Abstract
Genetic variants in CYP2C9 and VKORC1 strongly affect steady-state warfarin dose. However, these variants also affect early international normalized ratio (INR) values during warfarin initiation. We examined whether CYP2C9/VKORC1 genotypes provide information about warfarin sensitivity additional to that provided by early INR responses. In 214 patients starting warfarin with INR-guided dose adjustments, we determined whether CYP2C9 and VKORC1 genotypes were associated with early measures of warfarin sensitivity (time to INR ≥ lower limit of therapeutic range; time to INR > 4; and first stable warfarin dose) after adjusting for early (days 4-6) and week 1 (days 7-9) INR values. Early INRs were associated with all outcomes (all P < .001) and were more informative than genotypes. For time to INR more than or equal to the lower limit of therapeutic range, adding either early INRs or genotypes to a baseline model (clinical variables only) increased the goodness-of-fit (R2) from 0.05 to 0.42 and 0.19, respectively (full model, R2 = 0.46). For first stable warfarin dose, adding either early INRs or genotypes to the baseline model increased the R2 from 0.08 to 0.32 and 0.27, respectively (full model, R2 = 0.40). After inclusion of week 1 INRs, CYP2C9 (P = .08) and VKORC1 (P = .30) were not associated with stable warfarin dose. Thus, much of the information provided by CYP2C9 and VKORC1 genotypes during warfarin initiation is captured by the early INR response.
Introduction
Oral anticoagulation with warfarin is the most common therapy for the treatment and prevention of thromboembolic events. However, warfarin therapy is associated with considerable morbidity and mortality because it has a narrow therapeutic range and at least 10-fold interindividual variability in drug sensitivity. Warfarin therapy is monitored and dose adjusted according to its pharmacodynamic effects on clotting, measured as the international normalized ratio (INR). The INR value is closely related to the risk of thrombosis with underanticoagulation, and that of bleeding with overanticoagulation.1-3
Warfarin therapy is usually started with initial doses of 5 to 10 mg on the first 2 to 3 days, and subsequently titrated according to the INR response with the goal of achieving an INR within the therapeutic range within 4 to 6 days.3 Because warfarin dose requirements vary widely, this standard regimen often results in subtherapeutic INR values in more resistant patients and overshooting the therapeutic range in more sensitive patients. The risk of adverse effects associated with overanticoagulation is high, especially in the first few weeks of therapy.2,4,5 Therefore, strategies to individualize the initial warfarin dose have been sought.
Several factors are associated with a person's sensitivity to warfarin and, thus, the dose he or she requires for stable anticoagulation. These include age, ethnicity, comorbidities, concomitant medication, and genetic variation.6-19 Two genes have been extensively studied: CYP2C9, encoding the enzyme (cytochrome P450 2C9) primarily responsible for the metabolism of the more active S-enantiomer of warfarin, and VKORC1, encoding the subunit 1 of the vitamin K epoxide reductase complex, the target of warfarin. Variants in these 2 genes affect warfarin dose requirements at steady state substantially6-17,20,21 ; moreover, at the start of warfarin therapy, VKORC1 and, to a lesser extent, CYP2C9 genotypes also affect early INR responses.22,23
The clinically useful contribution of genotype to individualizing warfarin dose will probably be greatest in the initiation phase of therapy, before a person's dose requirement has been determined empirically by titration according to INR response. However, the genetic contribution to warfarin dose requirements has most often been studied using steady-state dose requirements in patients already receiving stable doses of warfarin, without considering the information provided by the early INR responses during initiation of warfarin therapy. Indeed, in a recent study in orthopedic patients starting genotype-guided warfarin therapy, VKORC1 genotype did not contribute significantly to the prediction of early stable warfarin dose when the INR value on the fourth day of treatment was included in the prediction model.24
In 2007, the Food and Drug Administration updated the warfarin label to include information for prescribers about genotype,25 and there has been debate about the introduction of genotype-guided warfarin dosing into clinical practice.26-30 A fundamental question is whether genotyping provides information about warfarin sensitivity that is additional to that provided by the early INR response.29 This question is especially important because early INR values are routinely determined in current clinical practice, whereas genotyping before starting therapy would require extra time and cost. We therefore examined the hypothesis that VKORC1 and CYP2C9 genotypes predict warfarin sensitivity after taking into account the information provided by early INR responses.
Methods
Study design and subjects
Details of the study design and patient recruitment have been published.22 Briefly, this was a prospective, observational study conducted between 2002 and 2004 at 3 outpatient clinics (Pharmacy, Cardiology, and Arthritis and Joint Replacement Center) at Vanderbilt University Medical Center that provide anticoagulation services. The study was approved by the Vanderbilt University Institutional Review Board, and informed consent was obtained from all subjects in accordance with the Declaration of Helsinki. Consecutive patients starting warfarin were eligible if they were older than 18 years, had complete information on doses and dates of warfarin initiation, did not suffer from alcoholism, and were not receiving cancer chemotherapy. Ethnicity was determined by self-report. Because the great majority of patients were blacks or whites, we excluded patients of other ethnicities from analysis. The initial INR response is often measured on day 4 of warfarin treatment (ie, after 3 daily doses); however, in practice, a substantial fraction of patients will have their initial INR response measured on day 5 or 6 for logistic reasons. Thus, we studied subjects who had at least one INR value determined on days 4 to 6 of warfarin treatment.
Warfarin anticoagulation
The individual target INR range, frequency of INR measurements, and warfarin dosing were determined by the treating physicians who were unaware of CYP2C9 or VKORC1 genotypes during the whole treatment period.
Data collection and follow-up
Demographic characteristics (age, sex, ethnicity) and details of medical history (concomitant diseases and medications) and warfarin therapy (indication, start date, range of therapeutic INR, initial and subsequent doses, and INR values) were obtained through patient interviews and a review of electronic medical records by a trained study nurse. A venous blood sample was drawn for DNA extraction. Information on warfarin dose and INR measurements was collected from the initiation of warfarin until the end of continuous follow-up, defined as date of the last record for an INR and warfarin dose, provided that previous records were available at intervals less than 60 days.
Genotyping
CYP2C9 *2 and *3 alleles were determined by a fluorescent allele-specific oligonucleotide ligation assay.22 VKORC1 genotyping for the variants G-1639A (rs17878363), T497G (rs17882154), C1173T (rs9934438), G1542C (rs17886369), C2255T (rs2359612) was performed by TaqMan SNP assays with the 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA).
Outcomes
To assess the contribution of genotypes and early INR values to the early phase of anticoagulation, we assessed the following outcomes: (1) time to INR more than or equal to the lower limit of the therapeutic range; (2) time to first INR greater than 4.0; and (3) first stable warfarin dose, defined as the first unchanged daily dose given for more than or equal to 7 consecutive days that yielded an INR value within the therapeutic range.24
Data analysis and statistics
For the VKORC1, group A haplotype was assigned by using the statistical software PowerMarker (J. Liu, North Carolina State University, Raleigh, NC; http://statgen.ncsu.edu/powermarker) based on the 5 variants as described.22 All other haplotypes, which in white Americans belong almost entirely to haplotype group B, were termed haplotype non-A. For the analysis, VKORC1 genotype was coded as numbers of copies (0-2) of haplotype A, and CYP2C9 genotypes were coded as number of variant alleles (0-2) assuming additive allele effects.
Patients with at least one INR value on days 4 to 6 of warfarin treatment (day 1 = day of first warfarin dose) were included (n = 214). For analysis, we created a variable for the first recorded INR value after 3 warfarin doses, INRearly (for 162 patients on day 4, 27 patients on day 5, and 25 patients on day 6). We also examined the additional clinical benefit of considering information provided by subsequent INR values after the first week of treatment for the 145 patients who also had at least one INR value on days 7 to 9. Analogous to INRearly, INRweek1 represented the earliest available INR after a week of treatment (for 63 patients on day 7, 34 patients on day 8, and 48 patients on day 9). In secondary analyses, we also replaced the variable INRearly by 3 variables: INR4 (set equal to the INR value on day 4 if available, and 0 otherwise), INR5 (set equal to the INR value on day 5 if available but 0 if INR4 available or INR5 unavailable), and INR6 (day 6 INR value if available but 0 if either INR4 or INR5 available). This allowed us to evaluate the effects of INR4, INR5, and INR6 separately. Similarly, we defined variables INR7, INR8, and INR9 to replace INRweek1 to evaluate their effects separately. Cumulative warfarin dose was calculated as the sum of all warfarin doses from day 1 to the day before the respective INR measurement.
To assess the effect of early INR response and genotypes on the outcomes, we performed Cox regression analyses to determine the hazard ratio (HR) of achieving (1) an INR more than or equal to the lower limit of the therapeutic range and (2) a first INR more than 4. We also performed multiple linear regression analyses to assess the effect of genotypes and early INR values on the first stable warfarin dose. All models were adjusted for age, sex, ethnicity, use of amiodarone, target INR, and cumulative warfarin dose. In addition, to assess their independent contribution to the outcome variables, we sequentially excluded early INR measurements, genotypes, or both from the model and compared the adjusted R2 of the fitted models and carried out likelihood ratio tests to evaluate their significance. All analyses were performed in the statistical software R (www.r-project.org). All tests were 2-tailed, and a P value of less than .05 was considered significant.
Results
Subject characteristics
Of 325 patients who met the inclusion criteria, 297 patients were recruited.22 Eighty-one of them were excluded from analysis because they did not have an INR value recorded on days 4 to 6, and 2 were excluded because they were neither black nor white. Demographic and clinical data for the remaining 214 patients are presented in Table 1. For 210 patients (98%), the target INR range was between 1.7 and 3.0. Demographic, clinical, and genotype data for the excluded patients did not differ significantly from the included patients, except a small difference in target INR (mean difference, 0.11; 95% confidence interval [CI], 0.07-0.15; P < .001).
Patient characteristics
| Variable . | All patients . |
|---|---|
| Demographics | |
| Subjects, no. | 214 |
| Female, no. (%) | 104 (49) |
| European-American, no. (%) | 197 (92) |
| Black, no. (%) | 17 (8) |
| Mean age, y (SD) | 61 (14) |
| Indication, no. (%) | |
| Atrial fibrillation/atrial flutter | 57 (27) |
| Joint replacement | 113 (53) |
| Venous thromboembolism | 26 (12) |
| Other or multiple indications | 18 (8) |
| Concomitant amiodarone medication, no. (%) | 25 (12) |
| Median follow-up, d (range) | 23.5 (4-225) |
| Variable . | All patients . |
|---|---|
| Demographics | |
| Subjects, no. | 214 |
| Female, no. (%) | 104 (49) |
| European-American, no. (%) | 197 (92) |
| Black, no. (%) | 17 (8) |
| Mean age, y (SD) | 61 (14) |
| Indication, no. (%) | |
| Atrial fibrillation/atrial flutter | 57 (27) |
| Joint replacement | 113 (53) |
| Venous thromboembolism | 26 (12) |
| Other or multiple indications | 18 (8) |
| Concomitant amiodarone medication, no. (%) | 25 (12) |
| Median follow-up, d (range) | 23.5 (4-225) |
Genotyping results
VKORC1 and CYP2C9 genotype could not be determined in 1 and 2 subjects, respectively. Allele frequencies were in the range expected for the respective ethnic group, and genotype distributions conformed to Hardy-Weinberg equilibrium (Table 2).
VKORC1 haplotype and CYP2C9 genotype frequency
| Gene . | White (n = 197) . | Black (n = 17) . | All (n = 214) . | |||
|---|---|---|---|---|---|---|
| n . | % . | n . | % . | n . | % . | |
| VKORC1 haplotype | ||||||
| NonA/NonA | 86 | 43.9 | 13 | 76.5 | 99 | 46.5 |
| NonA/A | 89 | 45.4 | 4 | 23.5 | 93 | 43.7 |
| A/A | 21 | 10.7 | 0 | 0.0 | 21 | 9.9 |
| CYP2C9 genotype | ||||||
| *1/*1 | 127 | 64.8 | 16 | 100.0 | 143 | 67.5 |
| *1/*2;*1/*3 | 60 | 30.6 | 0 | 0 | 60 | 28.3 |
| *2/*2;*3/*3;*2/*3 | 9 | 4.6 | 0 | 0 | 9 | 4.2 |
| Gene . | White (n = 197) . | Black (n = 17) . | All (n = 214) . | |||
|---|---|---|---|---|---|---|
| n . | % . | n . | % . | n . | % . | |
| VKORC1 haplotype | ||||||
| NonA/NonA | 86 | 43.9 | 13 | 76.5 | 99 | 46.5 |
| NonA/A | 89 | 45.4 | 4 | 23.5 | 93 | 43.7 |
| A/A | 21 | 10.7 | 0 | 0.0 | 21 | 9.9 |
| CYP2C9 genotype | ||||||
| *1/*1 | 127 | 64.8 | 16 | 100.0 | 143 | 67.5 |
| *1/*2;*1/*3 | 60 | 30.6 | 0 | 0 | 60 | 28.3 |
| *2/*2;*3/*3;*2/*3 | 9 | 4.6 | 0 | 0 | 9 | 4.2 |
VKORC1 haplotype could not be determined in 1 white subject, and CYP2C9 genotype could not be determined in 1 white and 1 black subject.
Early INR values and outcomes
Of the 214 patients, 191 (89%) received an initial dosage of 5 mg, and 203 (95%) continued the initial daily dose unchanged at least until the first INR value. The mean INR4 was 1.60 plus or minus 0.72 (range, 0.90-5.00; n = 163), and the summary early INR response (INRearly) averaged 1.63 plus or minus 0.71 (range, 0.90-5.00). Age (P = .007) and VKORC1 haplotype (P < .001), but not CYP2C9 genotype (P = .18) or cumulative dose (P = .30), were significantly associated with INRearly. Four patients had an INRearly more than 4.0 (all on day 4); their ages ranged from 63 to 80 years, their warfarin dose on days 1 to 3 was 5 mg daily, and none of them received amiodarone. All 4 patients were heterozygous carriers of VKORC1 haplotype A, and 1 patient also carried one CYP2C9 *2 allele.
Early INR response was significantly associated with outcomes characterizing warfarin sensitivity. Of the 214 patients, 200 achieved an INR more than or equal to the lower limit of the therapeutic range after a median time of 8.5 days (interquartile range [IQR], 5-13 days). Time to INR more than or equal to the lower limit of the therapeutic range was strongly associated with early INR response (unadjusted HR for a 1-unit increase in INRearly, 3.07; 95% CI, 2.54-3.71; P < .001). This association remained significant after adjustment for age, sex, ethnicity, use of amiodarone, target INR, cumulative dose, and VKORC1 and CYP2C9 genotypes (HR for INRearly, 3.34; 95% CI, 2.67-4.18; P < .001; Table 3). Overanticoagulation, defined as INR more than 4, occurred in 59 patients after a median of 15 days (IQR, 10-22 days) of treatment. Time to overanticoagulation was strongly associated with early INR values both before (P < .001) and after adjustment for the covariates and VKORC1 and CYP2C9 genotypes (HR for INRearly, 2.12; 95% CI, 1.47-3.06; P < .001; Table 3).
Cox regression analysis for time to INR more than or equal to the lower limit of the therapeutic range and time to INR more than 4
| Covariate . | Time to INR within or above therapeutic range . | Time to INR > 4 . | ||||
|---|---|---|---|---|---|---|
| Hazard ratio . | 95% CI . | P . | Hazard ratio . | 95% CI . | P . | |
| Age | 1.00 | 0.99-1.01 | .76 | 1.02 | 1.00-1.04 | .023 |
| Male sex | 0.66 | 0.48-0.90 | .008 | 0.65 | 0.38-1.12 | .12 |
| White ethnicity | 0.70 | 0.40-1.23 | .22 | 0.70 | 0.25-1.93 | .49 |
| Target INR | 0.16 | 0.06-0.45 | < .001 | 8.20 | 1.23-54.8 | .030 |
| Amiodarone use | 0.96 | 0.61-1.51 | .87 | 1.01 | 0.49-2.06 | .98 |
| Cumulative warfarin dose* | 1.01 | 0.97-1.04 | .78 | 0.98 | 0.93-1.04 | .50 |
| INRearly | 3.34 | 2.67-4.18 | < .001 | 2.12 | 1.47-3.06 | < .001 |
| VKORC1 A haplotype | 1.64 | 1.29-2.11 | < .001 | 1.49 | 0.96-2.32 | .08 |
| CYP2C9, any variant | 1.10 | 0.84-1.44 | .48 | 1.01 | 0.61-1.67 | .97 |
| Covariate . | Time to INR within or above therapeutic range . | Time to INR > 4 . | ||||
|---|---|---|---|---|---|---|
| Hazard ratio . | 95% CI . | P . | Hazard ratio . | 95% CI . | P . | |
| Age | 1.00 | 0.99-1.01 | .76 | 1.02 | 1.00-1.04 | .023 |
| Male sex | 0.66 | 0.48-0.90 | .008 | 0.65 | 0.38-1.12 | .12 |
| White ethnicity | 0.70 | 0.40-1.23 | .22 | 0.70 | 0.25-1.93 | .49 |
| Target INR | 0.16 | 0.06-0.45 | < .001 | 8.20 | 1.23-54.8 | .030 |
| Amiodarone use | 0.96 | 0.61-1.51 | .87 | 1.01 | 0.49-2.06 | .98 |
| Cumulative warfarin dose* | 1.01 | 0.97-1.04 | .78 | 0.98 | 0.93-1.04 | .50 |
| INRearly | 3.34 | 2.67-4.18 | < .001 | 2.12 | 1.47-3.06 | < .001 |
| VKORC1 A haplotype | 1.64 | 1.29-2.11 | < .001 | 1.49 | 0.96-2.32 | .08 |
| CYP2C9, any variant | 1.10 | 0.84-1.44 | .48 | 1.01 | 0.61-1.67 | .97 |
n = 211 resulting from 3 missing genotypes.
INRearly indicates INR on days 4, 5, and 6 in 162, 27, and 25 patients, respectively.
In each patient, cumulative dose was calculated up to the day preceding the respective INR used in the analysis.
Early INR values were also associated with the first stable warfarin dose. Of the 214 subjects, 132 reached a first stable warfarin dose that yielded a therapeutic INR after more than or equal to 7 days of unchanged dosing. The median time to reach a stable warfarin dose was 18 days (IQR, 12-32 days; overall range, 8-140 days). After adjustment for age, sex, ethnicity, amiodarone use, target INR, cumulative warfarin dose, and VKORC1 and CYP2C9 genotypes, higher INRearly values predicted lower therapeutic warfarin doses (β-coefficient, −1.3 mg/day; 95% CI, −1.8 to −0.8 mg/day; P < .001; Table 4).
Multiple linear regression analysis for first stable warfarin dose,* including early and week 1 INR values
| Covariate . | Including early INR values (days 4-6) . | Including late INR values (days 7-9) . | ||||
|---|---|---|---|---|---|---|
| β-coefficient . | 95% CI . | P . | β-coefficient . | 95% CI . | P . | |
| Age | −0.03 | −0.05 to −0.01 | .007 | −0.02 | −0.05 to 0.01 | .16 |
| Male sex | 0.42 | −0.18 to 1.02 | .17 | 0.34 | −0.46 to 1.14 | .40 |
| White ethnicity | 0.57 | −0.52 to 1.67 | .30 | −0.12 | −1.46 to 1.21 | .86 |
| Target INR | 1.05 | −0.92 to 3.03 | .29 | 2.16 | −0.31 to 4.63 | .09 |
| Amiodarone use | −0.92 | −1.72 to −0.13 | .023 | −0.46 | −1.55 to 0.63 | .40 |
| Cumulative warfarin dose† | 0.03 | −0.04 to 0.09 | .42 | 0.07 | 0.01 to 0.12 | .013 |
| INR values | ||||||
| INRearly | −1.29 | −1.79 to −0.80 | < .001 | NA | NA | NA |
| INRweek1 | NA | NA | NA | −1.21 | −1.70 to −0.73 | < .001 |
| VKORC1 A haplotype | −0.71 | −1.19 to −0.23 | .004 | −0.35 | −1.02 to 0.32 | .30 |
| CYP2C9 dose−reducing variant | −0.87 | −1.42 to −0.32 | .002 | −0.65 | −1.39 to 0.08 | .08 |
| Covariate . | Including early INR values (days 4-6) . | Including late INR values (days 7-9) . | ||||
|---|---|---|---|---|---|---|
| β-coefficient . | 95% CI . | P . | β-coefficient . | 95% CI . | P . | |
| Age | −0.03 | −0.05 to −0.01 | .007 | −0.02 | −0.05 to 0.01 | .16 |
| Male sex | 0.42 | −0.18 to 1.02 | .17 | 0.34 | −0.46 to 1.14 | .40 |
| White ethnicity | 0.57 | −0.52 to 1.67 | .30 | −0.12 | −1.46 to 1.21 | .86 |
| Target INR | 1.05 | −0.92 to 3.03 | .29 | 2.16 | −0.31 to 4.63 | .09 |
| Amiodarone use | −0.92 | −1.72 to −0.13 | .023 | −0.46 | −1.55 to 0.63 | .40 |
| Cumulative warfarin dose† | 0.03 | −0.04 to 0.09 | .42 | 0.07 | 0.01 to 0.12 | .013 |
| INR values | ||||||
| INRearly | −1.29 | −1.79 to −0.80 | < .001 | NA | NA | NA |
| INRweek1 | NA | NA | NA | −1.21 | −1.70 to −0.73 | < .001 |
| VKORC1 A haplotype | −0.71 | −1.19 to −0.23 | .004 | −0.35 | −1.02 to 0.32 | .30 |
| CYP2C9 dose−reducing variant | −0.87 | −1.42 to −0.32 | .002 | −0.65 | −1.39 to 0.08 | .08 |
NA indicates not applicable; INRearly, INR on days 4, 5, and 6 in 162, 27, and 25 patients, respectively; INRweek1, INR on days 7, 8, and 9 in 63, 34, and 48 patients, respectively.
First stable dose was defined as the first unchanged daily dose given for more than or equal to 7 days that yielded an INR value within the therapeutic range.
In each patient, cumulative dose was calculated up to the day preceding the respective INR used in the analysis.
Incremental value of genotypes in prediction of outcomes
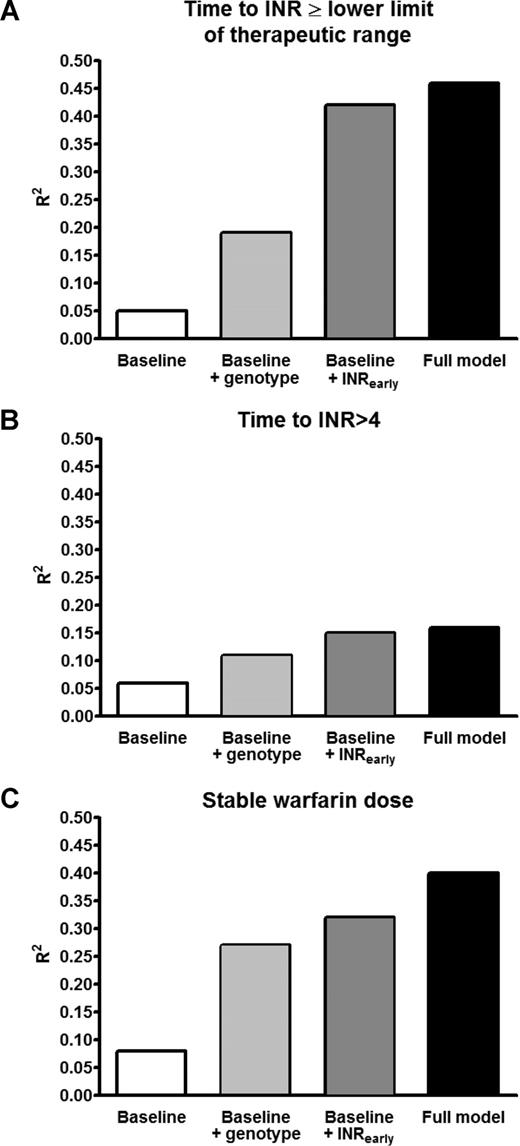
When VKORC1 and CYP2C9 genotypes were added to the regression models that included INRearly, VKORC1 haplotype A was significantly associated with a shorter time to the INR more than or equal to the lower limit of the therapeutic range (HR = 1.64, P < .001), whereas CYP2C9 genotype was not (HR = 1.10, P = .48; Table 3). However, the contribution of genotypes to the final model was modest and smaller than that of INRearly. Excluding genotypes from the final model reduced the goodness of fit only modestly (reduction in R2 from 0.46 to 0.42, P < .001; Figure 1A), whereas exclusion of INRearly reduced it substantially (reduction in R2 from 0.46 to 0.19, P < .001; Figure 1A).
Comparison of goodness of fit (R2) among 4 models for each of the 3 outcome variables. (A) Time to INR ≥ the lower limit of therapeutic range. (B) Time to INR > 4. (C) Early stable warfarin dose. In each panel, the first bar (□) represents the baseline model (including age, ethnicity, sex, amiodarone use, target INR, and cumulative warfarin dose); second bar ( ), the baseline model + VKORC1 / CYP2C9 genotypes; the third bar (
), the baseline model + VKORC1 / CYP2C9 genotypes; the third bar ( ), the baseline model + early INR values; fourth bar (■), the full model. The difference between the last 2 bars (■ and
), the baseline model + early INR values; fourth bar (■), the full model. The difference between the last 2 bars (■ and  ) in each panel represents the contribution of genotypes after adjustment for baseline covariates and INRearly.
) in each panel represents the contribution of genotypes after adjustment for baseline covariates and INRearly.
Comparison of goodness of fit (R2) among 4 models for each of the 3 outcome variables. (A) Time to INR ≥ the lower limit of therapeutic range. (B) Time to INR > 4. (C) Early stable warfarin dose. In each panel, the first bar (□) represents the baseline model (including age, ethnicity, sex, amiodarone use, target INR, and cumulative warfarin dose); second bar ( ), the baseline model + VKORC1 / CYP2C9 genotypes; the third bar (
), the baseline model + VKORC1 / CYP2C9 genotypes; the third bar ( ), the baseline model + early INR values; fourth bar (■), the full model. The difference between the last 2 bars (■ and
), the baseline model + early INR values; fourth bar (■), the full model. The difference between the last 2 bars (■ and  ) in each panel represents the contribution of genotypes after adjustment for baseline covariates and INRearly.
) in each panel represents the contribution of genotypes after adjustment for baseline covariates and INRearly.
When genotypes were included in the regression model for time to first INR more than 4, neither VKORC1 (P = .08) nor CYP2C9 genotype (P = .97) was associated with overanticoagulation (Table 3). In contrast, INRearly remained strongly associated with time to first INR more than 4 (P < .001; Table 3). Accordingly, excluding genotypes from the analysis did not significantly reduce goodness-of-model fit (reduction in R2 from 0.16 to 0.15; P = .21), whereas excluding INRearly reduced it from 0.16 to 0.11 (P < .001; Figure 1B).
When genotypes were included in the regression model predicting the first stable warfarin dose, both VKORC1 (P = .004) and CYP2C9 genotypes (P = .002) were significantly associated with the outcome (Table 4). Excluding genotypes from the analysis reduced the adjusted R2 from 0.40 to 0.32 (P < .001; Figure 1C). Again, exclusion of early INR response resulted in a larger reduction in goodness of fit (adjusted R2 from 0.40 to 0.27, P < .001; Figure 1C).
When the analyses were repeated using the 3 INR values separately (ie, INR4, INR5, and INR6 individually, rather than as the combined variable INRearly), the predictive value of the 3 INR variables was largely comparable in magnitude and statistical significance, suggesting that using any of the 3 values yielded concordant results. Similarly, using 3 separate INR variables did not appreciably affect the predictive value of genotypes on the outcomes.
INRweek1 values, genotypes, and therapeutic warfarin dose
First stable warfarin dose was achieved after a median of 18 days; therefore, we examined whether the predictive value of genotypes for the first stable warfarin dose was maintained after adjusting for INR values obtained at the end of the first week of treatment (INRweek1, ie, INR responses on days 7-9). In the full model, including age, sex, ethnicity, amiodarone use, target INR, cumulative dose, genotypes, and INRweek1, neither CYP2C9 (P = .08) nor VKORC1 (P = .30) genotype was significantly correlated with the first stable warfarin dose (Table 4). In contrast, in this model, there was a strong negative association between INRweek1 and the steady-state warfarin dose (P < .001; Table 4). This was also reflected in the R2 values for the different models. Excluding genotypes from the full model resulted in a nonsignificant reduction of R2 from 0.38 to 0.36 (P = .12), whereas excluding INRweek1 from the full model reduced R2 from 0.38 to 0.18 (P < .001).
Discussion
This study provides novel information about the value of pharmacogenetic determinants in the prediction of warfarin sensitivity beyond that provided by the pharmacodynamic measure of early INR response during warfarin induction by conventional dose titration. Our major findings are that a considerable part, but not all, of the predictive information provided by VKORC1 and CYP2C9 genotypes about warfarin sensitivity early in therapy was, indeed, reflected in the early INR response. After a week of dose titration guided by the INR response, pharmacogenetic factors did not contribute significantly to prediction of first stable dose of warfarin beyond what could be inferred from the clinical information provided by the relationship between warfarin dose and INR response.
Genetic variants in the enzyme that metabolizes warfarin, CYP2C9, and in the target of warfarin, VKORC1, strongly affect warfarin sensitivity and thus warfarin dose requirements at steady state, ie, when therapeutic INRs have been achieved after chronic stable dosing.31 Thus, rather than starting all patients on the same dose of warfarin, selection of an initial dose based on an individual's genotypes would be expected to improve INR control during warfarin initiation.32-37 Indeed, several prospective studies are underway to test this hypothesis.27,28 However, the pharmacodynamic measure of warfarin sensitivity, INR response, also differs early in therapy according to genotypes, particularly VKORC1 genotype,22,23 and it is possible that the information provided by genotypes about stable warfarin dose requirements is in large part captured by the information provided by the early INR response.29 If that were the case, one could argue that genotyping would contribute little additional information if one paid close attention to the early INR response and predicted future warfarin dose requirements accordingly.29 Moreover, the early INR response may also reflect additional unmeasured individual factors contributing to warfarin sensitivity that are not captured by genotype.29 The relative contributions of VKORC1/CYP2C9 genotypes and early INR response to prediction of sensitivity to warfarin have not been well characterized.
Millican et al studied patients receiving warfarin after orthopedic surgery.24 When the INR value after 3 doses of warfarin was included in a multiple linear regression model predicting first stable warfarin dose, CYP2C9 genotype remained a significant predictor, whereas the predictive information of the VKORC1 genotype was almost completely captured by the early INR value. In this cohort, however, the first 3 warfarin doses had already been individualized based on CYP2C9 and/or VKORC1 genotypes and other clinical predictors. In a recent study examining the relationship between early phenotypic markers of warfarin sensitivity and genotypes, the INR on day 4 explained 31% of the variability in warfarin dose at day 14, whereas CYP2C9-VKORC1 genotypes explained only 6.5%.38 However, this cohort consisted entirely of hospitalized patients with significant comorbidities receiving multiple medications, a clinical situation that is probable to have increased the contribution of nongenetic factors to variability in warfarin sensitivity and thus attenuated the contribution of genotype.
In our study, we systematically quantified the contribution of CYP2C9 and VKORC1 genotypes to the assessment of early warfarin sensitivity after adjustment for early INR response. In addition to the first stable warfarin dose, we chose several measures of warfarin sensitivity in the induction phase that are associated with clinically important endpoints. The time to a first INR value more than or equal to the lower limit of the therapeutic range affects the length of concomitant heparin therapy and, thus, the duration of hospitalization, for patients who require immediate anticoagulation. Time to INR more than 4 identifies patients with overanticoagulation who are at greater risk for bleeding. Finally, the early steady-state warfarin dose24 reflects subsequent stable warfarin steady-state dose requirements. For all outcomes, the predictive power of early INR values on days 4, 5, or 6 was greater than that of genotypes, and the additional contribution of genotypes to the predictive models was generally small.
After adjustment for early INR values (days 4-6), first stable warfarin dose was the only outcome to which genotypes made a contribution that was meaningful in magnitude. The independent contribution of genotyping to prediction of warfarin sensitivity probably decreases over time because it can be increasingly estimated from the relationship between INR responses and cumulative warfarin dose. Thus, we also examined whether genotypes maintain their predictive contribution to early steady-state warfarin dose after the first week of treatment. After adjustment for INR values on days 7 to 9, genotypes were no longer independent predictors of first stable warfarin dose. These findings suggest that a large part of the predictive value of CYP2C9 and VKORC1 genotypes is reflected in early INR measurements and that, as early as 1 week after the start of treatment, genotypes may contribute little additional predictive information about warfarin sensitivity beyond that reflected in INR and cumulative dose.
However, although much of a patient's warfarin sensitivity is reflected in the INR values obtained during the first week of treatment, in clinical practice it is often difficult to use this information optimally because this requires frequent INR determinations and dosing instructions by specifically trained healthcare providers, often in specialized anticoagulation clinics. Thus, several prospective clinical trials are planned or underway to test the value of CYP2C9/VKORC1-genotype guided warfarin therapy compared with conventional INR-guided dose titration during warfarin initiation. Our findings have important implications for these studies. Because much of the predictive information of genotypes is indeed reflected in early INR response, our results suggest that the benefit of genotype-guided therapy will be smallest in a clinical setting with closely monitored therapy, where expert dose titration includes close attention to the early INR response, and greatest in the clinical setting of nonexpert warfarin dosing, where the majority of warfarin-treated patients are followed. Second, algorithms for genotype-based warfarin dosing should allow for inclusion of early INR response to improve the prediction of warfarin sensitivity.28 Such an algorithm that provides dosing suggestions after accounting for clinical and genetic factors and early INR values is available (www.warfarindosing.org).21
For subjects who are very sensitive to warfarin, empiric dosing during days 1 to 3 may already result in overanticoagulation on day 4. In our study, 4 patients (1.9%) reached an INR value more than 4.0 (range, 4.6-5.0) after 3 daily warfarin doses of 5 mg. Clinical variables would not have predicted this sensitivity to warfarin. Yet, all 4 patients were carriers of genetic variants conferring increased sensitivity; thus, genotype-guided dosing may well have reduced the risk of overanticoagulation in these subjects.
Our study has several strengths, including the combination of several outcomes with potential clinical relevance to assess warfarin sensitivity, a heterogeneous study population in a real-life environment treated with similar warfarin starting doses, and a systematic statistical analysis of the predictive value of different variables for the assessment of warfarin sensitivity. As expected in an observational study, not all patients had INR determined on the same day, and dosing was regulated by different clinics. As we have pointed out, variability in management probably favors the contribution of genotype, thus enhancing the relevance of our findings. In addition, our study focuses on the induction phase of warfarin therapy and thus does not provide information about the predictive value of genotyping and early INR measurements on stability during long-term anticoagulation.
In conclusion, in this observational study, much of the information provided by CYP2C9 and VKORC1 genotype about warfarin sensitivity was captured by the early INR response. Prospective studies on the value of genotype-guided warfarin initiation should be evaluated against the background of the study's setting and include algorithms that incorporate early INR response.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by grants from the National Institutes of Health: a Pharmacogenetics Research Network Grant (U01 HL65962), HL04012, and GM31304.
Data presented here have been deposited at the Pharmacogenetics Knowledge Base (pharmgkb.org), which is supported by grant U01GM61374.
National Institutes of Health
Authorship
Contribution: C.L. designed the research, analyzed data, and edited the manuscript; U.I.S. and M.D.R. contributed to data collection and analysis and edited the manuscript; D.M.R. designed the research, revised the manuscript, and obtained funding; C.M.S. designed the research, supervised the study, obtained funding, and edited the manuscript; and D.K. conceived the research, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Daniel Kurnik, Division of Clinical Pharmacology and Toxicology, Sheba Medical Center, Tel Hashomer, Ramat Gan 52621, Israel; e-mail: daniel.kurnik@sheba.health.gov.il.