64 years) who were selected for having a normal complete blood count and being heterozygous at both the HUMARA and IDS loci. The methods used to isolate DNA and RNA from polymorphonuclear cells have been previously described.13 Herein, skewing was defined as an XCI ratio ≥ 3:1. This arbitrary definition has been used in several other articles.2,13-16
For the HUMARA assay, PCR amplification of the polymorphic CAG repeat at the HUMARA locus2,17 was performed in tandem on undigested (6-FAM–labeled primer) and on HpaII-digested (HEX-labeled primer) DNA. Amplification products were migrated on an ABI PRISM 3100 genetic analyzer (Applied Biosystems, Foster City, CA) to determine the area under the curve (AUC) for each allele. If the alleles are 1 or 2 repeat lengths different in size, shadow banding from the higher molecular weight allele (superior allele) can affect the measured AUC of the lower molecular- weight allele (inferior allele).18 To compensate for this, homozygous samples and samples with more than 5 repeat lengths between the 2 alleles were used to measure the percentage of shadow banding for each repeat length, and this percentage was subtracted from the inferior allele. The XCI ratios determined by the HUMARA assay are reported using the percentage of cells with the superior allele active (%Sup), and is calculated as follows:
(A and A′ represent the AUC of the superior HUMARA allele from the digested and undigested sample, respectively. The AUC of the inferior HUMARA allele for the digested and undigested samples are represented by a and a′, respectively.)
A %Sup of 50% indicates no skewing, a %Sup of 100% indicates that the superior allele is always active, and a %Sup of 0% indicates the opposite. Skewing is said to be present if %Sup is equal to or greater than 75% (in cases where the superior allele is the predominant allele) or equal to or less than 25% (in cases where the inferior allele is the predominant allele).
We used 2 transcription-based assays in this study. The first was the TaqMan SNP assay,11 which is based on the detection of a exonic SNP of the IDS gene (reference ID: rs1141608). For this assay, cDNA was synthesized from PMN RNA with the use of random hexamers and TaqMan Reverse Transcription Reagents (Applied Biosystems), after which the aforementioned SNP was detected with the use of the TaqMan SNP assay (Applied Biosystems). This assay included 2 probes capable of distinguishing each allele of the SNP in real time in the same PCR and was conducted on an ABI Prism 7000 machine (Applied Biosystems). The second transcription-based assay was the suppressive PCR assay at the IDS locus. For this assay, cDNA was synthesized from PMN RNA by use of the Omniscript Reverse Transcriptase kit (QIAGEN, Mississauga, ON). PCRs were performed in separate tubes for the C and T alleles with the cIDS-R reverse primer and either the cIDS-LNA-C-F or the cIDS-LNA-T-F forward primer (Integrated DNA Technologies, Coralville, IA), as described by Swierczek et al.9 Amplified products were detected using the Platinum SYBR green PCR Supermix (Invitrogen, Mississauga, ON). Real-time PCR was performed on a Bio-Rad iCycler IQ system (Bio-Rad Laboratories, Mississauga, ON). PCR parameters were 50°C for 2 minutes, 95°C for 10 minutes, and 50 cycles of 94°C for 10 seconds, 45°C for 20 seconds, and 72°C for 30 seconds. Ct values were determined by use of iCycler software (Bio-Rad Laboratories). Allele frequencies were calculated from a standard curve generated from cDNA prepared from cell mixtures (2 men hemizygous for the different alleles [C vs T]). Subject samples were measured in triplicate, and mean ΔCt value used to calculate allele frequency. A second standard curve was obtained from mixtures of PCR fragments obtained from hemizygous men using cIDS-R and a forward primer located 30 bases upstream of the SNP (cIDS-F). For both transcription-based assays, samples in which 75% or more of cells express the same IDS allele are said to be skewed. This is in line with our definition of skewing (XCI ratio ≥ 3:1).
Statistical analysis (Pearson correlation, χ2) was conducted with the use of NCSS 2004 (NCSS, Kaysville, UT). Agreement and repeatability coefficients were determined by use of the Bland-Altman method.19
Results and discussion
Repeatability of the 3 methods
The reproducibility of results obtained with each of the 3 assays was evaluated by the “coefficient of repeatability” described by Bland and Altman,19 which includes 95% of the differences between 2 repeated measurements. The coefficient of repeatability19 was 0.0332 (3.3%) for the HUMARA assay, 0.0640 (6.4%) for the TaqMan SNP assay, and 0.2302 (23.0%) for the suppressive PCR assay. Thus, the results obtained with the suppressive PCR assay were less reproducible than those obtained with the other 2 assays.
Incidence of skewing with 3 methods
We evaluated %Sup in the blood cells of 100 women by using HUMARA. Adequate RNA samples were available for 90 of them when we performed the TaqMan SNP assay and for 67 when we performed the suppressive PCR assay. The incidence of skewing (%Sup ≥ 75% or ≤ 25%; percentage of 1 of the 2 IDS alleles ≥ 75%) was similar with all assays: 42/100 (42%) with HUMARA, 34/90 (38%) with the TaqMan SNP assay, and 27/67 (40%) with the suppressive PCR assay. There was no statistically significant difference when comparing any 2 of these assays, or the group of 3 (χ2; P = .84). The incidence also was similar in the subgroup of women older than 65 years (mean age, 71 years): 16 of 40 (40%) with HUMARA, 15 of 36 (41.7%) with TaqMan SNP assay, and 9 of 22 (40.9%) with the suppressive PCR assay.
Intraindividual concordance of XCI patterns obtained with different methods
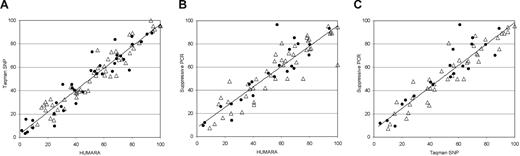
XCI patterns obtained with HUMARA were compared with those obtained with 2 transcription-based assays (Figure 1A,B). We observed highly concordant results between HUMARA and TaqMan SNP: mean of differences was 0.5% (standard deviation [SD] ± 6.7%), mean of the absolute value of the differences was 5.4%, and correlation was 96%. Similarly, HUMARA and suppressive PCR methods gave concordant results: mean difference was 1.6% (SD ± 12.2%), mean of the absolute value of the difference was 8.5%, and correlation was 90%. TaqMan SNP and suppressive PCR methods also were concordant (Figure 1C).
Correlation of XCI patterns obtained with the use of 3 different methods. (A) HUMARA assay versus TaqMan SNP assay at the IDS locus. For each individual, the %Sup measured by HUMARA was compared with the percentage of IDS allele obtained with the TaqMan SNP assay, which was quantitatively closer to the HUMARA value. Data from both assays were available for 90 women. (B) HUMARA assay versus suppressive PCR assay at the IDS locus. The %Sup obtained by HUMARA was compared with the percentage of IDS allele obtained with the suppressive PCR assay, which was quantitatively closer to the HUMARA value. Data from both assays were available for 67 women. (C) TaqMan SNP assay versus suppressive PCR assay. The percentage of IDS allele obtained with the TaqMan SNP assay was compared with that obtained with the suppressive PCR assay. Data from both assays were available for 60 women. Those aged 44 to 65 years are represented by ▵, whereas those older than 65 years are represented by ●.
Correlation of XCI patterns obtained with the use of 3 different methods. (A) HUMARA assay versus TaqMan SNP assay at the IDS locus. For each individual, the %Sup measured by HUMARA was compared with the percentage of IDS allele obtained with the TaqMan SNP assay, which was quantitatively closer to the HUMARA value. Data from both assays were available for 90 women. (B) HUMARA assay versus suppressive PCR assay at the IDS locus. The %Sup obtained by HUMARA was compared with the percentage of IDS allele obtained with the suppressive PCR assay, which was quantitatively closer to the HUMARA value. Data from both assays were available for 67 women. (C) TaqMan SNP assay versus suppressive PCR assay. The percentage of IDS allele obtained with the TaqMan SNP assay was compared with that obtained with the suppressive PCR assay. Data from both assays were available for 60 women. Those aged 44 to 65 years are represented by ▵, whereas those older than 65 years are represented by ●.
Confirmation that XCI skewing documented by HUMARA is valid
We show that measurements of XCI patterns by HUMARA are highly concordant with those obtained with 2 transcription-based methods, including the suppressive PCR assay. Our data should dissipate concerns about potential age-associated changes in methylation patterns that could influence HUMARA results. In so doing, the data presented herein validate the use of HUMARA for the study of X-inactivation in elderly females.
Although we observed a poorer repeatability of the suppressive PCR assay (compared with the other 2 assays), in our hands, the suppressive PCR assay gave results that correlated well with previously established XCI determination methods. Therefore, we find it difficult to explain the discordant data reported by Swierczek et al.9 Unlike Swierczek et al,9 we used SYBR green instead of the generic TaqMan probe (nonallele specific) for the real-time quantification. It is possible that an interaction between the TaqMan probe and the suppressive primers played a role in the discordant results. Moreover, the suppressive approach has no significant advantage over the TaqMan SNP assays, which use 2 different allele-specific probes in the same reaction, ensuring identical amplification conditions for the 2 alleles. The suppressive PCR approach, which uses allele-specific primers to amplify the 2 different SNP alleles in separate PCRs,9 is per se more subject to amplification biases. In fact, the standard curve obtained with cIDS-F primer indicated that with the PCR conditions used, the cIDS-LNA-T-F primer amplified only 1.2% of the T allele present in the reaction tube, whereas the cIDS-LNA-C-F amplified 2.9% of the C allele, indicating a strong bias in amplification efficiency for the 2 alleles. It is also important to note that the analysis of pure clonal or homozygous patients9 is not sufficient as validation or standardization for this assay.
Support for the acquired skewing phenomenon
Although this study was designed to ascertain the validity of the HUMARA method for determining XCI patterns in aging women, it indirectly validates previous studies in which HUMARA assay documented age-dependent increases in skewing incidence in females.2-4,6,20 Furthermore, if only transcription based analysis were to be taken into account, the results obtained in this cohort of aging females and our recently published data obtained in neonates13 document that the incidence of skewing triples from birth to older age. Further studies on age-dependent skewing may help understand the biology of the aging hematopoiesis, and the pathobiology of hematopoietic malignancies of the elderly.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
L.M. is a Fonds de la Recherche en Santé du Québec (FRSQ, Montréal, QC) scholar.
Authorship
Contribution: L.B. designed research and wrote the paper; Y.P. performed research; S.P. analyzed data; D.-C.R. designed research; R.L.L. reviewed the paper; L.M. designed the study and wrote the paper; and D.G.G. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lambert Busque, MD, FRCPC, Département d'Hématologie, Hôpital Maisonneuve-Rosemont, 5415, Boulevard de l'Assomption, Montréal, QC, Canada H1T 2M4; e-mail: lbusque.hmr@ssss.gouv.qc.ca.

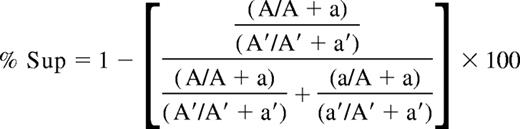
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal