Abstract
Erythropoiesis is primarily controlled by erythropoietin (Epo), which stimulates proliferation, differentiation, and survival of erythroid precursors. We have previously shown that the tyrosine kinase Lyn is critical for transducing differentiation signals emanating from the activated Epo receptor. A yeast 2-hybrid screen for downstream effectors of Lyn identified a novel protein, Liar (Lyn-interacting ankyrin repeat), which forms a multiprotein complex with Lyn and HS1 in erythroid cells. Interestingly, 3 of the ankyrin repeats of Liar define a novel SH3 binding region for Lyn and HS1. Liar also contains functional nuclear localization and nuclear export sequences and shuttles rapidly between the nucleus and cytoplasm. Ectopic expression of Liar inhibited the differentiation of normal erythroid progenitors, as well as immortalized erythroid cells. Significantly, Liar affected Epo-activated signaling molecules including Erk2, STAT5, Akt, and Lyn. These results show that Liar is a novel Lyn-interacting molecule that plays an important role in regulating intracellular signaling events associated with erythroid terminal differentiation.
Introduction
Binding of erythropoietin (Epo) to its cognate receptor initiates a series of intracellular signaling cascades within immature red blood cells, including JAK/STAT, ras/Raf/Erk, and PI-3 kinase/Akt pathways.1-6 Activation of these pathways is important in maintaining cell viability and promoting proliferation and differentiation of erythroid precursors.7-9 JAK2 is the primary kinase associated with phosphorylation of the Epo receptor;1 however, recent studies have shown Lyn, a member of the Src kinase family, is important as a secondary kinase for Epo receptor signaling.10-13 Lyn is an intracellular membrane-associated tyrosine kinase expressed predominantly in hemopoietic cells and is involved in the transmission of signals from several hemopoietic receptors, including the Epo receptor.10-14
The J2E erythroid cell line was used initially to decipher the role for Lyn in Epo-induced signaling.12,13,15 A mutant subclone of J2E cells lacking Lyn failed to differentiate in response to Epo.12 However, enforced expression of active Lyn restored the differentiation capacity of these mutant cells; conversely, expression of a dominant-negative Lyn suppressed Epo-induced differentiation of the parental J2E cells.12,13 Several subsequent studies have shown that Lyn physically associates with the Epo receptor and phosphorylates several key intracellular signaling proteins such as STAT5, CrkL, PLCγ, and Erk, as well as the Epo receptor itself.10,12,16-18
The effects of Lyn on erythroid cells are not limited to in vitro systems. Close scrutiny of the erythroid compartment of Lyn−/− mice demonstrated that these animals display extramedullary erythropoiesis, a compensatory mechanism to overcome intrinsic erythroid defects.19-21 Importantly, these knockout mice eventually become anemic.20,21 Recent ex vivo studies with erythroblasts from Lyn−/− mice have confirmed that Lyn plays an important role in terminal differentiation.22
To define the molecular mechanisms involved in Lyn signaling pathways, a yeast-2 hybrid screen was performed to identify potential downstream effectors. HS1, a known Lyn interactor, was identified in this screen.18,23 We demonstrated that Lyn and HS1 interact in erythroid cells and that this association is critical for Epo-induced erythroid differentiation.18 Furthermore, disruption of the Lyn/HS1 interaction resulted in a block in Epo-induced differentiation, as the cells failed to mature morphologically or synthesize hemoglobin.12,18
In addition to HS1, several other molecules were identified in the Lyn yeast 2-hybrid screen, including a novel ankyrin repeat protein. In this study we describe the characterization of this novel nucleocytoplasmic shuttling protein designated Liar (Lyn-interacting ankyrin repeat protein). Our data show that Liar forms a multiprotein complex with Lyn and HS1 in erythroid cells and that it plays an important role in Epo-induced erythroid differentiation.
Methods
Cell culture, inductions, and colony assays
J2E and J2E-NR cells12,15 were maintained in Dulbecco modified Eagle medium (DMEM) with 5% fetal calf serum (FCS), while COS-7, MEL707, F4N, M1, JM1, JM3, 1Bra, 1Bra-raf, 2Mes-raf, 3Mes, 3Mes-raf, W265, P815, EL4, 70-Z-3, ψ2, and PA317 cells were cultured in DMEM/10% FCS as described elsewhere.24 B-cell linage cells (70Z-3, 1Bra, 3Mes) were supplemented with 5.35 μM 2-mecaptoethanol. J2E cells were stimulated with 5 U/mL Epo in media containing T3-depleted FCS,24 and hemoglobin production was detected by benzidine staining.12 J2E subclones expressing myc-Liar or myc-Liar Δ nuclear localization signal (ΔNLS) were generated by infection with amphotropic retroviruses produced by PA317 packaging cells transfected with the pMSCV2.2Neo constructs; multiple unique clonal lines were isolated using methylcellulose cultures as previously described.24 Cell proliferation was measured using the MTT assay (Roche, Penzberg, Germany) as described by the manufacturer.
The use of mice in this study was approved by the Royal Perth Hospital Animal Ethics Committee. Murine fetal liver progenitor cells were obtained from 12-day-old fetuses of CBA mice as described previously.25 Cell suspensions were harvested and infected with ecotropic retroviruses produced by ψ2 packaging cells transfected with pMSCV2.2Neo constructs by cocultivation for 2 days; 5000 (CFU-E) or 20 000 (BFU-E; 0.4 mg/mL G418) cells were then plated per dish as previously described.25 Benzidine-positive colonies were counted 2 or 7 days later, to determine CFU-E and BFU-E, respectively.
Statistical analysis of enumerated cell and colony counts were undertaken by 2-way analysis of variance (ANOVA) using Excel version 11.4.1 (Microsoft, Redmond, WA).
Plasmid construction, Northern blot, and in situ hybridization
All plasmid constructs were generated by site-directed mutagenesis using oligonucleotides (sequences available upon request), subcloning in-frame into the appropriate vector, and confirmation by sequencing. Lyn-, LynY397F-, and LynY508F-expressing plasmids have previously been described.12 Full-length Liar cDNA was generated by incorporation of a 5′ rapid amplification of cDNA ends (RACE) product and a cDNA isolated from a J2E library using site-directed mutagenesis; the resultant 1925-bp cDNA was used as the template for all subsequent Liar constructs. Myc-tagged Liar was subcloned into the mammalian expression vector pcDNA3.1 (Invitrogen, Carlsbad, CA), which was then used as a substrate for site-directed mutagenesis to generate the NLS (LiarKKRL111AALA, LiarRR98AA, LiarΔ98-114) and nuclear export signal (NES; LiarLSL289ASA) mutants.
Total RNA was extracted from cells by the method of Chomczynski and Sacchi,26 and Northern blots were performed essentially as described previously,27 on poly(A)+ RNA isolated using the poly(A)-Tract system (Promega, Madison, WI). Northern blot analyses were quantitated using Quantity One (Bio-Rad Laboratories, Hercules, CA).
Whole-mount in situ hybridization was performed on mouse embryos (day E10, CBA) using sense and antisense digoxigenin-labeled probe prepared from the full-length Liar cDNA, essentially as previously described.28
Yeast 2-hybrid library screening and analysis
The yeast 2-hybrid system used18,29 utilized the Saccharomyces cerevisiae L40 strain. Wild-type Lyn (Lyn) and a dominant-negative Lyn (Y397F) cDNAs were subcloned into the LexA fusion vector pBTM116 and used to screen a yeast 2-hybrid VP16-fusion library derived from the lymphohemopoietic progenitor cell line EMLC.130 as described previously.18 Wild-type Liar was also screened against the EMLC.1 library. Plasmids expressing LexA-pBTM116 were fusions of the Lyn unique, SH2, SH3, kinase domains and with the kinase domain deleted (ΔKin).12,18 Plasmids expressing LexA and VP16 fusions of C-terminal deletion (amino acids [aa] 1-219), N-terminal only (aa 1-120), ankyrin repeats (aa 138-240), N-terminal deletion (aa 119-299), and C-terminal alone (aa 218-299) domains of Liar for the yeast 2-hybrid system were generated by subcloning and site-directed mutagenesis.
In vitro–binding assay
A glutathione S transferase (GST) Lyn fusion protein was generated, expressed, and purified as previously described.18,31 The GST-Lyn or GST protein attached to glutathione agarose beads (Sigma-Aldrich, St Louis, MO) was quantified, and equal amounts were added to 5 μL rabbit reticulocyte lysates (TnT system; Promega) with pcDNA3.1-Liar in the presence of [35S]-methionine (Sigma-Aldrich). After incubation in Lysis buffer (120 mM NaCl, 50 mM Tris-HCl, pH 8.0, 1% Nonidet P-40 [NP-40], 1 mM ethylenediaminetetraacetic acid [EDTA], 1 mM ethyl-eneglycoltetraacetic acid [EGTA], 25 mM sodium fluoride, 1 mM phenylmethanesulfonyl fluoride [PMSF], 1 μg/mL aprotinin, 25 mM β-glycerophosphate, 1 mM Na3VO4, 1 mM benzamidine) for 3 hours with rotation at 4°C, the beads were washed 4 times with lysis buffer. Bound proteins were eluted in sodium dodecyl sulfate (SDS) sample buffer and were resolved by SDS–polyacrylamide gel electrophoresis (SDS-PAGE); radiolabeled protein was then detected using a Molecular Dynamics 445SI Phosphorimager (GE Healthcare, Little Chalfont, United Kingdom), and GST-tagged proteins were identified by Western blot analysis with anti-GST antibodies (Santa Cruz Biotechnology, Santa Cruz, CA).
Antibodies, transfections, immunoprecipitation, and Western blot analysis
A GST fusion protein consisting of the C terminus (aa 236-299) of Liar was expressed in bacteria using pGEX2T.31 The fusion protein was purified, and rabbit antisera directed against GST-Liar were raised according to the protocol previously described.25
COS-7 cells were transiently transfected using Lipofectamine 2000 (Invitrogen) and harvested 24 hours after transfection. Protein lysates were prepared in 1% NP-40, 0.1% SDS, 150 mM NaCl, 50 mM Tris-HCl, pH 8.0, 25 mM sodium fluoride, 1 mM PMSF, 1 μg/mL aprotinin, 25 mM β-glycerophosphate, 1 mM Na3VO4, 1 mM benzamidine for 30 minutes on ice, followed by centrifugation at 10 000g for 20 minutes. Protein concentration was estimated using the Dc protein assay according to manufacturer's instructions (Bio-Rad Laboratories), using bovine serum albumin as a standard.
For immunoprecipitation, cell lysates were incubated with antibodies to Liar, Lyn (SC-15), HS1,23 hemagglutinin (HA)–epitope tag (HA.11; BabCo, Berkley, CA), or myc-epitope tag (9E10 ascites32 ) for 2 hours at 4°C, then collected with protein G–Sepharose beads (Sigma-Aldrich) for 16 hours before washing and analyzing by Western blot analysis. Additional antibodies used in Western analysis included phoshotyrosine (PY99), STAT5a/b (sc-835), Erk2 (sc-154), GATA1 (sc-265), phospho-Erk2 (sc-7383), phospho-Akt (9271; Cell Signaling Technology, Danvers, MA), Akt (9272; Cell Signaling Technology), phospho-STAT5 (sc-1176; Santa Cruz Biotechnology), hemoglobin (Cappel, Durham, NC), and GST.23 Secondary antibodies were coupled to horseradish peroxidase and detected by enhanced chemiluminescence (both from GE Healthcare).
Immunofluorescent microscopy
Indirect immunofluorescence was performed on either transiently transfected COS-7 seeded onto glass coverslips (prewashed with 1 M HCl and 70% ethanol) in 12-well trays or J2E cells cytocentrifuged onto slides. Cells were fixed and permeabilized in ice-cold 50% methanol/50% acetone for 5 minutes at 4°C and stored at −80°C. Protein localization was detected using antibodies against Liar, Lyn, HS1, or a myc-epitope tag, followed by an anti-immunoglobulin G (IgG) Alexa Fluor 488 secondary antibody or an anti-IgG Alexa Flour 546 secondary antibody (Invitrogen). DNA was counterstained with Hoechst 33 258. Slides were mounted in 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.05% Tween 20, 2.5% 1,4-diaabicyclo-[2.2.2] octane and visualized on an MRC 1024 UV laser-scanning confocal microscope (Bio-Rad Laboratories). Images were acquired using a Nikon Diaphot 300 microscope fitted with a 40×/1.15 NA oil objective and analyzed using Confocal Assistant version 4.02 (Bio-Rad Laboratories) and ImageJ version 1.40c (National Institutes of Health [NIH], Bethesda, MD). Further analysis was undertaken using Adobe Photoshop CS2 version 9.02 (Adobe Systems, San Jose, CA).
For cell-cycle analysis, COS-7 cells transfected with green fluorescent protein (GFP)–tagged Liar were either synchronized with 2 mM thymidine (for 16 hours), then released and tracked for 12 hours, or unsynchronized and individual cells were tracked over 24 hours using an IN Cell Analyzer 1000 (GE Healthcare). Synchronized cells were also analyzed by flow cytometry to determine cell-cycle stage after thymidine block release.
Results
Liar, a novel Lyn-interacting protein
Several Lyn-binding proteins were identified via a yeast 2-hybrid screen (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). These molecules included the Src family kinase substrate HS1,18,23,33 the Src adaptor Cbp,34 thyroid hormone receptor interacting protein Trip 1,24 ARAP3,35 and several uncharacterized proteins. One of these novel molecules, Liar, was isolated in 2 separate yeast 2-hybrid screens using kinase-inactive and wild-type Lyn as the baits. This indicated that the possible Lyn/Liar interaction was independent of Lyn's enzymatic activity.
The Liar fragment from the yeast 2-hybrid screen was used to isolate a full-length murine cDNA encoding a 299-aa protein with a predicted molecular mass of 32.5 kDa (Figure 1A). The human and murine orthologues of Liar share 92% sequence identity and 95% similarity.
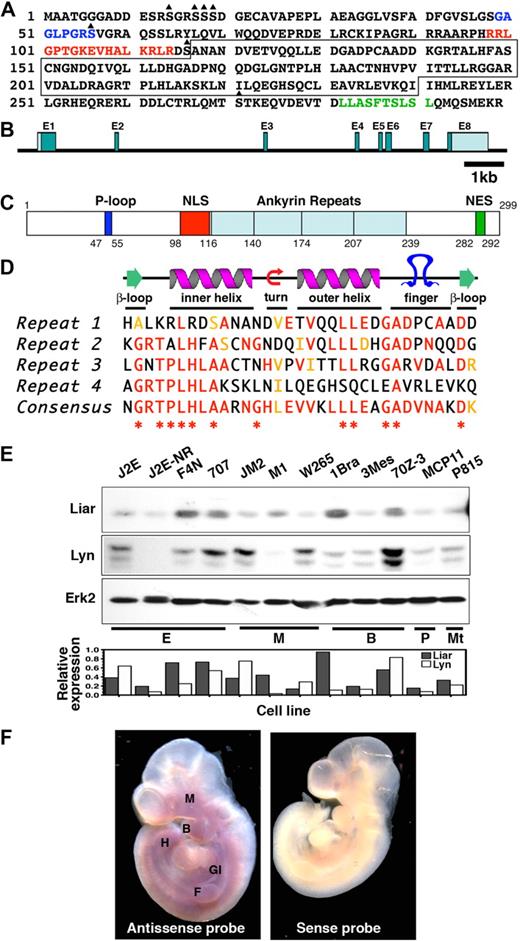
The Liar sequence. (A) The full-length amino acid sequence of Liar, illustrating the P-loop (blue letters), NLS (red letters), NES (green letters), potential serine phosphorylation sites (triangle), and the ankyrin repeats (boxed). (B) The genomic organization of Liar showing the relative size/location of the introns and exons (E1-8). (C) Representation of Liar domain organization (P-loop, dark blue; NLS, red; ankyrin repeats, light blue; NES, green); amino acids at the domain boundaries are numbered. (D) Alignment of the 4 Liar ankyrin repeats with the ankyrin consensus, illustrating the degree of identity (red) and homology (yellow). *Residues highly conserved among all ankyrin repeats. (E) Western blot analysis of Liar and Lyn protein expression in hemopoietic cell lines, quantitated relative to Erk2 levels (E, erythroid; M monocyte; Mt, mast cell; B, B cell; T, T cell). (E) In situ hybridization showing expression of Liar mRNA during embryonic development. Mouse embryos at day E10 were probed with sense and antisense probes. Intense Liar expression was detected in the fore (F) and hind (H) limb buds, the maxillary process (M), the branchial arches (B), and the gastrointestinal tract (GI).
The Liar sequence. (A) The full-length amino acid sequence of Liar, illustrating the P-loop (blue letters), NLS (red letters), NES (green letters), potential serine phosphorylation sites (triangle), and the ankyrin repeats (boxed). (B) The genomic organization of Liar showing the relative size/location of the introns and exons (E1-8). (C) Representation of Liar domain organization (P-loop, dark blue; NLS, red; ankyrin repeats, light blue; NES, green); amino acids at the domain boundaries are numbered. (D) Alignment of the 4 Liar ankyrin repeats with the ankyrin consensus, illustrating the degree of identity (red) and homology (yellow). *Residues highly conserved among all ankyrin repeats. (E) Western blot analysis of Liar and Lyn protein expression in hemopoietic cell lines, quantitated relative to Erk2 levels (E, erythroid; M monocyte; Mt, mast cell; B, B cell; T, T cell). (E) In situ hybridization showing expression of Liar mRNA during embryonic development. Mouse embryos at day E10 were probed with sense and antisense probes. Intense Liar expression was detected in the fore (F) and hind (H) limb buds, the maxillary process (M), the branchial arches (B), and the gastrointestinal tract (GI).
Database analyses (http://www.informatics.jax.org/) revealed the murine Liar gene spans approximately 9.8 kb, with 8 exons and 7 introns (Figure 1B). Exon 1 contains the 5′ untranslated region and the initiating methionine, while exon 8 contains a stop codon and 3′ untranslated region. The murine Liar gene maps to chromosome 15, while human Liar is located on chromosome 22q13.1. Comparison of the genomes demonstrated that these regions are syntenic, and several hereditary disorders have been linked to this region, including leukemias and cancers. Liar has also been identified as a highly expressed transcript in ciliated cells and was designated Ankrd54.36
One major feature of Liar protein is the 4 ankyrin repeats in the core of the protein (Figure 1C), which are known to mediate protein-protein interactions.37 The predicted structure of the 33 residue ankyrin repeats consists of a pair of antiparallel α-helices stacked side by side, connected by a series of intervening β-hairpin motifs (Figure 1D). In addition, Liar has a potential ATP-binding P-loop, a bipartite NLS and NES, and several potential sites for serine phosphorylation.
Examination of the murine Liar promoter region identified several putative transcription factor binding sites, including motifs for hemopoietic-specific transcription factors such as GATA-1, Oct6, and MZF1. Several ubiquitously expressed transcription factor binding sites (E47, Sp1, E2f5, and RFX1) were also identified, as well as a cAMP responsive element.
Expression of Liar
Liar mRNA is expressed as a single transcript of approximately 2.0 kb in a variety of hemopoietic cell lines and tissues. It was abundant in erythroid cell lines (Figure S1A), with intermediate/low expression in other hemopoietic cell lines. Endogenous Liar protein and its partner Lyn were present in all cell lines examined; however, expression levels were not uniform (Figure 1E).
In adult tissues, Liar was expressed widely, with the highest levels in testis, (Figure S1B). In addition, in situ hybridization studies showed expression of Liar in brachial arches, maxillary process, fore and hind limb buds, and the developing gastrointestinal tract of day 10 embryos (Figure 1F), consistent with the adult tissue expression pattern.
Liar contains a functional NLS and NES
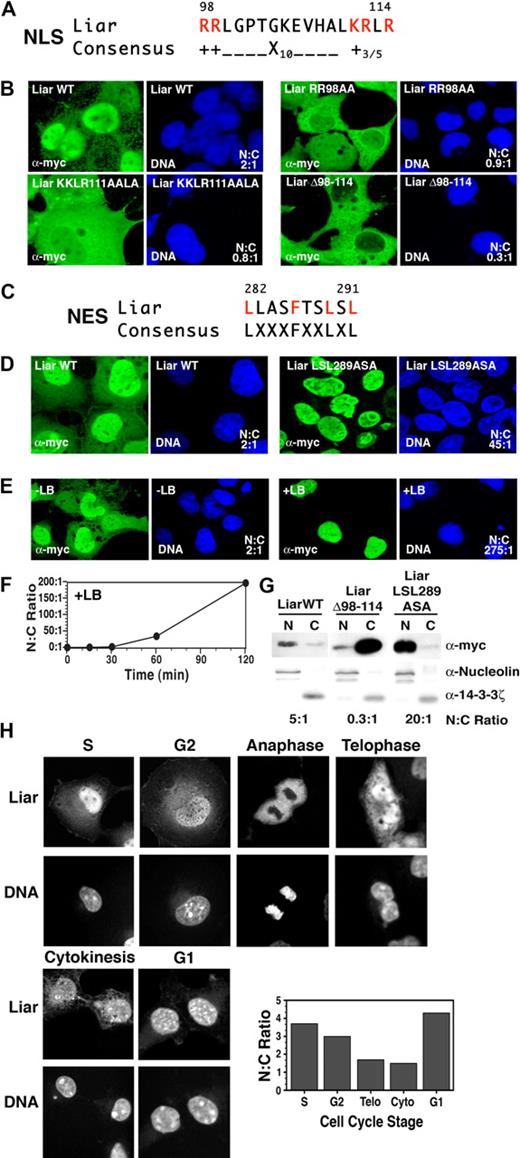
Liar contains a putative NLS (Figures 1C, 2A). While confocal microscopy revealed that Liar was present in the nucleus and cytoplasm of transiently transfected COS-7 cells (Figure 2B), the majority of the protein was located in the nucleus (nuclear:cytoplasmic [N:C] ratio 2:1). To establish whether the NLS was responsible for the nuclear import of Liar, each of the basic amino acid clusters was mutated. Site-directed mutagenesis of the first basic amino acid cluster RR 98 to AA increased the cytoplasmic Liar content appreciably (N:C ratio 0.9:1), as did mutation of the second cluster KRLR 111 to AALA (N:C ratio 0.8:1). Strikingly, deletion of the entire NLS (Δ98-114) resulted in a marked exclusion of Liar from the nucleus (N:C ratio 0.3:1). These results demonstrate that Liar possesses a functional NLS and that both clusters of basic amino acids are important for nuclear import.
Nuclear import and export of Liar. (A) Alignment of the NLS of Liar with the consensus NLS, critical arginine (R) and lysine (K) residues are highlighted in red. (B) Localization analysis of Liar NLS mutants. Wild-type Liar (Liar WT), NLS point mutants (Liar RR98AA, Liar KKLR111AALA), and the NLS-deleted Liar (LiarΔ98-114) were expressed as myc-tagged fusion proteins in COS-7 cells, and their subcellular localization was analyzed by confocal microscopy/immunofluorescence staining using anti-myc antibodies. The nucleus was identified by DNA counterstaining with Hoechst 33 258. (C) Alignment of the NES of Liar with the consensus NES; critical leucine (L) and phenylalanine (F) residues are highlighted in red. (D) Localization analysis of Liar the NES mutant. Wild-type Liar (Liar WT) and the NES point mutant (Liar LSL289ASA) were expressed and analyzed as described in panel B top. (E) Wild-type Liar-expressing cells were analyzed after treatment with Leptomycin B (+LB) for 2 hours (0.4 ng/mL). (F) Quantitation of nuclear Liar after treatment with Leptomycin B. (G) Subcellular fractionation analysis of Liar. Nuclear (N) and cytoplasmic (C) fractions of COS-7 cells expressing myc-tagged wild-type Liar (Liar WT), the NLS mutant (LiarΔ98-114), and the NES mutant (LiarLSL289ASA) of Liar were analyzed by Western blot analysis using anti-myc antibodies. Nuclear/cytoplasmic fractionation was confirmed by blotting for nucleolin (nuclear) and 14-4-4ζ (cytoplasm). (H) Nuclear and cytoplasmic distribution of Liar during the cell cycle. COS-7 cells expressing wild-type myc-tagged Liar were synchronized by thymidine block, then analyzed and quantitated as in panel B after release. Cell-cycle stages were determined by flow cytometry (propidium iodine staining) and morphology.
Nuclear import and export of Liar. (A) Alignment of the NLS of Liar with the consensus NLS, critical arginine (R) and lysine (K) residues are highlighted in red. (B) Localization analysis of Liar NLS mutants. Wild-type Liar (Liar WT), NLS point mutants (Liar RR98AA, Liar KKLR111AALA), and the NLS-deleted Liar (LiarΔ98-114) were expressed as myc-tagged fusion proteins in COS-7 cells, and their subcellular localization was analyzed by confocal microscopy/immunofluorescence staining using anti-myc antibodies. The nucleus was identified by DNA counterstaining with Hoechst 33 258. (C) Alignment of the NES of Liar with the consensus NES; critical leucine (L) and phenylalanine (F) residues are highlighted in red. (D) Localization analysis of Liar the NES mutant. Wild-type Liar (Liar WT) and the NES point mutant (Liar LSL289ASA) were expressed and analyzed as described in panel B top. (E) Wild-type Liar-expressing cells were analyzed after treatment with Leptomycin B (+LB) for 2 hours (0.4 ng/mL). (F) Quantitation of nuclear Liar after treatment with Leptomycin B. (G) Subcellular fractionation analysis of Liar. Nuclear (N) and cytoplasmic (C) fractions of COS-7 cells expressing myc-tagged wild-type Liar (Liar WT), the NLS mutant (LiarΔ98-114), and the NES mutant (LiarLSL289ASA) of Liar were analyzed by Western blot analysis using anti-myc antibodies. Nuclear/cytoplasmic fractionation was confirmed by blotting for nucleolin (nuclear) and 14-4-4ζ (cytoplasm). (H) Nuclear and cytoplasmic distribution of Liar during the cell cycle. COS-7 cells expressing wild-type myc-tagged Liar were synchronized by thymidine block, then analyzed and quantitated as in panel B after release. Cell-cycle stages were determined by flow cytometry (propidium iodine staining) and morphology.
In addition, Liar contains a putative leucine-rich NES, characterized by the presence of 4 closely spaced hydrophobic residues (Figures 1C and 2C). To determine whether the putative NES was functional, the leucines at positions 289 and 291 were mutated to alanine (Figure 2D); this resulted in substantial build up of Liar within the nucleus (N:C ratio 45:1). Cells were then treated with Leptomycin B (Figure 2E), a specific inhibitor of Crm1-dependent nuclear export, which induced considerable Liar relocation to the nucleus (N:C ratio 275:1). Time-course studies (Figure 2F) revealed appreciable nuclear accumulation of Liar 1 hour after treatment (N:C ratio 45:1), followed by almost exclusive nuclear localization within 2 hours (N:C ratio > 200:1). These data demonstrate that the NES is required to transport out of the nucleus.
Cellular fractionation studies confirmed that deletion of the NLS (Δ98-114) resulted in preferential cytoplasmic localization, while mutation of the NES (LSL 289 to ASA) significantly enhanced nuclear accumulation (Figure 2G). Collectively, these data show shuttling of Liar between nuclear and cytoplasmic compartments requires the NLS and NES, and occurs over a relatively short time frame.
During the cell cycle, Liar displayed interesting fluctuations in subcellular compartmentalization (Figures 2H, S2, and S3). Strong nuclear accumulation was observed in S, G2, and G1 stages. During M phase, Liar was excluded from the partitioning chromosomes (anaphase), before reaccumulation in the nucleus after the nuclear envelope reformed (telophase). A proportion of Liar was also detected in the midbody during cytokinesis. Together these observations demonstrate that Liar is an active nucleocytoplasmic shuttling protein, which oscillates between compartments during the cell cycle.
Liar and Lyn interact and colocalize in cells
To examine the interaction between Liar and Lyn, COS-7 cells were cotransfected with plasmids expressing full-length Liar and Lyn. Figure 3A shows the coimmunoprecipitation of Liar and Lyn. In addition, Liar coimmunoprecipitated with inactive (Y397F) Lyn and constitutively active (Y508F) Lyn (Figure 3B), demonstrating that Liar associates with Lyn in an activation-independent manner. This interaction was direct as determined by in vitro binding assays (Figure 3C). However, despite the interaction between these 2 molecules, Lyn did not phosphorylate Liar on either of its 2 tyrosines, using both in vitro and in vivo kinase assays (data not shown).
Liar interacts with Lyn. (A) Lyn and Liar coimmunoprecipitate. Western blot analysis of Lyn and Liar immunoprecipitates (IP) from COS-7 cells transfected with Lyn and myc-tagged Liar. Blots were probed with anti-Lyn and anti-myc antibodies. (B) Lyn-Liar coimmunoprecipitation is not dependent upon kinase activity. Western blot analysis of Lyn immunoprecipitates (IP) from COS-7 cells transfected with Lyn wild-type (LynWT), kinase inactive (LynY397F), constitutively active (LynY508F), and myc-tagged Liar. Blots were probed with anti-Lyn and anti-myc antibodies. (C) Liar and Lyn interact directly. GST pull-down assays using purified GST/GST-Lyn and in vitro–translated [35S]-radiolabeled Liar. Radiolabeled Liar was detected on Western blot analyses using a PhosphorImager, then probed with anti-Lyn and anti-GST antibodies. (D) Overlapping subcellular localization of Liar and Lyn. COS-7 cells transfected with Lyn and myc-tagged Liar were analyzed by confocal microscopy and immunofluorescence staining using anti-myc and anti-Lyn antibodies.
Liar interacts with Lyn. (A) Lyn and Liar coimmunoprecipitate. Western blot analysis of Lyn and Liar immunoprecipitates (IP) from COS-7 cells transfected with Lyn and myc-tagged Liar. Blots were probed with anti-Lyn and anti-myc antibodies. (B) Lyn-Liar coimmunoprecipitation is not dependent upon kinase activity. Western blot analysis of Lyn immunoprecipitates (IP) from COS-7 cells transfected with Lyn wild-type (LynWT), kinase inactive (LynY397F), constitutively active (LynY508F), and myc-tagged Liar. Blots were probed with anti-Lyn and anti-myc antibodies. (C) Liar and Lyn interact directly. GST pull-down assays using purified GST/GST-Lyn and in vitro–translated [35S]-radiolabeled Liar. Radiolabeled Liar was detected on Western blot analyses using a PhosphorImager, then probed with anti-Lyn and anti-GST antibodies. (D) Overlapping subcellular localization of Liar and Lyn. COS-7 cells transfected with Lyn and myc-tagged Liar were analyzed by confocal microscopy and immunofluorescence staining using anti-myc and anti-Lyn antibodies.
Because Liar and Lyn associate in the yeast-2 hybrid system, coimmunoprecipitate, and bind in vitro (Table S1 and Figure 3A-C), the interaction was then assessed by confocal microscopy. Data presented in Figure 3D revealed that Liar and Lyn occupied distinct, but overlapping, subcellular compartments in transiently transfected COS-7 cells. As shown earlier, Liar localized primarily in the nucleus, whereas Lyn displayed diffuse cytoplasmic and plasma membrane staining. Importantly, a proportion of Liar protein present in the cytoplasm and cell membrane overlapped with Lyn (Figure 3D inset).
Ankyrin repeats of Liar bind SH3 domains
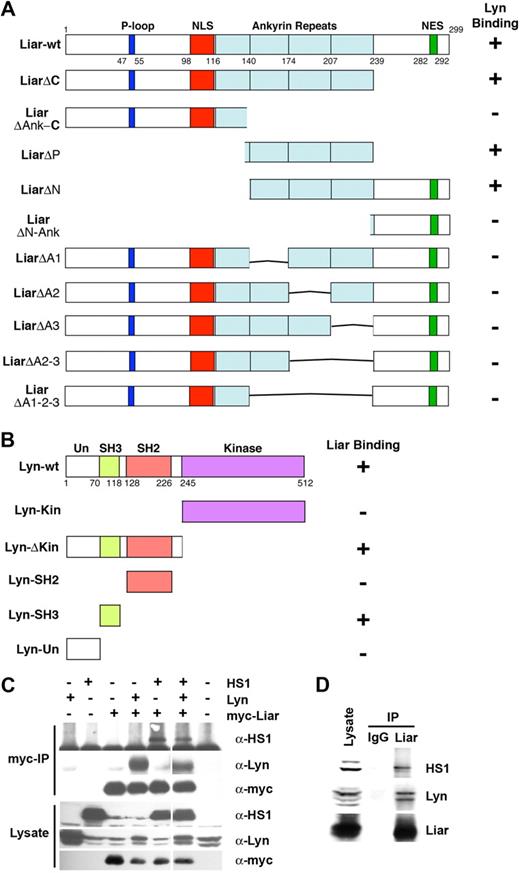
To characterize the domains required for Liar and Lyn binding, several deletion mutants were generated. Significantly, amino acids 140 to 240, spanning 3 of the 4 ankyrin repeat motifs, were required to bind Lyn (Figure 4A). Deletion of individual repeats abrogated Lyn binding, demonstrating that all 3 repeats were needed for this association. These data suggest the structure of Liar and the fold of the ankyrin repeat region are important for Liar binding activity.
Liar, Lyn, and HS1 interaction analysis. (A) Three of the 4 ankyrin repeats of Liar bind Lyn. Full-length Liar and deletion mutants, were tested for interaction with Lyn in a yeast 2-hybrid assay. (B) The SH3 domain of Lyn binds Liar. Full-length Lyn and its domains (Un-unique, SH3, SH2, and kinase) were tested for interaction with Liar in a yeast 2-hybrid assay. (C) Liar, Lyn, and HS1 coimmunoprecipitated. COS-7 cells transfected with Lyn, HS1, and myc-tagged Liar were analyzed by Western blot analysis after immunoprecipitation (IP) of Liar. Blots were probed with anti-Lyn, and-HS1 and anti-myc antibodies. (D) Liar, Lyn, and HS1 form a complex in erythroid cells. Liar was immunoprecipitated (IP) from J2E cells, compared with nonspecific IgG immunoprecipitates. Blots were probed with anti-Lyn, anti-Liar, and anti-HS1 antibodies.
Liar, Lyn, and HS1 interaction analysis. (A) Three of the 4 ankyrin repeats of Liar bind Lyn. Full-length Liar and deletion mutants, were tested for interaction with Lyn in a yeast 2-hybrid assay. (B) The SH3 domain of Lyn binds Liar. Full-length Lyn and its domains (Un-unique, SH3, SH2, and kinase) were tested for interaction with Liar in a yeast 2-hybrid assay. (C) Liar, Lyn, and HS1 coimmunoprecipitated. COS-7 cells transfected with Lyn, HS1, and myc-tagged Liar were analyzed by Western blot analysis after immunoprecipitation (IP) of Liar. Blots were probed with anti-Lyn, and-HS1 and anti-myc antibodies. (D) Liar, Lyn, and HS1 form a complex in erythroid cells. Liar was immunoprecipitated (IP) from J2E cells, compared with nonspecific IgG immunoprecipitates. Blots were probed with anti-Lyn, anti-Liar, and anti-HS1 antibodies.
Several Lyn deletion constructs were then examined for their ability to bind Liar. Only constructs bearing the Src homology 3 (SH3) domain of Lyn were able to interact with Liar, indicating that this domain was responsible for the interaction between Liar and Lyn (Figure 4B). As the Liar ankyrin repeat domains do not contain a consensus SH3-binding PXPXXP motif,38 these data suggest that the SH3 domain of Lyn associates with the Liar ankyrin repeats via a novel mechanism.
To determine whether other potential binding partners of Liar also possessed SH3 domains, a yeast 2-hybrid screen was performed using full-length Liar as bait. Liar also bound several other intracellular signaling proteins that possess SH3 domains (Table S2). Deletion analyses demonstrated that the SH3 domains of these proteins were responsible for binding to the ankyrin repeat region of Liar. These results show that the Liar ankyrin repeats are able to mediate binding to a variety of SH3 domains.
The interaction between Liar and HS1 was intriguing (Table S2), since we have previously shown that HS1 binds Lyn and plays a critical role in erythroid differentiation.18 Cotransfection experiments in COS-7 cells revealed that all 3 proteins could be detected in an immunoprecipitable complex (Figure 4C). Importantly, endogenous Lyn, HS1, and Liar from J2E cells also coimmunoprecipitated (Figure 4D), demonstrating that these proteins also exist in a multiprotein complex within erythroid cells.
Liar expression in erythroid cells
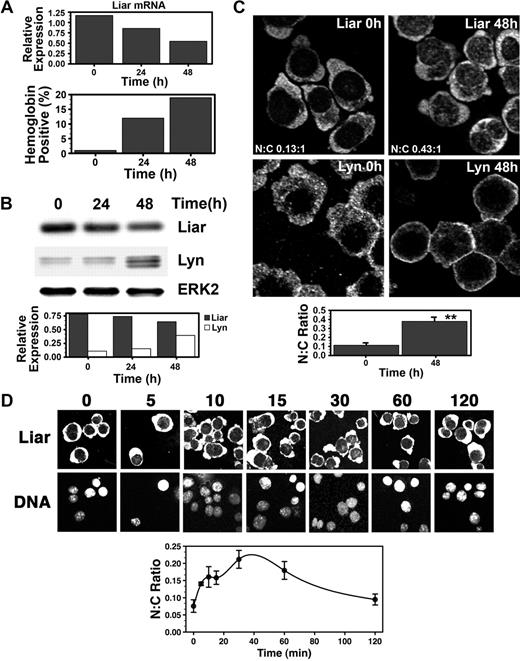
Expression of endogenous Liar was then examined during Epo-induced differentiation of J2E erythroid cells. Figure 5A shows that Liar mRNA decreased as the erythroid cells matured into hemoglobin-producing cells. Liar protein levels also fell, in contrast with the rise in Lyn content (Figure 5B).
Dynamics of Liar expression and localization during Epo-induced erythroid differentiation. (A) Liar mRNA expression during erythroid differentiation. J2E cells were stimulated with Epo and analyzed for Liar mRNA levels by quantitative reverse transcription–polymerase chain reaction (RT-PCR) at 0, 24, and 48 hours after stimulation. (B) Liar and Lyn protein expression during erythroid differentiation. Cells prepared as for panel A were analyzed by Western blot analysis with anti-Liar, anti-Lyn, and anti-Erk2 antibodies. Liar and Lyn levels expressed relative to Erk2. (C) Liar translocates to the nucleus during erythroid differentiation. J2E cells stimulated with Epo were analyzed by confocal microscopy and immunofluorescence staining using anti-Liar and anti-Lyn antibodies. (D) Liar translocates to the nucleus shortly after Epo stimulation. J2E cells stimulated with Epo were analyzed by confocal microscopy and immunofluorescence staining using anti-Liar antibodies. The nucleus was identified by DNA counterstaining with Hoechst 33 258.
Dynamics of Liar expression and localization during Epo-induced erythroid differentiation. (A) Liar mRNA expression during erythroid differentiation. J2E cells were stimulated with Epo and analyzed for Liar mRNA levels by quantitative reverse transcription–polymerase chain reaction (RT-PCR) at 0, 24, and 48 hours after stimulation. (B) Liar and Lyn protein expression during erythroid differentiation. Cells prepared as for panel A were analyzed by Western blot analysis with anti-Liar, anti-Lyn, and anti-Erk2 antibodies. Liar and Lyn levels expressed relative to Erk2. (C) Liar translocates to the nucleus during erythroid differentiation. J2E cells stimulated with Epo were analyzed by confocal microscopy and immunofluorescence staining using anti-Liar and anti-Lyn antibodies. (D) Liar translocates to the nucleus shortly after Epo stimulation. J2E cells stimulated with Epo were analyzed by confocal microscopy and immunofluorescence staining using anti-Liar antibodies. The nucleus was identified by DNA counterstaining with Hoechst 33 258.
Somewhat surprisingly, endogenous Liar localized primarily in the cytoplasm of J2E cells (Figure 5C). This contrasted markedly with the transient transfection studies conducted with COS-7 cells (Figures 2,3). As a consequence, much greater overlap in subcellular localization was observed between Liar and Lyn in erythroid cells. Interestingly, a 4-fold increase in Liar was detected in the nucleus 48 hours after Epo induction, particularly within punctate nuclear bodies (Figure 5C). This indicated that translocation and accumulation of Liar within the nuclear compartment was associated with erythroid terminal differentiation.
Although Liar was not tyrosine phosphorylated after Epo induction (data not shown), a proportion of Liar rapidly relocated to the nucleus after hormonal stimulation (Figure 5D). Within 30 minutes of Epo stimulation, a 3-fold increase in the nuclear content of Liar was detected. However, by 120 minutes, the N:C ratio of Liar had almost returned to prestimulation levels. These data show that Epo initiates a rapid transient translocation of Liar to the nucleus, which is resolved after 2 hours (Figure 5D); however, a subsequent nuclear buildup of Liar occurs as the cells undergo terminal differentiation (Figure 5C).
Exogenous Liar inhibits erythroid differentiation
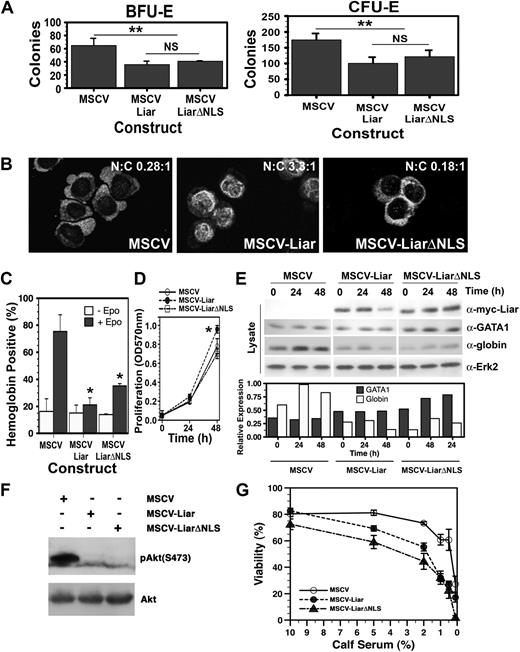
In an attempt to determine the biologic effects of Liar, either full-length Liar or a mutant lacking the NLS (LiarΔNLS), were expressed in erythroid progenitors via retroviral infection. Figure 6A shows that expression of either Liar or LiarΔNLS suppressed the development of both erythroid burst–forming units (BFU-E) and erythroid colony–forming units (CFU-E). Thus, appropriate levels and correct subcellular compartmentalization of Liar at distinct maturation stages are essential for the normal development of erythroid progenitors.
Overexpression of Liar inhibits erythroid differentiation. (A) Myc-tagged Liar (MSCV-Liar) and LiarΔNLS (MSCV-LiarΔNLS) overexpression inhibit BFU-E and CFU-E formation. Methylcellulose was used for erythroid colony–forming assays using fetal liver cells infected with retroviruses expressing Liar (MSCV-Liar), LiarΔNLS (MSCV-LiarΔNLS), or vector only (MSCV). The number of Epo-responsive benzidine-positive colonies (CFU-E and BFU-E) is shown. The mean ± standard deviation (SD) is presented (n = 3). **Statistically significant (2-way ANOVA) values (P ≤ .01). (B) J2E cells were infected with retroviruses expressing myc-tagged Liar (MSCV-Liar), LiarΔNLS (MSCV-LiarΔNLS), or vector only (MSCV) and analyzed for Liar subcellular localization by confocal microscopy and immunofluorescence staining using anti-myc antibodies. (C) Effects of Liar/LiarΔNLS expression on Epo-induced hemoglobin production detected by benzidine staining. (D) Effects of Liar/LiarΔNLS expression on J2E proliferation measured by a MTT-based assay after Epo stimulation. The mean (± SD) is presented (n = 3). *Statistically significant (2-way ANOVA) values (P ≤ .05). (E) Liar/LiarΔNLS expression reduce globin protein levels. Expression of exogenous Liar/LiarΔNLS (anti-myc), GATA-1, globin, and Erk2 were analyzed by Western blot analysis. GATA-1 and globin levels are expressed relative to Erk2 expression. (F) Western blot analysis of J2E cells overexpressing Liar (MSCV-Liar), LiarΔNLS (MSCV-LiarΔNLS), and control cells (MSCV) for phospo-Akt levels relative to total Akt. (G) Effect on viability of reducing the serum levels in media for the cell lines as in panel F.
Overexpression of Liar inhibits erythroid differentiation. (A) Myc-tagged Liar (MSCV-Liar) and LiarΔNLS (MSCV-LiarΔNLS) overexpression inhibit BFU-E and CFU-E formation. Methylcellulose was used for erythroid colony–forming assays using fetal liver cells infected with retroviruses expressing Liar (MSCV-Liar), LiarΔNLS (MSCV-LiarΔNLS), or vector only (MSCV). The number of Epo-responsive benzidine-positive colonies (CFU-E and BFU-E) is shown. The mean ± standard deviation (SD) is presented (n = 3). **Statistically significant (2-way ANOVA) values (P ≤ .01). (B) J2E cells were infected with retroviruses expressing myc-tagged Liar (MSCV-Liar), LiarΔNLS (MSCV-LiarΔNLS), or vector only (MSCV) and analyzed for Liar subcellular localization by confocal microscopy and immunofluorescence staining using anti-myc antibodies. (C) Effects of Liar/LiarΔNLS expression on Epo-induced hemoglobin production detected by benzidine staining. (D) Effects of Liar/LiarΔNLS expression on J2E proliferation measured by a MTT-based assay after Epo stimulation. The mean (± SD) is presented (n = 3). *Statistically significant (2-way ANOVA) values (P ≤ .05). (E) Liar/LiarΔNLS expression reduce globin protein levels. Expression of exogenous Liar/LiarΔNLS (anti-myc), GATA-1, globin, and Erk2 were analyzed by Western blot analysis. GATA-1 and globin levels are expressed relative to Erk2 expression. (F) Western blot analysis of J2E cells overexpressing Liar (MSCV-Liar), LiarΔNLS (MSCV-LiarΔNLS), and control cells (MSCV) for phospo-Akt levels relative to total Akt. (G) Effect on viability of reducing the serum levels in media for the cell lines as in panel F.
Liar and LiarΔNLS were then introduced into J2E cells. In contrast with the cytoplasmic localization of endogenous Liar in J2E cells, raising the levels of Liar resulted in increased nuclear occupancy (Figure 6B)—this observation is similar to the overexpression of Liar in COS-7 cells (Figures 2,3). Conversely, ΔNLS-Liar was restricted to the cytoplasm.
Hemoglobin synthesis after Epo-induced differentiation was reduced in J2E cells expressing either Liar or LiarΔNLS (Figure 6C); however, the decrease in differentiation (as demonstrated by hemoglobin production) was more pronounced in cells expressing Liar than LiarΔNLS. Interestingly, the Liar overexpressing cells also had a significantly higher rate of proliferation (Figure 6D). Immunoblot analyses displayed in Figure 6E show that the levels of globin protein were reduced in cells expressing Liar or LiarΔNLS, thereby contributing to the decrease in hemoglobin synthesis in these cells. However, neither Liar nor LiarΔNLS decreased GATA-1, the key transcriptional regulator of globin synthesis39,40 ; thus, the inhibitory effects of Liar upon globin protein content are not likely to be mediated by GATA-1 levels.
J2E cells overexpressing Liar or LiarΔNLS also showed a considerable reduction in the levels of phospho-Akt (Figure 6F). As Akt phosphorylation in linked with apoptosis, the effects of exogenous Liar and LiarΔNLS on cell viability were examined. Significantly, these cells displayed reduced viability when placed in media with lower serum levels (Figure 6G). It is noteworthy that COS-7 cells overexpressing Liar at high levels underwent mitotic failure and subsequent apoptosis (Figure S4).
Exogenous Liar affects Epo signaling
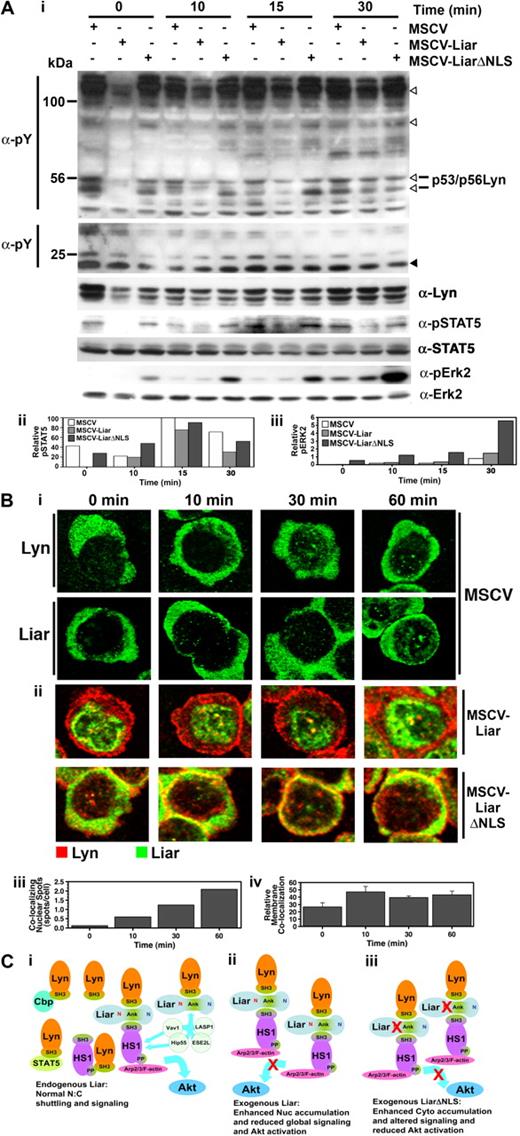
The effects of overexpressing Liar or LiarΔNLS were then examined immediately after Epo stimulation of J2E cells. Data presented in Figure 7A demonstrate that these constructs had a noticeable affect upon Epo-initiated signaling events. Interestingly, in unstimulated cells overexpressing Liar a decrease in tyrosine phosphorylation of some proteins was observed (open arrowheads); in particular, Lyn phosphorylation was markedly reduced. The reduction in phosphorylated Lyn may account for the decrease in other tyrosine phosphorylated proteins seen in these cells, because Lyn is the predominant Src family kinase expressed.12 The delayed activation of STAT5 in Liar cells is also consistent with reduced Lyn activity, as Lyn has been shown to influence the phosphorylation of STAT5.10 However, the overall reduction in tyrosine phosphorylation in these cells was resolved 30 minutes after Epo stimulation (Figure 7A). In contrast, global tyrosine phosphorylation appeared relatively normal in the LiarΔNLS cells (Figure 7A), although, elevated phosphorylation of Erk2 was detected, even in the absence of Epo stimulation. These data demonstrate that Liar plays an important role in modulating the phosphorylation patterns and kinetics of Epo-induced signal transduction pathways.
Liar regulates Epo-induced signaling. (A) Altered intracellular signaling caused by overexpression of Liar and LiarΔNLS. Lysates of J2E cells expressing Liar (MSCV-Liar) or LiarΔNLS (MSCV-LiarΔNLS) were analyzed after Epo stimulation by Western blot analysis (i) with anti-phosphotyroine, anti–phospho-STAT5, anti-STAT5, anti-Lyn, anti–phospho-Erk2, and anti-Erk2 (loading control) antibodies. Phosphotyrosine proteins reduced in Liar overexpressing cells are indicated by open arrowheads, compared with an unchanged protein indicated by a closed arrowhead. Quantitation of phospho-STAT5 (ii) and phospho-Erk2 (iii) levels relative to total STAT5, Erk2. (B) Overexpression of Liar or LiarΔNLS alters Epo-induced Lyn subcellular localization. J2E cells described in panel A were analyzed by confocal microscopy and immunofluorescence staining using anti-myc antibodies over 60 minutes of Epo-stimulation (i). A representative cell from each time point is illustrated, depicting the nuclear localization of Lyn and Liar in control (MSCV) cells, nuclear colocalizing spots in MSCV-Liar cells, and membrane colocalization in MSCV-LiarΔNLS cells. Changes in Epo-induced Liar/Lyn colocalization in nuclear spots in the Liar-expressing cells were enumerated (ii), as was the relative membrane colocalization of Lyn and LiarΔNLS (iii). (C) Model of potential Liar interactions and its effects upon shuttling/signaling. (i) Endogenous Liar dynamically interacts with Lyn and HS1 and potentially other molecules (Vav1, Hip55, LASP1, ESE2L), allowing normal nuclear:cytoplasmic shuttling and signaling to HS1 pathways (eg, Arp2/3/F-actin) and facilitating Akt activation. (ii) Overexpression of wild-type Liar enhances the nuclear accumulation of Liar/Liar complexes and reduces global Epo-induced signaling events and inhibiting Akt activation. (iii) Overexpression of NLS-mutated Liar (LiarΔNLS) enhances the cytoplasmic accumulation of Liar/Liar complexes altering Epo-induced erythroid signaling including inhibiting Akt activation. The NLS (red N), NES (blue N), and ankyrin repeats (Ank) are depicted.
Liar regulates Epo-induced signaling. (A) Altered intracellular signaling caused by overexpression of Liar and LiarΔNLS. Lysates of J2E cells expressing Liar (MSCV-Liar) or LiarΔNLS (MSCV-LiarΔNLS) were analyzed after Epo stimulation by Western blot analysis (i) with anti-phosphotyroine, anti–phospho-STAT5, anti-STAT5, anti-Lyn, anti–phospho-Erk2, and anti-Erk2 (loading control) antibodies. Phosphotyrosine proteins reduced in Liar overexpressing cells are indicated by open arrowheads, compared with an unchanged protein indicated by a closed arrowhead. Quantitation of phospho-STAT5 (ii) and phospho-Erk2 (iii) levels relative to total STAT5, Erk2. (B) Overexpression of Liar or LiarΔNLS alters Epo-induced Lyn subcellular localization. J2E cells described in panel A were analyzed by confocal microscopy and immunofluorescence staining using anti-myc antibodies over 60 minutes of Epo-stimulation (i). A representative cell from each time point is illustrated, depicting the nuclear localization of Lyn and Liar in control (MSCV) cells, nuclear colocalizing spots in MSCV-Liar cells, and membrane colocalization in MSCV-LiarΔNLS cells. Changes in Epo-induced Liar/Lyn colocalization in nuclear spots in the Liar-expressing cells were enumerated (ii), as was the relative membrane colocalization of Lyn and LiarΔNLS (iii). (C) Model of potential Liar interactions and its effects upon shuttling/signaling. (i) Endogenous Liar dynamically interacts with Lyn and HS1 and potentially other molecules (Vav1, Hip55, LASP1, ESE2L), allowing normal nuclear:cytoplasmic shuttling and signaling to HS1 pathways (eg, Arp2/3/F-actin) and facilitating Akt activation. (ii) Overexpression of wild-type Liar enhances the nuclear accumulation of Liar/Liar complexes and reduces global Epo-induced signaling events and inhibiting Akt activation. (iii) Overexpression of NLS-mutated Liar (LiarΔNLS) enhances the cytoplasmic accumulation of Liar/Liar complexes altering Epo-induced erythroid signaling including inhibiting Akt activation. The NLS (red N), NES (blue N), and ankyrin repeats (Ank) are depicted.
The subcellular localization of Lyn in Liar, LiarΔNLS, and control (murine stem cell virus [MSCV]) cells after Epo stimulation was then examined (Figure 7B). In control cells and those overexpressing Liar, Lyn concentrated around the cell membrane immediately after Epo activation, then became diffuse throughout the cytoplasm by 60 minutes after stimulation. Interestingly, increased punctate nuclear colocalization of Lyn and Liar was detected after exposure to Epo in control and Liar-overexpressing cells. In contrast, considerable colocalization of Lyn and LiarΔNLS occurred at the cell membrane upon hormonal activation; increased amounts of Lyn were also detected in the nucleus after Epo stimulation (Figure 7B). These observations suggest that, as well as affecting intracellular signaling, Liar also has an impact upon the distribution of Lyn within cells after Epo stimulation.
Discussion
Lyn is the most abundant Src family kinase present in erythroid cells, and its involvement in Epo-signaling cascades is gradually being elucidated10,12,13,16-18,21,41,42 —it has been implicated in the phosphorylation of the Epo receptor, STAT5, CrkL, PLCγ, and Erk2/MAP kinase.10,12,16-18 We have shown the importance of (1) Lyn activation after Epo stimulation12,13,18 and (2) the subsequent down-regulation of Lyn orchestrated via Cbp, followed by SOCS1-mediated proteasomal degradation.34 In addition, we have demonstrated an interaction between Lyn and Trip1, which links the Epo pathway with thyroid hormone receptor signaling,24 another vital component in erythropoiesis.43 These studies underscore the significance of Lyn signaling pathways for correct erythroid development.
Here we describe the association between Lyn and the previously uncharacterized protein, Liar. The bond between Lyn and Liar defines a novel interaction between the Lyn SH3 domain and 3 of the Liar ankyrin domains, which do not contain a classic PXPXXP motif.38 The Liar ankyrin repeats also associated with other SH3 domain-containing proteins (namely, HS1, ESE2L, Vav1, Hip55, and LASP1), indicating that these modules have a generalized capacity to bind SH3 domains. Recently, other nonconsensus motifs have been described that bind SH3 domains.44,45 Significantly, the association between Lyn and Liar is independent of the activation status of Lyn, and Lyn does not phosphorylate Liar.
We have previously demonstrated that Lyn and HS1 interact in erythroid cells,18 and in this manuscript we show that Lyn, Liar, and HS1 are able to form a multiprotein complex (Figure 7C). HS1 has been shown to influence proliferation and apoptosis by recruiting molecules (eg, Vav1, PLCγ1) into signaling complexes associated with the F-actin/Arp2/3 complex.33,46-49 It is noteworthy that Liar is able to interact with Vav1 and Hip55 (Table S2), which are also involved with HS1 signaling to the cytoskeleton.46,49 Because erythroid terminal differentiation is characterized by major morphologic alterations after Epo stimulation,50 Liar could play a significant role in coordinating this process through its links with Lyn and HS1. As we have observed previously that Lyn−/− erythroblasts have morphologic abnormalities,21 this may be due to a failure to form appropriate Lyn-Liar interactions which influence downstream signaling to the cytoskeleton.
Altering the levels and subcellular compartmentalization of Liar had a significant effect upon Epo-induced intracellular signaling. Tyrosine phosphorylation of several proteins was affected, including STAT5 and Erk2/MAP kinase, as well as serine phosphorylation of Akt. Because Lyn has been shown to affect numerous signaling molecules,10,16-18,51 some of the changes in tyrosine phosphorylation are probably due to the impact of Liar upon Lyn activity. Liar, therefore, plays an important role in modulating Lyn-initiated signals within erythroid cells. Altering the stoichiometry of Liar in multiprotein complexes (Figure 7C) is likely to interfere with the delicate balance required for accurate transmission of intracellular signals.
The presence of functional NLS and NES demonstrate that Liar is a molecule that shuttles between nuclear and cytoplasmic compartments. Endogenous Liar in erythroid cells is located primarily in the cytoplasm where it colocalizes with Lyn. Increasing the levels of Liar redirects the molecule into the nucleus—this appears to be a concentration-dependent phenomenon, as overexpression of Liar in both J2E and COS-7 cells resulted in nuclear accumulation. The relocation of Liar to the nucleus immediately after Epo stimulation is reminiscent of STAT5 nuclear translocation,52 albeit in the absence of tyrosine phosphorylation of Liar. Furthermore, Liar accumulated in the nucleus of differentiating erythroid cells, the significance of which awaits further elucidation.
While the role of Liar in the nucleus is yet to be defined, other ankyrin repeat proteins have been shown to act as transcriptional regulators by controlling transport to the nucleus (eg, ankyrin repeat protein IκB regulates subcellular localization of NF-κB).53-55 It is possible that Liar may be involved in translocating Lyn into the nucleus—there have been previous reports of Lyn in the nucleus, although its role there is unclear.56 Perhaps nuclear Lyn is involved in phosphorylating specific proteins within that compartment.
The biologic consequences of changing Liar levels and location were quite marked. Development of normal erythroid progenitors (both BFU-E and CFU-E) were suppressed when Liar levels were elevated. In addition, ectopic expression of Liar and LiarΔNLS in immortalized erythroid cells affected differentiation (hemoglobin synthesis), proliferation and pAkt levels/viability in low serum—this is likely to be a consequence of altered Epo-induced signaling. Collectively, these data indicate that appropriate levels and compartmentalization of Liar (ie, Epo-induced nuclear accumulation) are essential for normal erythroid maturation. As the genetic locus where human Liar resides is implicated in leukemias, alterations in Liar expression could conceivably play a role in lineage determination and leukemogenesis.
In this manuscript, we have described the identification and characterization of Liar, a novel Lyn-binding protein. This previously undescribed protein plays a significant role in modulating Epo-induced intracellular signaling. As Liar is capable of rapid nucleocytoplasmic shuttling, it may transmit signals emanating from an activated Epo receptor to the nucleus. Moreover, because Liar is able to interact with signaling components known to form multiprotein complexes (ie, Lyn, HS1, Vav1, Hip55), Liar is at the intersection of several signaling pathways involved in regulating proliferation, differentiation, and cell death.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants 303101, 403987, and 513714 from the National Health and Medical Research Council of Australia, the Medical Research Foundation of Royal Perth Hospital, the University of Western Australia, and the Center for Food and Genomic Medicine.
Authorship
Contribution: A.L.S. designed and performed experiments, analyzed data, and contributed to writing the manuscript; S.P.K. supported the research, designed experiments, analyzed results, and contributed to writing the manuscript; and E.I. designed and supported the research, designed and undertook experiments, analyzed data, and contributed to writing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Evan Ingley, Cell Signaling Group, Laboratory for Cancer Medicine, Western Australian Institute for Medical Research, Level 6, MRF Building, Rear 50 Murray St, Perth WA 6000, Australia; e-mail: eingley@waimr.uwa.edu.au.

![Figure 3. Liar interacts with Lyn. (A) Lyn and Liar coimmunoprecipitate. Western blot analysis of Lyn and Liar immunoprecipitates (IP) from COS-7 cells transfected with Lyn and myc-tagged Liar. Blots were probed with anti-Lyn and anti-myc antibodies. (B) Lyn-Liar coimmunoprecipitation is not dependent upon kinase activity. Western blot analysis of Lyn immunoprecipitates (IP) from COS-7 cells transfected with Lyn wild-type (LynWT), kinase inactive (LynY397F), constitutively active (LynY508F), and myc-tagged Liar. Blots were probed with anti-Lyn and anti-myc antibodies. (C) Liar and Lyn interact directly. GST pull-down assays using purified GST/GST-Lyn and in vitro–translated [35S]-radiolabeled Liar. Radiolabeled Liar was detected on Western blot analyses using a PhosphorImager, then probed with anti-Lyn and anti-GST antibodies. (D) Overlapping subcellular localization of Liar and Lyn. COS-7 cells transfected with Lyn and myc-tagged Liar were analyzed by confocal microscopy and immunofluorescence staining using anti-myc and anti-Lyn antibodies.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/16/10.1182_blood-2008-04-153452/7/m_zh80090931630003.jpeg?Expires=1767766441&Signature=G2PkR9wHNv0VErKv2DNL2Ss5lwNGlTULOkzJnBZbRGMQDGH06nutS0O9KXJnTYYRLB3LINkPRpyGNgzkPpVbLf~F1dAXQRaMKp~QP-Qfk1CqjTKLXTB6zMMu5lj-GzmAetQ-tRGsMV0ZqhJhavXbsnkkTxso9z1jmIz-25GY6BdNgqtoBbZmJfr5qIuH2T2OiwaHrauUblQ5BQGiVVgMXvSFb2jSb-syo3bne0uN~~SWCmVp2u4nQFrA-LIlgT7rDD~kab1jfRU07LvMbcG9P5WlqAN6P9wL-rryVe2jkS3lBqLwR7-OvSINu8ZJwofIqgZI2f7V8g5EXrb-j0L3VQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal