Abstract
Many somatic genetic abnormalities have been identified in T-cell acute lymphoblastic leukemia (T-ALL) but each individual abnormality accounts for a small proportion of cases; therapeutic stratification consequently still relies on classical clinical markers. NOTCH1 and/or FBXW7 mutations both lead to activation of the NOTCH1 pathway and are among the most frequent mutations in T-ALL. We screened 141 adult diagnostic T-ALL samples from patients treated on either the Lymphoblastic Acute Leukemia in Adults (LALA)-94 (n = 87) or the GRAALL-2003 (n = 54) trials. In 88 cases (62%) there were demonstrated NOTCH1 mutations (42% heterodimerization [HD], 10% HD+proline glutamate serine threonine [PEST], 6% PEST, 2% juxtamembrane mutations, 2% transactivation domain [TAD]) and 34 cases (24%) had FBXW7 mutations (21 cases had both NOTCH1 and FBXW7 mutations); 40 cases (28%) were wild type for both. There was no significant correlation between NOTCH1 and/or FBXW7 mutations and clinico-biologic features. Median event-free survival (EFS) and overall survival (OS) were 36 versus 17 months (P = .01) and not reached versus 32 months (P = .004) in patients with NOTCH1 and/or FBXW7 mutations versus other patients, respectively. Multivariate analysis showed that the presence of NOTCH1/FBXW7 mutations was an independent good prognostic factor for EFS and OS (P = .02 and P = .01, respectively). These data demonstrate that NOTCH1 pathway activation by either NOTCH1 or FBXW7 mutation identifies a large group of patients with a favorable outcome that could justify individual therapeutic stratification for T-ALL.
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) accounts for 25% of adult ALL cases. Although outcome has been improved with current therapy, survival rates remain between 50% and 60% at 5 years.1-3 The few initial prognostic factors used for therapeutic stratification are predominantly clinical: age, WBC count, and central nervous system (CNS) involvement. Although recent advances have led to spectacular progress in understanding of T-ALL oncogenesis,4 molecular markers are so numerous that they are poorly adapted to therapeutic stratification of major discriminatory patient subgroups. The only common subgroup markers with prognostic significance are TLX1 and TLX3 overexpression, considered respectively as favorable and adverse genetic markers in adult French and German trials; however, both are present in only 10% to 15% of cases.5-8 There is therefore a clear need for prognostic markers that identify larger patient subgroups that require specific treatment, notably but not exclusively, the use of allogeneic hematopoietic stem cell transplantation (HSCT) in first complete remission (CR).
The NOTCH1 signaling pathway has been shown to be an essential factor in normal and pathologic T-lymphoid development.9,10 In T-ALL, NOTCH1 abnormalities were first identified in the t(7;9)(q34;q34.3), which juxtaposes the C-terminal region of human NOTCH1 to the TCRβ enhancer, leading to aberrant expression of a truncated dominant active and ligand independent form of NOTCH1 (called TAN-1) in rare patients (< 1%).11 In mouse models, constitutive activation of NOTCH1 signaling induced T-ALL, as did transplantation with TAN1-expressing hematopoietic progenitor cells.12-15 In 2004, activating NOTCH1 mutations were reported in approximately 50% of childhood T-ALLs.16 These mutations involve either the heterodimerization (HD) domain, when they probably facilitate cleavage of the NOTCH receptor and/or the negative regulatory proline glutamateserine threonine (PEST) domain, when they probably increase the half-life of intracellular NOTCH (ICN). A large series of pediatric T-ALLs confirmed this incidence and suggested a favorable outcome overall and within the cortical subgroup.17 More recently, rare NOTCH1 mutations of the juxtamembrane domain have been reported as a mechanism of marked NOTCH1 pathway activation in T-ALL.18 An alternative mechanism of increased NOTCH1 activation by loss-of-function mutations of FBXW7, leading to inhibition of ubiquitin-mediated degradation of the activated form of NOTCH1,19-21 has also been reported recently but their prognostic impact is unknown in both pediatric and adult T-ALL. The incidence and clinical impact of NOTCH1 mutations has only been reported in a very limited number of adult cases.22,23 We therefore evaluated the incidence and prognostic impact of NOTCH1 and FBXW7 mutations in 141 adult T-ALLs from the Lymphoblastic Acute Leukemia in Adults (LALA)–94 and Group for Research on Adult Acute Lymphoblastic Leukemia (GRAALL)–2003 trials.
Methods
LALA-94 and GRAALL-2003 trials
The present study is based on analysis of a total of 141 adults more than 15 years of age treated on the LALA-94 (n = 87) and GRAALL-2003 (n = 54) prospective multicenter trials. The CR rate, survival, outcome, and follow-up of the 87 LALA-94 T-ALL patients with available material did not differ significantly from the 236 T-ALL patients included in this protocol:24 their 3-year overall survival rates were 45% (95% confidence interval [CI], 34-55) and 41% (95% CI, 34-47), respectively. The GRAALL-2003 protocol was a pediatric-inspired phase 2 trial that enrolled 224 adults with Ph-negative ALL between November 2003 and November 2005.25 We report here on 54 patients with T-ALL from whom material was available for single center analysis. These 54 patients were representative of the overall population because their outcome did not differ from the overall 74 patients with T-ALL. The 3-year overall survival rates for these groups were 68% (95% CI, 54%-79%) and 67% (95% CI, 55%-77%), respectively.
Furthermore, the patients from the LALA-94 and the GRAALL-2003 trials did not differ with respect to male-to-female sex ratio (71/16 and 47/7, respectively), age (median, range: 28 years, 15-55 years and 27 years, 15-58 years, respectively), WBC (median, range: 71 109/L, 1.4-620 and 31 109/L, 0.9-399, respectively) and initial complete remission rate (92% and 98%, respectively). The median follow-up rates for LALA-94 and GRAALL-2003 are 90 and 38 months, respectively, with a median follow-up of 47 months for the cohort presented here. The number of patients who received allogeneic HSCT in first CR was 10 of 87 (11%) in the LALA-94 group versus 15 of 54 (28%) in the GRAALL-2003 protocol.
Samples and diagnostic analysis
Diagnostic peripheral blood or bone marrow samples from 141 adult patients with T-ALL, defined by expression of CD7 and cytoplasmic and/or surface CD3 and negativity for CD19 and MPO, were analyzed, with informed consent according to the Declaration of Helsinki. Details of patient classification, immunophenotype, T-cell receptor (TCR) analysis, fusion transcript detection (SIL-TAL1 and CALM-AF10), and oncogenic transcript quantification (TLX1 and TLX3) are as described.5
NOTCH1 and FBXW7 mutation screening
DNA and RNA were extracted from cryopreserved tissue samples, as described.5 Direct sequencing was performed centrally at the Necker Hospital for the 141 patients with T-ALL from 121 DNA and 20 cDNA samples. Exons 26 (HD N-terminal), 27 (HD C-terminal), 28 (juxtamembrane domain), and 34 (transactivation domain TAD and the PEST domain) of NOTCH1 and exons 9, 10, and 12 of WD40 domain of FBXW7 were sequenced. Primers used for NOTCH1 and FBXW7 amplification are reported in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Polymerase chain reaction (PCR) was performed in a 50-μL reaction containing 100 ng DNA (or equivalent cDNA), 0.25 μL HotStarTaq DNA polymerase (QIAGEN, Courtabeauf, France), and 20 μM forward and reverse primer with the GeneAmp 2700 (Applied Biosystems, Foster City, CA). PCR products were run on 1.5% agarose gels, purified (QIAGEN), sequenced in both directions, and analyzed using SeqMan software (DNA). Before sequencing, all the NOTCH1 and FBXW7 single nucleotide polymorphisms (SNPs) were identified in the NCBI SNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/). Diagnostic samples were classified on the basis of their NOTCH1 and FBXW7 status as mutated or wild-type (WT), based on analysis of the aforementioned gene segments.
Statistical analysis
Patient characteristics and CR rates were compared using the Fisher exact test and median comparisons were performed with the Mann-Whitney test. Overall survival (OS) was calculated from the date of randomization until the date of death or last contact. Event-free survival (EFS) was calculated from the date of randomization until the date of induction failure, first relapse, death, or last contact. OS and EFS were estimated by the Kaplan-Meier method,26 then compared by the log-rank test.27 For OS and EFS estimations and comparisons, all patients who received an allogeneic HSCT were analyzed with and without censoring at the time of HSCT. Multivariate analyses were performed using the Cox model and tested by the log likelihood ratio test. The following covariates were tested in univariate analyses for both EFS and OS endpoints: age as continuous variable, WBC as log (WBC+1), GRAALL-2003 versus LALA-94 trial, cortical phenotype, and NOTCH1 or NOTCH1/FBXW7 mutational status. Only covariates with a P value of .10 or less in univariate analysis were retained for multivariate models. Cortical phenotype was not retained for this reason. As TLX1 expression levels were studied in a subset of 127 patients only, all multivariate analyses were performed first in the whole population of 141 patients, not taking this factor into account, and repeated in these 127 patients but taking TLX1 expression level into account. Hazard ratios (HRs) are given with 95% confidence intervals (CIs). All calculations were performed using the STATA software, version 9.0 (Stata, College Station, TX).
Results
Incidence and type of NOTCH1 and FBXW7 mutations in adult T-ALL
Overall, NOTCH1 mutations were identified in 88 (62%) of the 141 adult patients with T-ALL whereas 53 cases (38%) were wild type (WT) for the HD, TAD, and PEST domains and the recently reported juxtamembrane mutation (Table 1). PEST mutations were only detected in 9 cases as a unique mutation and in 16 cases in association with HD mutation (15 cases) or duplication (1 case). HD mutations alone (59 cases), in association with PEST (15 cases), or TAD mutations (1 case) were the most frequent NOTCH1 mutations (Table 2). The juxtamembrane mutation was detected in 3 of 141 (2%) adult cases, but was only found as an isolated abnormality in one case, being associated with HD or TAD mutation in the other 2 cases. The translated proteins of all identified NOTCH1 mutations are shown in Table S2.
Clinical, immunophenotypic, and genotypic characteristics of adult T-ALL as a function of NOTCH1/FBXW7 status
| . | Total (%) . | NOTCH1 mutation . | FBXW7 mutation . | NOTCH1 and/or FBXW7 mutation . | |||
|---|---|---|---|---|---|---|---|
| Mutant, n (%) . | Wild-type, n (%) . | Mutant, n (%) . | Wild-type, n (%) . | Mutant, n (%) . | Wild-type, n (%) . | ||
| 141 | 88 (62) | 53 (38) | 34 (24) | 107 (76) | 101 (72) | 40 (28) | |
| TCR subsets analyzed | |||||||
| Immature | 42 | 25 (60) | 17 (40) | 6 (14) | 36 (86) | 27 (64) | 15 (36) |
| Pre-αβ | 44 | 30 (68) | 14 (32) | 13 (30) | 31 (70) | 35 (80) | 9 (20) |
| TCR+ | 29 | 13 (45) | 15 (53) | 7 (24) | 22 (76) | 19 (65) | 10 (34) |
| ND | 26 | 19 (73) | 7 (27) | 8 (31) | 18 (70) | 20 (77) | 6 (23) |
| EGIL | |||||||
| 1-2 | 43 | 25 (58) | 18 (42) | 6 (14) | 37 (86) | 27 (63) | 16 (37) |
| 3 | 73 | 52 (71) | 21 (29) | 24 (33) | 49 (67) | 59 (81) | 14 (19) |
| 4 | 15 | 7 (47) | 8 (53) | 4 (27) | 11 (73) | 11 (73) | 4 (27) |
| ND | 10 | 4 (40) | 6 (60) | 0 (0) | 10 (100) | 4 (40) | 6 (60) |
| Genotype subsets analyzed | |||||||
| CALM-AF10 | 7 | 3 (43) | 4 (57) | 4 (57) | 3 (43) | 5 (71) | 2 (29) |
| SIL-TAL1 | 10 | 4 (40) | 6 (60) | 1 (10) | 9 (90) | 5 (50) | 5 (50) |
| HOX11 | 21 | 18 (86) | 3 (14) | 5 (24) | 16 (76) | 20 (95) | 1 (5) |
| HOX11L2 | 13 | 7 (54) | 6 (46) | 4 (31) | 9 (69) | 10 (77) | 3 (23) |
| None of above | 76 | 44 (58) | 32 (42) | 17 (22) | 59 (78) | 48 (63) | 28 (37) |
| ND | 14 | 12 (86) | 2 (14) | 3 (21) | 11 (79) | 13 (93) | 1 (7) |
| Clinical subsets analyzed | |||||||
| Median age, y | 28 | 28 | 28 | 28 | 31 | 27 | 29 |
| Age >35 | 46 (33) | 30 (34) | 16 (30) | 10 (29) | 36 (34) | 33 (33) | 13 (33) |
| Median WBC | 54 | 51 | 94 | 59 | 94 | 51 | 104 |
| WBC >100g/L | 52 (37) | 28 (31) | 24 (45) | 7 (21) | 45 (42) | 31 (31) | 21 (53) |
| High-risk MRC-ECOG | 85 (60) | 50 (57) | 35 (66) | 16 (47) | 69 (64) | 56 (56) | 29 (73) |
| Mediastinal involvement | 66 (47) | 43 (48) | 23 (43) | 20 (59) | 46 (43) | 51 (52) | 15 (38) |
| CNS involvement | 12 (9) | 4 (5) | 8 (15) | 2 (6) | 10 (9) | 6 (6) | 6 (15) |
| . | Total (%) . | NOTCH1 mutation . | FBXW7 mutation . | NOTCH1 and/or FBXW7 mutation . | |||
|---|---|---|---|---|---|---|---|
| Mutant, n (%) . | Wild-type, n (%) . | Mutant, n (%) . | Wild-type, n (%) . | Mutant, n (%) . | Wild-type, n (%) . | ||
| 141 | 88 (62) | 53 (38) | 34 (24) | 107 (76) | 101 (72) | 40 (28) | |
| TCR subsets analyzed | |||||||
| Immature | 42 | 25 (60) | 17 (40) | 6 (14) | 36 (86) | 27 (64) | 15 (36) |
| Pre-αβ | 44 | 30 (68) | 14 (32) | 13 (30) | 31 (70) | 35 (80) | 9 (20) |
| TCR+ | 29 | 13 (45) | 15 (53) | 7 (24) | 22 (76) | 19 (65) | 10 (34) |
| ND | 26 | 19 (73) | 7 (27) | 8 (31) | 18 (70) | 20 (77) | 6 (23) |
| EGIL | |||||||
| 1-2 | 43 | 25 (58) | 18 (42) | 6 (14) | 37 (86) | 27 (63) | 16 (37) |
| 3 | 73 | 52 (71) | 21 (29) | 24 (33) | 49 (67) | 59 (81) | 14 (19) |
| 4 | 15 | 7 (47) | 8 (53) | 4 (27) | 11 (73) | 11 (73) | 4 (27) |
| ND | 10 | 4 (40) | 6 (60) | 0 (0) | 10 (100) | 4 (40) | 6 (60) |
| Genotype subsets analyzed | |||||||
| CALM-AF10 | 7 | 3 (43) | 4 (57) | 4 (57) | 3 (43) | 5 (71) | 2 (29) |
| SIL-TAL1 | 10 | 4 (40) | 6 (60) | 1 (10) | 9 (90) | 5 (50) | 5 (50) |
| HOX11 | 21 | 18 (86) | 3 (14) | 5 (24) | 16 (76) | 20 (95) | 1 (5) |
| HOX11L2 | 13 | 7 (54) | 6 (46) | 4 (31) | 9 (69) | 10 (77) | 3 (23) |
| None of above | 76 | 44 (58) | 32 (42) | 17 (22) | 59 (78) | 48 (63) | 28 (37) |
| ND | 14 | 12 (86) | 2 (14) | 3 (21) | 11 (79) | 13 (93) | 1 (7) |
| Clinical subsets analyzed | |||||||
| Median age, y | 28 | 28 | 28 | 28 | 31 | 27 | 29 |
| Age >35 | 46 (33) | 30 (34) | 16 (30) | 10 (29) | 36 (34) | 33 (33) | 13 (33) |
| Median WBC | 54 | 51 | 94 | 59 | 94 | 51 | 104 |
| WBC >100g/L | 52 (37) | 28 (31) | 24 (45) | 7 (21) | 45 (42) | 31 (31) | 21 (53) |
| High-risk MRC-ECOG | 85 (60) | 50 (57) | 35 (66) | 16 (47) | 69 (64) | 56 (56) | 29 (73) |
| Mediastinal involvement | 66 (47) | 43 (48) | 23 (43) | 20 (59) | 46 (43) | 51 (52) | 15 (38) |
| CNS involvement | 12 (9) | 4 (5) | 8 (15) | 2 (6) | 10 (9) | 6 (6) | 6 (15) |
No statistically significant differences were observed.
Combinations of NOTCH1 mutations HD (heterodimerization) and PEST (proline, glutamatic acid, serine, threonine), and FBXW7 mutations
| NOTCH1/FBXW7 mutation . | HD only, n (%) . | HD + PEST, n (%) . | PEST only, n (%) . | Others, n (%) . |
|---|---|---|---|---|
| GL | 41 (29) | 13 (9) | 9 (6) | 4* (3) |
| Mutated | 18 (13) | 2 (1) | 0 (0) | 1† (1) |
| Total | 59 (42) | 15 (10) | 9 (6) | 5 (4) |
| NOTCH1/FBXW7 mutation . | HD only, n (%) . | HD + PEST, n (%) . | PEST only, n (%) . | Others, n (%) . |
|---|---|---|---|---|
| GL | 41 (29) | 13 (9) | 9 (6) | 4* (3) |
| Mutated | 18 (13) | 2 (1) | 0 (0) | 1† (1) |
| Total | 59 (42) | 15 (10) | 9 (6) | 5 (4) |
Duplication, PEST+duplication, TAD+duplication, TAD.
HD+TAD
FBXW7 mutations were present in 34 cases (24%), alone in 13 cases and in association with NOTCH1 mutations in 21 cases (Table 1). Of these mutations, 30 of 34 correspond to the 4 previously reported arginine substitutions at R479, R465, R505, and R689.21 The 4 remaining FBXW7 mutations (G423V, G477S, S516G, and a stop insertion) were all found in association with an HD NOTCH1 mutation.
Overall, 101 cases were classified as NOTCH1 and/or FBXW7 mutated and 40 cases (28%) as WT for both. The proportion of NOTCH1/FBXW7 mutated cases was similar within the 2 protocols: 39 of 54 (72%) and 62 of 87 (71%) in the GRAALL-2003 and the LALA-94, respectively.
Clinical, phenotypic, and oncogenic features of NOTCH1 and NOTCH1/FBXW7 mutated T-ALL
Clinical features of patients were analyzed according to the presence or absence of NOTCH1 and/or FBXW7 mutations (Table 1). The median age of the patients and the incidence of mediastinal involvement were similar in all groups. There was a trend for a higher WBC count and more frequent CNS involvement in patients demonstrating WT NOTCH1 and FBXW7 (P = .08). Similarly, high-risk Medical Research Council/Eastern Cooperative Oncology Group (MRC/ECOG) features28 (age > 35 years and WBC count > 100 g/L) were found in 55% of patients with mutated NOTCH1 and/or FBXW7, compared with 72.5% of patients with WT NOTCH1 and FBXW7 status, but this difference was not statistically significant (P = .085).
In contrast to pediatric T-ALL, there was no statistical correlation between the presence of NOTCH1 and/or FBXW7 mutation and stage of maturation arrest, although mutations tended to be more frequent in pre-αβ/EGILIII immunophenotypic subtypes. With respect to oncogenetic subtypes, NOTCH1/FBXW7 mutations were more frequent in cases expressing TLX1 and less frequent in SIL-TAL1+ cases. All but one of the 21 TLX1-expressing cases (95%) were NOTCH1/FBXW7 mutated, as compared with only 64% of cases not expressing TLX1 (P = .004).
Prognostic value of NOTCH1 and FBXW7 mutations within the adult LALA-94 and GRAALL-2003 therapeutic protocols
Results of prognostic analyses are summarized in Table 3. Basically, we first analyzed the prognosis of patients with mutated NOTCH1 (n = 88) with those with WT NOTCH1 (n = 53), without taking into account FBXW7 status. We then compared patients with a WT genotype for both NOTCH1 and FBXW7 (n = 40) with those with either a NOTCH1 and/or FBXW7 mutation (n = 101). As TLX1 expression level was a significant prognostic factor in this cohort of patients, but could not be tested in all of them, Table 3 also presents multivariate analyses in the subset of 127 patients tested for TLX1 expression.
Univariate and multivariate prognostic analysis
| . | NOTCH1 . | NOTCH1/FBXW7 . | ||||
|---|---|---|---|---|---|---|
| Univariate P (95% CI) . | Multivariate HR (95% CI) . | P . | Multivariate HR (95% CI) . | P . | ||
| For EFS | ||||||
| In all patients | ||||||
| Age* | .22 | NA | NA | NA | NA | |
| WBC† | .08 | 1.11 (0.92-1.34) | .29 | 1.10 (0.91-1.32) | .33 | |
| GRAALL-2003 trial | < .001 | 0.44 (0.26-0.75) | .002 | 0.43 (0.25-0.72) | .001 | |
| NOTCH mutation | .03 | 0.68 (0.44-1.06) | .9 | NA | NA | |
| NOTCH1/FBXW7 mutation | .01 | NA | NA | 0.58 (0.37-0.92) | .02 | |
| In the 127 pateints tested for TLX1 expression | ||||||
| Age* | .27 | NA | NA | NA | NA | |
| WBC† | .05 | 1.12 (0.92-1.36) | .28 | 1.11 (0.92-1.35) | .28 | |
| GRAALL-2003 trial | < .001 | 0.42 (0.24-0.73) | .002 | 0.39 (0.22-0.69) | .001 | |
| High TLX1 expression | .03 | 0.72 (0.48-1.08) | .11 | 0.73 (0.49-1.09) | .12 | |
| NOTCH1 mutation | .02 | 0.72 (0.45-1.16) | .17 | NA | NA | |
| NOTCH1/FBXW7 mutation | .015 | NA | NA | 0.63 (0.39-1.02) | .06 | |
| For OS | ||||||
| In all patients | ||||||
| Age* | .03 | 1.03 (1.01-1.05) | .015 | 1.03 (1.01-1.05) | .03 | |
| WBC† | .09 | 1.16 (0.95-1.41) | .15 | 1.14 (0.94-1.39) | .19 | |
| GRAALL-2003 trial | .01 | 0.57 (0.33-0.99) | .05 | 0.55 (0.31-0.87) | .03 | |
| NOTCH1 mutation | .03 | 0.63 (0.39-1.01) | .06 | NA | NA | |
| NOTCH1/FBXW7 mutation | .004 | NA | NA | 0.54 (0.33-0.87) | .01 | |
| In the 127 patients tested for TLX1 expression | ||||||
| Age* | .08 | 1.03 (1.01-1.06) | .02 | 1.03 (1.01-1.05) | .04 | |
| WBC† | .08 | 1.17 (0.95-1.44) | .13 | 1.16 (0.94-1.42) | .16 | |
| GRAALL-2003 trial | .006 | 0.53 (0.29-0.96) | .35 | 0.50 (0.27-0.90) | .02 | |
| High TLX1 expression | .02 | 0.62 (0.39-0.99) | .05 | 0.64 (0.40-1.03) | .07 | |
| NOTCH1 mutation | .03 | 0.71 (0.43-1.18) | .19 | NA | NA | |
| NOTCH1/FBXW7 mutation | .005 | NA | NA | 0.61 (0.36-1.01) | .06 | |
| . | NOTCH1 . | NOTCH1/FBXW7 . | ||||
|---|---|---|---|---|---|---|
| Univariate P (95% CI) . | Multivariate HR (95% CI) . | P . | Multivariate HR (95% CI) . | P . | ||
| For EFS | ||||||
| In all patients | ||||||
| Age* | .22 | NA | NA | NA | NA | |
| WBC† | .08 | 1.11 (0.92-1.34) | .29 | 1.10 (0.91-1.32) | .33 | |
| GRAALL-2003 trial | < .001 | 0.44 (0.26-0.75) | .002 | 0.43 (0.25-0.72) | .001 | |
| NOTCH mutation | .03 | 0.68 (0.44-1.06) | .9 | NA | NA | |
| NOTCH1/FBXW7 mutation | .01 | NA | NA | 0.58 (0.37-0.92) | .02 | |
| In the 127 pateints tested for TLX1 expression | ||||||
| Age* | .27 | NA | NA | NA | NA | |
| WBC† | .05 | 1.12 (0.92-1.36) | .28 | 1.11 (0.92-1.35) | .28 | |
| GRAALL-2003 trial | < .001 | 0.42 (0.24-0.73) | .002 | 0.39 (0.22-0.69) | .001 | |
| High TLX1 expression | .03 | 0.72 (0.48-1.08) | .11 | 0.73 (0.49-1.09) | .12 | |
| NOTCH1 mutation | .02 | 0.72 (0.45-1.16) | .17 | NA | NA | |
| NOTCH1/FBXW7 mutation | .015 | NA | NA | 0.63 (0.39-1.02) | .06 | |
| For OS | ||||||
| In all patients | ||||||
| Age* | .03 | 1.03 (1.01-1.05) | .015 | 1.03 (1.01-1.05) | .03 | |
| WBC† | .09 | 1.16 (0.95-1.41) | .15 | 1.14 (0.94-1.39) | .19 | |
| GRAALL-2003 trial | .01 | 0.57 (0.33-0.99) | .05 | 0.55 (0.31-0.87) | .03 | |
| NOTCH1 mutation | .03 | 0.63 (0.39-1.01) | .06 | NA | NA | |
| NOTCH1/FBXW7 mutation | .004 | NA | NA | 0.54 (0.33-0.87) | .01 | |
| In the 127 patients tested for TLX1 expression | ||||||
| Age* | .08 | 1.03 (1.01-1.06) | .02 | 1.03 (1.01-1.05) | .04 | |
| WBC† | .08 | 1.17 (0.95-1.44) | .13 | 1.16 (0.94-1.42) | .16 | |
| GRAALL-2003 trial | .006 | 0.53 (0.29-0.96) | .35 | 0.50 (0.27-0.90) | .02 | |
| High TLX1 expression | .02 | 0.62 (0.39-0.99) | .05 | 0.64 (0.40-1.03) | .07 | |
| NOTCH1 mutation | .03 | 0.71 (0.43-1.18) | .19 | NA | NA | |
| NOTCH1/FBXW7 mutation | .005 | NA | NA | 0.61 (0.36-1.01) | .06 | |
HR indicates hazard ratio; and NA, not applicable.
Considered as a continuous variable.
Entered as log (WBC+1).
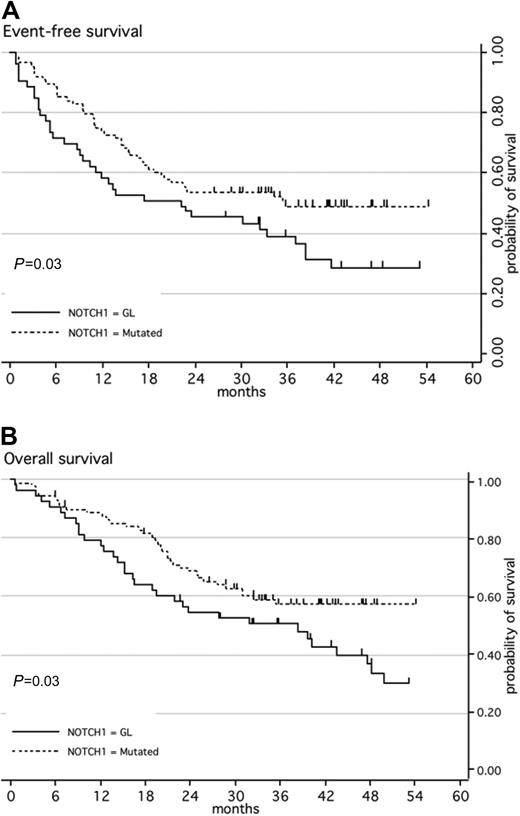
When first considering NOTCH1 mutation only, the CR rate was similar in patients with NOTCH1 mutations as compared with those with WT NOTCH1 (97% vs 91% respectively; P = .15). The median EFS was 22 months for patients with WT NOTCH1 versus 36 months for the group with mutated NOTCH1 (P = .03; Figure 1A). The median OS was 38 months for patients with WT NOTCH1 but had not been reached for cases with NOTCH1/FBXW7 mutations (P = .03; Figure 1B). In multivariate analysis, this good prognostic impact of the presence of a NOTCH1 mutation did not reach statistical significance, although trends were observed for both EFS and OS (Table 3).
Kaplan-Meier estimates of event-free survival or overall survival according to NOTCH1 mutation. (A) Event-free survival (EFS); (B) overall survival (OS). Patients with ALL with a wild-type genotype of NOTCH1 (black line; n = 53) have a significantly higher risk of event or death than patients with ALL with mutated NOTCH1 (dashed line; n = 88).
Kaplan-Meier estimates of event-free survival or overall survival according to NOTCH1 mutation. (A) Event-free survival (EFS); (B) overall survival (OS). Patients with ALL with a wild-type genotype of NOTCH1 (black line; n = 53) have a significantly higher risk of event or death than patients with ALL with mutated NOTCH1 (dashed line; n = 88).
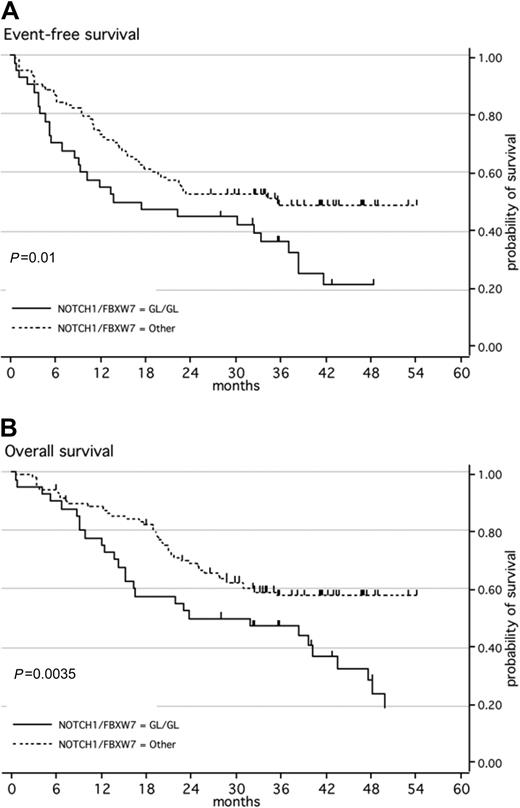
When both NOTCH1 and FBXW7 mutations were taken into account, CR rates remained comparable in patients with a WT genotype for both NOTCH1 and FBXW7 as compared with those with either a NOTCH1 and/or FBXW7 mutation (95% and 92.5%, respectively; P = .69). Median EFS for the WT group was only 17 months, compared with 36 months for the mutated group (Figure 2A, P = .01). An impact from HSCT in first CR is unlikely since the proportion of patients who underwent transplantation was 6 of 40 (15%) in the WT group as compared with 19 of 101 (19%) in the mutated group (P = .81). With respect to OS, median OS was not reached for the mutated group compared with only 32 months for the WT group (P = .004; Figure 2B). The 21 patients with both NOTCH1 and FBXW7 mutations demonstrated the same OS and EFS as patients with mutations of one or the other (data not shown). In the whole patient population, only 2 independent good prognostic factors were found for EFS (GRAALL-2003 trial and NOTCH1/FBXW7 mutation; P = .001 and .02, respectively) and 3 independent good prognostic factors for OS (younger age, GRAALL-2003 trial, and NOTCH1/FBXW7 mutation; P = .03, .03, and .01, respectively; Table 3). Despite the fact that most patients with high TLX1 expression had NOTCH1/FBXW7 mutation (Table 1), trends for a favorable impact of NOTCH1/FBXW7 mutation were still observed in multivariate analysis performed in the subset of 127 patients tested for TLX1 expression (P = .06 for both EFS and OS; Table 3). On the other hand, patients with NOTCH1/FBXW7 mutation and no high TLX1 expression still displayed a better outcome than those without NOTCH1/FBXW7 mutation (P = .04 and .02 for EFS and OS, respectively).
Kaplan-Meier estimates of event-free survival or overall survival according to NOTCH1 and/or FBXW7 mutations. (A) Event-free survival (EFS); (B) overall survival (OS). Patients with ALL with a wild-type genotype of NOTCH1 and FBXW7 (black line; n = 40) have a significantly higher risk of event or death than patients with ALL with a mutated NOTCH1 and/or FBXW7 (dashed line; n = 110).
Kaplan-Meier estimates of event-free survival or overall survival according to NOTCH1 and/or FBXW7 mutations. (A) Event-free survival (EFS); (B) overall survival (OS). Patients with ALL with a wild-type genotype of NOTCH1 and FBXW7 (black line; n = 40) have a significantly higher risk of event or death than patients with ALL with a mutated NOTCH1 and/or FBXW7 (dashed line; n = 110).
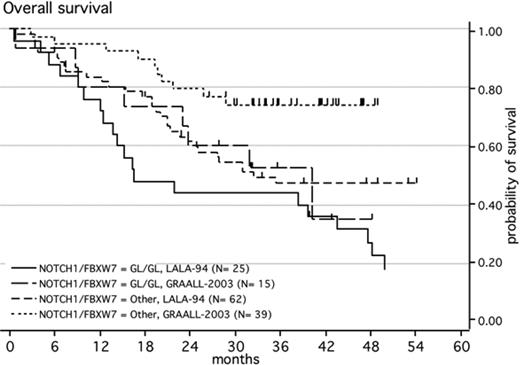
When each protocol was analyzed separately, patients with NOTCH1/FBXW7 mutations treated on the most recent GRAALL-2003 trial have a 3-year OS of 74% (95% CI, 57-85) compared with 52.5% (95% CI, 25-74) for those with a WT status (P = .05) confirming that NOTCH1/FBXW7 status is of major prognostic significance even in modern trials that give overall good results for T-ALL (Figure 3).
Kaplan-Meier estimates of overall survival of patients with ALL according to the presence of a NOTCH1 and/or FBXW7 mutations and according to the chemotherapeutic protocol. Survival of patients with a wild-type genotype of NOTCH1 and FBXW7, treated with the LALA-94 protocol (n = 25); survival of patients with a wild-type genotype of NOTCH1 and FBXW7, treated with the GRAALL-2003 protocol (n = 15); patients with a mutated genotype of NOTCH1 and/or FBXW7, treated with the LALA-94 protocol (n = 62); patients with a mutated genotype of NOTCH1 and/or FBXW7, treated with the GRAALL-2003 protocol (n = 39).
Kaplan-Meier estimates of overall survival of patients with ALL according to the presence of a NOTCH1 and/or FBXW7 mutations and according to the chemotherapeutic protocol. Survival of patients with a wild-type genotype of NOTCH1 and FBXW7, treated with the LALA-94 protocol (n = 25); survival of patients with a wild-type genotype of NOTCH1 and FBXW7, treated with the GRAALL-2003 protocol (n = 15); patients with a mutated genotype of NOTCH1 and/or FBXW7, treated with the LALA-94 protocol (n = 62); patients with a mutated genotype of NOTCH1 and/or FBXW7, treated with the GRAALL-2003 protocol (n = 39).
Finally, when censoring patients at the time of HSCT, NOTCH1/FBXW7 mutations retained their significant favorable impact on EFS and OS in the multivariate analysis (P = .04 and .025, respectively; data not shown).
Taken overall, these data demonstrate that detection of FBXW7 mutations adds significant prognostic value to assessment of NOTCH1 status alone and allows identification of a majority (72%) of relatively good prognosis patients with a NOTCH1/FBXW7 mutated T-ALL, which cannot be identified by more classical, clinical, immunophenotypic or oncogenic markers.
Discussion
The pediatric concept of risk-adapted therapy of ALL has recently been extended to adult protocols, leading to marked improvements in outcome.25 This strategy relies on the identification of risk factors at diagnosis or in response to cortico- or chemotherapy during induction. For high-risk adult patients, classical trials favor treatment intensification based on allogeneic HSCT in first CR. For B-cell ALL, a variety of numerical or structural chromosomal abnormalities identified at diagnosis are used for therapeutic stratification.29,30 In contrast, corresponding abnormalities in T-ALL, such as TLX3, TLX1, CALM-AF10, NUP214-ABL, or SIL-TAL1 are either found in a small percentage of cases or are of controversial clinical significance.6-8 Consequently, most adult T-ALL trials use simple clinical features such as age, CNS involvement, or WBC count to define high-risk patients, with relatively limited impact on prognosis.31
The identification of a widely expressed genetic marker that could stratify adult patients with T-ALL at diagnosis will allow judicious use of the increasing number of therapeutic alternatives. We here report that NOTCH1 mutations are present in more than 60% of a large series of adult T-ALL. FBXW7 is mutated in 24%, alone in 9%, and in association with NOTCH1 in 15%. Mutation detection allows identification of a major subgroup of 72% of patients with a favorable impact on both EFS and OS.
In the pediatric ALL Berlin-Frankfurt-Munster (BFM) 2000 protocol, NOTCH1 mutations were found in 52% of cases and were associated with a good prognosis, including within the favorable cortical thymic subgroup,17 whereas in a predominantly pediatric Chinese study, NOTCH1 mutation was associated with a poor prognosis.23 Little information is available to date regarding the prognostic impact of FBXW7 mutation but the same synergic good prognosis of NOTCH1 and FBXW7 mutations was described in 26 pediatric cases of T-ALL.32 In this first large series of adult T-ALL, we confirm the good prognosis of NOTCH1 found in the ALL-BFM series, but more importantly, show that the addition of functionally convergent FBXW7 mutations strikingly increases prognostic impact. This suggests that NOTCH1 pathway constitutive activation is more important than the type of somatic genetic abnormality and is globally associated with cortico/chemo-responsive T-ALL, which could potentially be further improved by the addition of novel agents such as gamma-secretase inhibitors (GSIs). Conversely, the significant side effects of GSIs as used to date may indicate cautious use in this good prognostic subgroup.33,34 Furthermore, the proportion of patients with NOTCH1 mutated T-ALL who have mutated phosphate and tensin homolog (PTEN) will not respond to GSIs.35 Whether patients with NOTCH1 and/or FBXW7 mutations should be considered for HSCT should be evaluated in a prospective trial since their OS within the GRAALL-2003 trial, adapted to the T-cell lineage, was more than 70% at 3 years. The minor subgroup of NOTCH1/FBXW7 WT patients have a 52.5% OS rate at 3 years within the GRAALL-2003 trial. As such, these patients are likely to be appropriate candidates for intensive treatment, including HSCT and/or specific targeted therapy. The relatively small size of the NOTCH1/FBXW7 WT group prevented retrospective analysis of the impact of HSCT within this group, which will probably require retrospective meta-analysis within different clinical trials.
Compared with previously identified oncogenic or immunophenotypic markers, NOTCH1 and/or FBXW7 mutations identify 2 major subgroups, which justifies prospective screening and therapeutic stratification. Exhaustive diagnostic strategies should involve sequencing NOTCH1 exons 26 to 28 and 34 (allowing detection of HD, TAD, and PEST mutations and juxtamembrane mutations) and FBXW7 for all patients. This is justifiable if linked to cognitive analysis of NOTCH pathways in T-ALL, but is relatively onerous if the main priority is individual patient stratification. In the latter situation, since double NOTCH1/FBWX7 mutated cases do not have a different prognosis from single mutated cases, a more restrictive approach would involve initial sequencing for NOTCH1 HD and PEST mutations (exons 26, 27, and 34), followed by FBXW7 sequencing and specific detection of NOTCH1 ITD by fluorescent PCR sizing for NOTCH1 WT cases (approximately 40%). Overall, approximately 10% of additional cases will be classified as NOTCH1 mutated based on their FBXW7 status and less than 1% on their NOTCH1 juxtamembrane mutation status. Since TLX1 expression retains its prognostic impact in patients with NOTCH1/FBXW7 mutations in this series, molecular analyses of a limited number of genes are likely to be necessary for optimal prognostic classification of T-ALL.
Therapeutic stratification based on mutational status is increasingly used for individual patient management in CLL (IgH)36 and AML (NPM, FLT3, and CEBPα).37 The present data justify an extension of this approach to T-ALL. The synergic prognostic impact of NOTCH1 and FBXW7 mutations data is in favor of oncogenically significant gain-of-function “driver,” rather than “passenger” mutation, as recently identified in AML.38
To date, this is the first target for prognostic stratifying diagnostic sequencing in ALL, since to our knowledge, PAX5 mutations have not yet been shown to have prognostic impact. Judicious use of the increasing range of therapeutic options, including HSCT, will largely justify such an initial diagnostic investment.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We especially thank A. Delannoy, J. Soulier, and C. Graux for their constructive comments on the manuscript and V. Lheritier for precious help in collecting samples and data. We thank all participants in the LALA-94 and the GRAALL-2003 trials for collecting and providing data and samples: Hôpital Edouard Herriot, Lyon (X. Thomas, C, Dumontet, S. Hayette, V. Lhéritier); Hôpital Saint-Louis, Paris (H. Dombret, J.M. Cayuela, D. Djordjidjevic); Hôpital du Haut Levêque, Pessac (T. Leguay, F. Lacombe, F. Perry); Hôpital Purpan, Toulouse (F. Huguet, E. Kulhein, C. Escriva); Hôpital Pitié-Salpétrière, Paris; (N. Dhédin, H. Merle-Beral, F. Davi); Institut Paoli Calmettes, Marseille (N. Vey, D. Sainty, G. Dridi); Hôpital Cochin, Paris (F. Dreyfus, F. Picard); Hôpital de l'Archet, Nice (N. Gratecos, P. Philip; I. Touitou); Hôpital Michallon, Grenoble (F. Garban, M.C. Jacob, V. Rolland); Institut Gustave Roussy, Villejuif (J.H. Bourhis, C. Bayle); Center Hospitalier, Caen (O. Reman, V. Salaun, E. Lepesant); HIA Percy, Clamart (T. De Revel, T. Sanson); Hôpital Dupuytren, Limoges (P. Turlure, F. Trimoreau, C. Tisseuil); Center Hospitalier, Avignon (O. Boulat, M. Derre); Center Henri Becquerel, Rouen (St. Leprêtre, B. Lenormand, V. Tallon); Center Hospitalier, Lille (St. De Botton, P. Lepelley, H. Djeda); Hôpital André Mignot, Versailles (S. Castaigne, I. Garcia); Hôpital Beaujon, Clichy (C. Gardin, G. Leroux, B. Beve); Center Hospitalier Lapeyronie, Montpellier (N. Fegueux, J. Taib, M. Leroux); Center Hospitalier, Perpignan (X. Vallantin); Center René Huguenin, St Cloud (M. Janvier, A. Bourgignat); Center Hospitalier, Nantes (P. Chevallier, R. Garand, B. Saulquin); Center Hospitalier, Rennes (M. Escoffre, M. Rousset, C. Picouleau); Institut de Cancérologie de la Loire, St Priest en Jarez (D. Guyotat, L. Campos, S. Marchand); Center Hospitalier, Angers (M. Hunault, L. Baranger, C. Marie); Center Hospitalier, Besancon (F. Legrand, F. Garnache-Ottou); Center Hospitalier Mulhouse (M. Ojeda-Uribe, C. Gervais, S. Iglarz); Center Hospitalier Régional, Orléans (M. Alexis).
This work was supported by grants from the Association Cent Pour Sang La Vie (Convention N°8-Asnafi) and the Association Laurette Fugain (RAF ALF N°06-03), in part by PHRC no. 94-95-97.02, Soutien des therapeutiques innovatrices coûteuses–Réseau de Biologie Innovatrice en Ouco-hématologie (RuBIH), France (LALA-94), by Promoteur Hospices Civils de Lyon (GRAALL-2003), and by grants P0200701 and P030425/AOM03081 from Le Programme Hospitalier de Recherche Clinique, Ministère de l'Emploi et de la Solidarité (Paris, France).
Authorship
Contribution: V.A., A.B., H.D., and E.A.M. wrote the manuscript; S.L.N., F.B., and A.S. performed NOTCH1/FBXW7 sequencing; S.L.N., V.A., and K.H.B. supervised and analyzed molecular data; O.R., F.W., T.R., E.T., P.T., T.L., F.H., J.P.V., F.D., M.C.B., N.I., and X.T. contributed to supervision of clinical research and data collection; H.D. and E.A.M. oversaw conceptual development of the project.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Elizabeth Macintyre, Hôpital Necker-Enfants Malades, Tour Pasteur 2e étage, Laboratoire d'hématologie, 149 rue de Sèvres, 75015 Paris, France; e-mail: elizabeth.macintyre@nck.aphp.fr.
References
Author notes
*V.A., A.B., and S.L.N. contributed equally to this report.