Abstract
The Rac1 and Rac2 GTPases play important roles in many processes including cytoskeletal reorganization, proliferation, and survival, and are required for B-cell development. Previous studies had shown that deficiency in Rac2 did not affect T-cell development, whereas the function of Rac1 in this process has not been investigated. We now show that simultaneous absence of both GTPases resulted in a very strong developmental block at the pre-TCR checkpoint and in defective positive selection. Unexpectedly, deficiency of Rac1 and Rac2 also resulted in the aberrant survival of thymocytes lacking expression of TCRβ, showing hallmarks of hyperactive Notch signaling. Furthermore, we found a similar novel phenotype in the absence of Vav1, Vav2, and Vav3, which function as guanine nucleotide exchange factors for Rac1 and Rac2. These results show that a pathway containing Vav and Rac proteins may negatively regulate Notch signaling during early thymic development.
Introduction
T-cell development in the thymus proceeds through a series of developmental stages, with transitions controlled by signals from the T-cell antigen receptor (TCR) and pre-TCR.1 The Vav1 protein transduces signals from these receptors and the absence of Vav1 results in blocks at these developmental checkpoints.2 In particular, Vav1 deficiency results in a partial block at the pre-TCR checkpoint, which is made much stronger when all 3 Vav-family proteins (Vav1, Vav2, and Vav3) are absent.3,4 In contrast, deficiency of Vav1 alone is sufficient to cause a strong block in TCR-driven positive and negative selection of CD4+CD8+ double-positive (DP) thymocytes.4 Because Vav1 is a guanine nucleotide exchange factor (GEF) for the Rac-family of GTPases (Rac1, Rac2, and Rac3), it has been proposed that part of Vav1's developmental function is to transduce (pre-)TCR signals to Rac proteins. These GTPases in turn are important signal transducing molecules in a broad range of cellular processes including cytoskeletal reorganization, proliferation, and survival.5,6
Despite the recognized importance of Rac GTPases, and a demonstrated requirement for Rac1 and Rac2 in B-cell development,7 relatively little is known about the function of these proteins in the T-cell lineage. Ectopic expression of constitutively active Rac1 or Rac2 proteins during thymic development substituted for a pre-TCR signal, and redirected thymocytes from positive to negative selection,8-10 suggesting that Rac1 and Rac2 have the potential to participate in these pre-TCR– and TCR-driven developmental transitions. Despite this, lack of Rac2 did not affect T-cell development.11,12 Similar analysis of the highly related Rac1 gene has been hampered by the early embryonic lethality of mice deficient in Rac1.13
Further support for the view that Rac proteins are likely to play an important role in T-cell biology has come from studies of mice deficient in 2 other Rac-specific GEFs: DOCK2 and IBP (also known as SLAT). Deletion of the Dock2 gene results in mild impairment of thymic selection and T-cell activation, but a strong defect in chemokine-induced T-cell migration.14-16 In the absence of IBP, mice show defective thymic development, and T cells respond poorly to TCR stimulation.17,18
In this report, we directly examine the roles of Rac1 and Rac2 in T-cell development, using a conditional allele of Rac17 to investigate the effects of deleting Rac1 alone or in combination with Rac2. We show that the 2 GTPases have overlapping redundant functions, and that in the absence of both GTPases, T-cell development is completely blocked, probably due to a critical role for the proteins in transducing pre-TCR and TCR signals. Unexpectedly, we also found that the Rac GTPases and the Vav GEFs may negatively regulate Notch function during early thymic development.
Methods
Mice
Mice bearing a conditional loxP-flanked allele of Rac1 (Rac1flox/flox)7 and Rac2-deficient mice (Rac2−/−)19 were crossed with a transgenic strain carrying the Cre recombinase under the control of the human CD2 promoter (hCD2-iCre),20 to generate mice deficient in Rac1 (Rac1flox/floxhCD2-iCre; Rac1T), Rac2 (Rac2−/−hCD2-iCre), or both (Rac1flox/floxRac2−/−hCD2-iCre; Rac1TRac2−/−). Mice carrying the hCD2-iCre transgene alone were used as control wild-type (WT) mice. In all mice analyzed, the hCD2-iCre transgene was always heterozygous. These 4 strains were further crossed to mice bearing the F5 TCR transgene21 that were also deficient in Rag1 (Rag1−/−),22 and to a transgenic strain carrying the GFP gene under the control of the human CD2 promoter (hCD2-GFP).20 Mice deficient in Vav1, Vav2, and Vav3 (Vav1−/−Vav2−/−Vav3−/−) on a B10.BR background23 were compared with wild-type B10.BR mice. All mice were bred and maintained in accordance with United Kingdom Home Office regulations.
Radiation chimeras
To make competitive radiation chimeras, bone marrow from either Rac1+/+-Rac2+/+hCD2-iCre (WT) or Rac1flox/floxRac2−/−hCD2-iCre (Rac1TRac2−/−) mice (both Ly5.2+) and bone marrow from B6.SJL mice (Ly5.1+) were mixed at a 4:1 ratio, and injected 2.5 × 106 cells/mouse intravenously into B6.SJL mice that had received 2 doses of 4.75 Gy total body irradiation from a 137Cs source, administered 3 hours apart to minimize gastrointestinal tract damage. For analysis of DN thymocyte localization, bone marrow from either Rac1+/+Rac2+/+Rag1−/−hCD2-iCre/hCD2-GFP or Rac1flox/floxRac2−/−Rag1−/−hCD2-iCre/hCD2-GFP and bone marrow from C57BL/6 mice were mixed at a 4:1 ratio and injected 2.5 × 106 cells/mouse intravenously into C57BL/6 mice that had been irradiated as described previously in this paragraph. Mice were treated with Baytril (Bayer HealthCare, Uxbridge, United Kingdom) in their drinking water for at least 4 weeks after transfer and were analyzed 6 to 8 weeks after the transfer.
Flow cytometric analysis and cell sorting
Biotin and fluorophore-conjugated antibodies against CD4, CD8, CD44, CD25, B220, Mac1, Dx5, Gr1, TCRβ, TCRγδ, Thy1.2, CD3, IL7Rα, Bcl2, and Bax (BD Biosciences [Oxford, United Kingdom], eBioscience [San Diego, CA], and Tebu-bio [Peterborough, United Kingdom]) were used in standard flow cytometric procedures to phenotypically characterize and sort cell populations. For analysis of DN subsets, thymocytes were stained with antibodies to lineage (Lin) markers (CD4, CD8, CD3, TCRβ, TCRγδ, B220, Mac1, Gr1, Dx5), Thy1.2, CD44, and CD25, to allow identification of DN1 (Lin−Thy1.2+CD44+CD25−), DN2/3 (Lin−Thy1.2+CD25+), DN3 (Lin−Thy1.2+CD44−CD25+), and DN4 (Lin−Thy1.2+CD44−CD25−) fractions. Cell sorting was performed on a MoFlo cytometer (Dako UK, Ely, United Kingdom) and analysis was carried out on FACSCalibur, LSR, or LSRII cytometers (BD Biosciences).
For intracellular staining with antibodies against TCRβ or CD3, cells were first stained with antibodies to cell-surface proteins to define thymocyte subsets, as well as with a saturating amount of anti-TCRβ or anti-CD3ϵ antibodies to block cell-surface receptors. Cells were fixed for 20 minutes at room temperature with 1% paraformaldehyde. After washing in PBS, autofluorescence was quenched by incubating the cells for 10 minutes with PBS, 10 mM glycine. Cells were then permeabilized using PBS, 0.5% wt/vol saponin, 5% FCS, 10 mM Hepes (pH 7.4) for 10 minutes at room temperature and stained with FITC-conjugated antibodies specific for TCRβ or CD3 (H57-597 and 17A2, respectively; BD Biosciences) for 45 minutes at room temperature. Intracellular staining of Bcl2 and Bax was carried out in a similar manner, except that the antibodies were revealed with goat anti–mouse IgG-FITC and goat anti–rabbit-FITC (Jackson ImmunoResearch, West Grove, PA), respectively.
To measure proliferation of thymocytes, mice were injected intraperitoneally with 5-bromo-2-deoxyuridine (BrdU; 1 mg in PBS; Sigma-Aldrich, Poole, United Kingdom). Four hours later, the thymus was harvested, cells were stained with antibodies to cell-surface proteins, and BrdU incorporation into DNA was measured using a BrdU Flow Kit (BD Biosciences) according to the manufacturer's instructions. Apoptosis was measured using the TUNEL procedure with the In Situ Cell Death Detection Kit (Roche Diagnostics, Burgess Hill, United Kingdom) according to the manufacturer's instructions.
PCR to detect Rac1 gene deletion
DNA from the relevant thymocyte or T-cell subset was isolated and analyzed by polymerase chain reaction (PCR) using 3 oligonucleotides: 5′-ATTTTGTGCCAAGGACAGTGACAAGCT-3′, 5′-GAAGGAGAAGAAGCTGACTCCCATC-3′, and 5′-CAGCCACAGGCAATGACAGATGTTC-3′. PCR was carried out for 30 cycles with a 30-second annealing time at 54°C. PCRs were analyzed by agarose gel electrophoresis. The reaction gave rise to a 328-bp fragment from the Rac1flox allele and a 172-bp fragment from the deleted Rac1− allele.
Fetal thymic organ culture
Fetal thymic lobes isolated from embryonic day 15.5 embryos were transferred onto nucleopore polycarbonate filters (Millipore, Watford, United Kingdom) and cultured at 37°C, in RPMI 1640 (Invtirogen, Paisley, United Kingdom), 10% FCS, 2 mM l-glutamine, 1 mM sodium pyruvate, penicillin, and streptomycin. After 5 days, the filters were transferred to fresh cultures containing 10 μM of the agonist NP68 peptide (ASNENMDAM) or a control gag peptide (SQVTNPANI) or no peptide at all and cultured for 11 hours. Thymocytes were harvested for analysis by gently disrupting the thymic lobes and analyzed by staining with annexin V (BD Biosciences), and antibodies to CD4 and CD8.
Quantitation of mRNA levels
RNA was isolated from sorted DN3 or DN4 thymocytes using the RNeasy Kit (QIAGEN, Crawley, United Kingdom), contaminating DNA was removed using the Ambion DNA-free Kit (Applied Biosystems, Warrington, United Kingdom), and cDNA was prepared using SuperScript II Reverse transcriptase (Invitrogen). To quantitate mRNA levels, the cDNA samples were analyzed in triplicate by Real-Time PCR using TaqMan Gene Expression Assays for the appropriate genes, on an ABI Prism 7000 Sequence Detector (Applied Biosystems). Transcript levels of targets were normalized to levels of Hprt1 mRNA.
Confocal microscopy
Thymic lobes were fixed in PBS, 3% paraformaldehyde for 1 hour, and embedded in 8% agarose, PBS. Vibratome sections (150 μm) were blocked in PBS, 0.15% Triton X-100, 2% FCS, 0.5% BSA and stained overnight with anti–GFP-Alexa 647 (Invitrogen), anti–pan-cytokeratin (Sigma-Aldrich), and MTS10 (BD Biosciences). Sections were washed in PBS, 0.15% Triton X-100, 2% FCS, 0.5% BSA and stained overnight with anti–mouse IgG1–Alexa 546 and anti–rat IgM–Alexa 488 (Invitrogen). After washing in PBS, 0.15% Triton X-100, sections were dehydrated in methanol and treated with benzyl benzoate/benzyl alcohol (1:1) before mounting on a slide using a Gene Frame coverslip (ABgene, Epsom, United Kingdom). Sections were analyzed on a Leica SP2 confocal microscope (Leica, Milton Keynes, United Kingdom) using sequential scanning and a 40×/1.0 NA oil objective.
Statistical analysis
All statistical comparisons were carried out using the nonparametric 2-tailed Mann-Whitney test.
Results
Complete block in T-cell development in the absence of Rac1 and Rac2
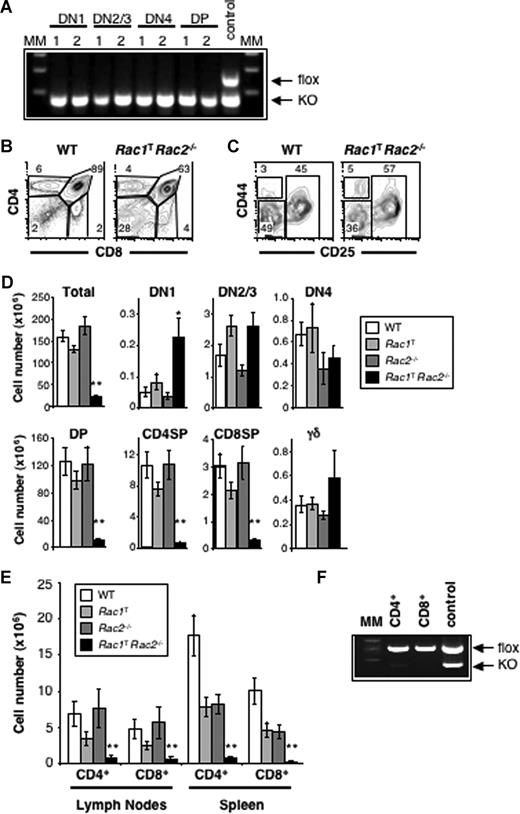
Measurement of mRNA levels showed that thymocytes express both Rac1 and Rac2, but no detectable Rac3 (not shown), thus in subsequent analyses we focused only on Rac1 and Rac2. Mice bearing a conditional loxP-flanked allele of Rac1 (Rac1flox)7 were crossed to transgenics expressing the CD2-Cre transgene, which expresses the Cre recombinase at early stages of T-cell development in the thymus,20 to eliminate expression of Rac1 in the T-cell lineage (Figure 1A). These Rac1flox/floxCD2-Cre (hereafter Rac1T) mice were also further crossed to a Rac2-deficient strain, and thymic T-cell development was assessed in mice deficient in both GTPases (Rac1TRac2−/−). Whereas absence of either Rac1 or Rac2 alone had no effect on thymic development, deficiency of both GTPases resulted in a very small thymus, with greatly reduced numbers of CD4+CD8+ double-positive (DP), CD4+CD8− single-positive (4SP), and CD4−CD8+ single-positive (8SP) thymocytes (Figure 1B,D). The number of CD4−CD8− double-negative (DN) thymocytes, the earliest thymic population, was largely unaffected, as was the number of cells in DN subsets 2 to 4 as separated by expression of CD25 and CD44 (Figure 1C,D). However there was an increase in the number of DN1 and TCRγδ+ thymocytes in double-deficient mice. As would be expected from the reduced number of 4SP and 8SP thymocytes, the number of CD4+ and CD8+ T cells in the spleen and lymph nodes was also greatly reduced in Rac1/Rac2-deficient mice (Figure 1E). Indeed, analysis of deletion of the Rac1 gene showed that these very few peripheral T cells had all failed to delete the gene, in contrast to the efficient deletion seen in DN and DP thymocytes, demonstrating that there is strong selection against loss of Rac1 (compare Figure 1A,F). These results show that there is redundancy of function between Rac1 and Rac2, and that there is a requirement for these GTPases in the developmental transition from the DN to DP thymic compartment and subsequent maturation to T cells.
Impaired T-cell development in the absence of Rac1 and Rac2. (A) Gel electrophoretic analysis showing PCR products diagnostic for the Rac1flox (flox) and Rac1− (KO) alleles amplified from DNA extracted from the indicated sorted thymocyte subsets (DN1, DN2/3, DN4, and DP) from 2 individual Rac1TRac2−/− mice (lanes 1,2) and, as a control for efficient amplification, from total thymocytes from a Rac1flox/−Rac2−/− (control) mouse that did not carry the hCD2-iCre transgene. (B) Flow cytometric analysis of CD4 and CD8 expression on thymocytes from Rac1+/+Rac2+/+hCD2-iCre (WT) and Rac1flox/floxRac2−/−hCD2-iCre (Rac1TRac2−/−) mice. Gates identify CD4−CD8− double-negative (DN), CD4+CD8+ double-positive (DP), and CD4+CD8− and CD4−CD8+ single-positive (CD4SP and CD8SP) cells. Numbers indicate percentage of cells falling into the gates. (C) Flow cytometric analysis of CD44 and CD25 expression on thymocytes from WT and Rac1TRac2−/− mice, which were Thy1+ but negative for expression of the lineage (Lin) markers (CD4, CD8, CD3ϵ, TCRβ, TCRγδ, B220, Gr1, Mac1, Dx5; not shown). Gates show DN1 (CD44+CD25−), DN2 and DN3 (DN2/3, CD44+/−CD25+), and DN4 (CD44−CD25−) populations. Numbers indicate percentage of cells falling into the gates. (D) Graphs showing mean (± SEM) total thymocyte cell numbers or numbers of cells in populations gated as in panels B and C. Also shown are the number of thymocytes expressing TCRγδ (γδ). Thymocytes were analyzed from WT, Rac1flox/floxRac2+/+hCD2-iCre (Rac1T), Rac1+/+Rac2−/−hCD2-iCre (Rac2−/−), and Rac1TRac2−/− mice. (E) Graph showing mean (± SEM) CD4+ or CD8+ T-cell numbers in the lymph nodes and spleen of mice of the indicated genotypes. (F) Gel electrophoretic analysis showing PCR products diagnostic for the Rac1flox (flox) and Rac1− (KO) alleles amplified from DNA extracted from CD4+ or CD8+ T cells isolated from the lymph nodes of Rac1TRac2−/− mice, or from tail DNA of a control Rac1flox/−Rac2−/− mouse that did not carry the hCD2-iCre transgene. MM indicates molecular weight markers. Statistically significant differences between WT and Rac1TRac2−/− mice are indicated (*P < .01; **P < .001).
Impaired T-cell development in the absence of Rac1 and Rac2. (A) Gel electrophoretic analysis showing PCR products diagnostic for the Rac1flox (flox) and Rac1− (KO) alleles amplified from DNA extracted from the indicated sorted thymocyte subsets (DN1, DN2/3, DN4, and DP) from 2 individual Rac1TRac2−/− mice (lanes 1,2) and, as a control for efficient amplification, from total thymocytes from a Rac1flox/−Rac2−/− (control) mouse that did not carry the hCD2-iCre transgene. (B) Flow cytometric analysis of CD4 and CD8 expression on thymocytes from Rac1+/+Rac2+/+hCD2-iCre (WT) and Rac1flox/floxRac2−/−hCD2-iCre (Rac1TRac2−/−) mice. Gates identify CD4−CD8− double-negative (DN), CD4+CD8+ double-positive (DP), and CD4+CD8− and CD4−CD8+ single-positive (CD4SP and CD8SP) cells. Numbers indicate percentage of cells falling into the gates. (C) Flow cytometric analysis of CD44 and CD25 expression on thymocytes from WT and Rac1TRac2−/− mice, which were Thy1+ but negative for expression of the lineage (Lin) markers (CD4, CD8, CD3ϵ, TCRβ, TCRγδ, B220, Gr1, Mac1, Dx5; not shown). Gates show DN1 (CD44+CD25−), DN2 and DN3 (DN2/3, CD44+/−CD25+), and DN4 (CD44−CD25−) populations. Numbers indicate percentage of cells falling into the gates. (D) Graphs showing mean (± SEM) total thymocyte cell numbers or numbers of cells in populations gated as in panels B and C. Also shown are the number of thymocytes expressing TCRγδ (γδ). Thymocytes were analyzed from WT, Rac1flox/floxRac2+/+hCD2-iCre (Rac1T), Rac1+/+Rac2−/−hCD2-iCre (Rac2−/−), and Rac1TRac2−/− mice. (E) Graph showing mean (± SEM) CD4+ or CD8+ T-cell numbers in the lymph nodes and spleen of mice of the indicated genotypes. (F) Gel electrophoretic analysis showing PCR products diagnostic for the Rac1flox (flox) and Rac1− (KO) alleles amplified from DNA extracted from CD4+ or CD8+ T cells isolated from the lymph nodes of Rac1TRac2−/− mice, or from tail DNA of a control Rac1flox/−Rac2−/− mouse that did not carry the hCD2-iCre transgene. MM indicates molecular weight markers. Statistically significant differences between WT and Rac1TRac2−/− mice are indicated (*P < .01; **P < .001).
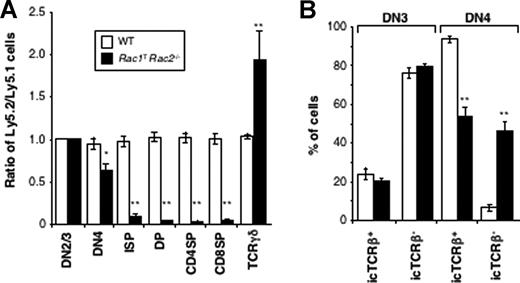
Although the conditional deletion of Rac1 is limited to the lymphoid lineages, the constitutive disruption of the Rac2 gene could affect all lineages. Thus the observed block in T-cell development in the double-deficient mice might have been due to a combined absence of Rac1 in the T-cell lineage, and of Rac2 in another cell type. To assess whether the requirement for both GTPases is cell autonomous, we generated competitive radiation chimeras reconstituted with bone marrow from CD2-Cre expressing wild-type (WT) or Rac1TRac2−/− mice mixed with allelelically marked (Ly5.1+) WT marrow. Analysis of these chimeras showed that T-lineage cells deficient in both Rac1 and Rac2 were also blocked in development at the DN-DP transition, resulting in very few DP, 4SP, or 8SP thymocytes or peripheral T cells, with, once again, somewhat elevated numbers of TCRγδ thymocytes (Figure 2A and data not shown). Thus the requirement for Rac1 and Rac2 in T-cell development is intrinsic to the T-cell lineage and cell autonomous.
Impaired thymic development and accumulation of aberrant TCRβ− DN4 thymocytes in the absence of Rac1 and Rac2 is intrinsic to the T-cell lineage. Mixed radiation chimeras were made using bone marrow from either Rac1+/+Rac2+/+hCD2-iCre (WT) or Rac1flox/floxRac2−/−hCD2-iCre (Rac1TRac2−/−) mice (both Ly5.2+) and bone marrow from B6.SJL mice (Ly5.1+). (A) Graph showing the mean (± SEM) ratio between Ly5.2+ and Ly5.1+ cells in each thymic developmental compartment, defined as in Figure 1, with the addition of intermediate single-positive (ISP) cells defined as CD4−CD8+TCRβ−. These are cells in transit between the DN4 and DP compartments. The ratios were all normalized to the ratio of Ly5.2+ to Ly5.1+ cells in the DN2/3 compartment, which was set to 1. (B) Graph showing mean (± SEM) percentage of cells that were either positive or negative for intracellular TCRβ (icTCRβ) in the Ly5.2+ DN3 or DN4 compartments of the mixed radiation chimeras. Colors indicate genotypes as in panel A. Statistically significant differences between Rac1TRac2−/− and WT chimeras are indicated (*P < .01; **P < .001).
Impaired thymic development and accumulation of aberrant TCRβ− DN4 thymocytes in the absence of Rac1 and Rac2 is intrinsic to the T-cell lineage. Mixed radiation chimeras were made using bone marrow from either Rac1+/+Rac2+/+hCD2-iCre (WT) or Rac1flox/floxRac2−/−hCD2-iCre (Rac1TRac2−/−) mice (both Ly5.2+) and bone marrow from B6.SJL mice (Ly5.1+). (A) Graph showing the mean (± SEM) ratio between Ly5.2+ and Ly5.1+ cells in each thymic developmental compartment, defined as in Figure 1, with the addition of intermediate single-positive (ISP) cells defined as CD4−CD8+TCRβ−. These are cells in transit between the DN4 and DP compartments. The ratios were all normalized to the ratio of Ly5.2+ to Ly5.1+ cells in the DN2/3 compartment, which was set to 1. (B) Graph showing mean (± SEM) percentage of cells that were either positive or negative for intracellular TCRβ (icTCRβ) in the Ly5.2+ DN3 or DN4 compartments of the mixed radiation chimeras. Colors indicate genotypes as in panel A. Statistically significant differences between Rac1TRac2−/− and WT chimeras are indicated (*P < .01; **P < .001).
Rac1 and Rac2 are required for positive selection of T cells
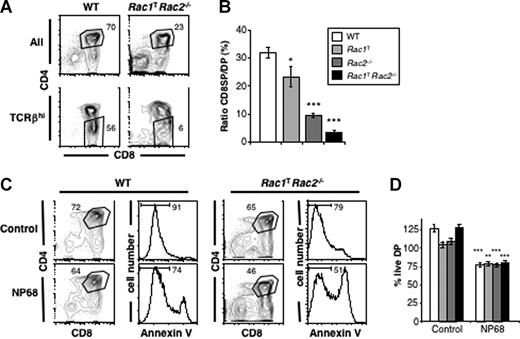
The transition from DP to 4SP and 8SP thymocytes is controlled by TCR signals that drive positive and negative selection.24 To evaluate the role of Rac1 and Rac2 in these processes in more detail, we crossed WT, Rac1T, Rac2−/−, or Rac1TRac2−/− mice to transgenics expressing the F5 TCR, an MHC class I–restricted receptor, which forces T-cell development exclusively into the CD8+ lineage.21 The mice were also deficient for Rag1 (F5Rag1−/−), to eliminate endogenous TCR gene rearrangement. Flow cytometric analysis showed that although absence of either Rac1 or Rac2 alone had no effect on positive selection of F5-expressing DP thymocytes into the 8SP lineage, deficiency in both GTPases resulted in almost no 8SP thymocytes or peripheral T cells (Figure 3A). To take account of the reduced number of DP thymocytes in double-deficient mice, we measured the ratio of 8SP to DP thymocytes in the 4 genotypes. This showed that although this ratio was around 32% in WT mice, it was reduced to 23%, 10%, and 3% in mice deficient in Rac1, Rac2, or both, respectively (Figure 3B), demonstrating a critical role for the GTPases in positive selection of cells expressing this class I–restricted TCR.
Rac1 and Rac2 are required for efficient positive, but not negative, selection in the thymus. (A) Flow cytometric analysis of CD4 and CD8 expression on thymocytes from F5Rag1−/− mice that were either Rac1+/+Rac2+/+hCD2-iCre (WT) or Rac1TRac2−/−. The top plots show all thymocytes, whereas the bottom plots show only cells expressing high levels of TCRβ. Gates indicate DP and CD8SP cells. Numbers indicate percentage of cells falling into the gates. (B) Graphs showing the mean (± SEM) ratio of CD8SP to DP cells in thymi from F5Rag1−/− mice of the indicated Rac1 and Rac2 genotypes. Statistically significant differences between WT and the 3 Rac mutant genotypes are indicated. (C) Flow cytometric analysis of CD4 and CD8 expression on cells from fetal thymic lobes from F5Rag1−/− mice that were either WT or Rac1TRac2−/−, cultured for 5 days, and then treated for 11 hours with a control peptide, or with NP68, an agonist peptide for the F5 TCR. Cell death was assessed by annexin V staining on DP cells, gated as shown in CD4/CD8 plots. Marker indicates live annexin V− cells. Numbers indicate percentage of cells in indicated gates or markers. (D) Graph showing mean (± SEM) percentage of live DP cells in fetal thymic organ cultures treated and analyzed as in panel C. Percentages were normalized to the number of live DP cells in WT F5Rag1−/− thymi cultured in medium alone (set to 100%). Shading of columns indicate Rac1 and Rac2 genotypes as in panel B. Statistically significant differences between cultures treated with NP68 or control peptides are indicated. No difference was seen in the response to NP68 between the 4 different genotypes. (*P < .05; **P < .01; ***P < .001.)
Rac1 and Rac2 are required for efficient positive, but not negative, selection in the thymus. (A) Flow cytometric analysis of CD4 and CD8 expression on thymocytes from F5Rag1−/− mice that were either Rac1+/+Rac2+/+hCD2-iCre (WT) or Rac1TRac2−/−. The top plots show all thymocytes, whereas the bottom plots show only cells expressing high levels of TCRβ. Gates indicate DP and CD8SP cells. Numbers indicate percentage of cells falling into the gates. (B) Graphs showing the mean (± SEM) ratio of CD8SP to DP cells in thymi from F5Rag1−/− mice of the indicated Rac1 and Rac2 genotypes. Statistically significant differences between WT and the 3 Rac mutant genotypes are indicated. (C) Flow cytometric analysis of CD4 and CD8 expression on cells from fetal thymic lobes from F5Rag1−/− mice that were either WT or Rac1TRac2−/−, cultured for 5 days, and then treated for 11 hours with a control peptide, or with NP68, an agonist peptide for the F5 TCR. Cell death was assessed by annexin V staining on DP cells, gated as shown in CD4/CD8 plots. Marker indicates live annexin V− cells. Numbers indicate percentage of cells in indicated gates or markers. (D) Graph showing mean (± SEM) percentage of live DP cells in fetal thymic organ cultures treated and analyzed as in panel C. Percentages were normalized to the number of live DP cells in WT F5Rag1−/− thymi cultured in medium alone (set to 100%). Shading of columns indicate Rac1 and Rac2 genotypes as in panel B. Statistically significant differences between cultures treated with NP68 or control peptides are indicated. No difference was seen in the response to NP68 between the 4 different genotypes. (*P < .05; **P < .01; ***P < .001.)
To evaluate the role of Rac1 and Rac2 in negative selection, we cultured fetal thymic lobes from F5Rag1−/− mice that were WT, Rac1T, Rac2−/−, or Rac1TRac2−/− in the presence of an agonist peptide for the F5 TCR. We found that this induced a similar amount of deletion irrespective of genotype, suggesting that Rac1 and Rac2 are not required for negative selection of DP thymocytes (Figure 3C,D).
Defective β-selection in Rac1- and Rac2-deficient thymocytes
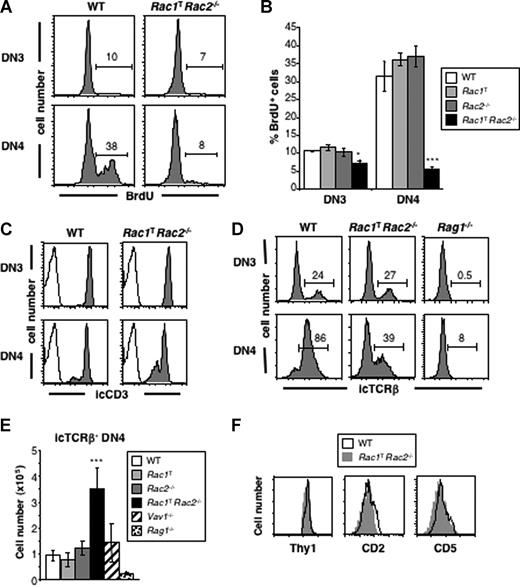
The developmental transition from the DN to DP stage of thymic development is dependent on successful rearrangement of the TCRβ genes, expression of TCRβ protein and its assembly with preTα and the CD3 complex into the pre-TCR. Signals from the pre-TCR at the DN3 stage of development result in cell proliferation and maturation into the DN4 and then DP compartments. The net result is a selective expansion of TCRβ-expressing cells and their maturation into DP thymocytes, a process referred to as β-selection. Because Rac1TRac2−/− mice are severely blocked at this transition, we investigated whether pre-TCR–driven β-selection may be defective in these mice. Measurement of proliferation showed that in WT, Rac1T, and Rac2−/− mice there was a low level of division in DN3 cells, which rose to approximately 30% in DN4 cells (Figure 4A,B). In contrast, in Rac1TRac2−/− mice there was a significant reduction in proliferation in the DN4 compartment, with no evident increase in proliferation from the DN3 to DN4 stages, suggesting that in the absence of Rac1 and Rac2, β-selection had failed.
Impaired pre-TCR–driven proliferation and accumulation of aberrant icTCRβ− DN4 thymocytes in the absence of Rac1 and Rac2. (A) Flow cytometric analysis for incorporation of BrdU into DN3 and DN4 thymocytes from WT or Rac1TRac2−/− mice injected 4 hours earlier with BrdU. Markers indicate cells positive for BrdU, and thus were dividing during the labeling period. Numbers indicate percentage of cells falling into the marker. (B) Graph showing mean (± SEM) percentage of BrdU+ DN3 or DN4 thymocytes from mice of the indicated genotypes, analyzed as in panel A. (C) Histograms showing expression of intracellular CD3 (icCD3) in DN3 and DN4 thymocytes from WT or Rac1TRac2−/− mice (shaded plots) or stained with an isotype control antibody (open plots). (D) Histograms showing expression of intracellular TCRβ (icTCRβ) on DN3 and DN4 thymocytes from mice of the indicated genotypes. Markers indicate cells positive for icTCRβ. Numbers indicate percentage of cells falling into the markers. (E) Mean (± SEM) number of icTCRβ− DN4 thymocytes from mice of the indicated genotypes. (F) Histograms showing expression of Thy1 on icTCRβ− DN4 thymocytes and CD2 and CD5 on DN4 thymocytes from mice of the indicated genotypes. Statistically significant differences between Rac1TRac2−/− and WT mice are indicated (*P < .005; ***P < .001).
Impaired pre-TCR–driven proliferation and accumulation of aberrant icTCRβ− DN4 thymocytes in the absence of Rac1 and Rac2. (A) Flow cytometric analysis for incorporation of BrdU into DN3 and DN4 thymocytes from WT or Rac1TRac2−/− mice injected 4 hours earlier with BrdU. Markers indicate cells positive for BrdU, and thus were dividing during the labeling period. Numbers indicate percentage of cells falling into the marker. (B) Graph showing mean (± SEM) percentage of BrdU+ DN3 or DN4 thymocytes from mice of the indicated genotypes, analyzed as in panel A. (C) Histograms showing expression of intracellular CD3 (icCD3) in DN3 and DN4 thymocytes from WT or Rac1TRac2−/− mice (shaded plots) or stained with an isotype control antibody (open plots). (D) Histograms showing expression of intracellular TCRβ (icTCRβ) on DN3 and DN4 thymocytes from mice of the indicated genotypes. Markers indicate cells positive for icTCRβ. Numbers indicate percentage of cells falling into the markers. (E) Mean (± SEM) number of icTCRβ− DN4 thymocytes from mice of the indicated genotypes. (F) Histograms showing expression of Thy1 on icTCRβ− DN4 thymocytes and CD2 and CD5 on DN4 thymocytes from mice of the indicated genotypes. Statistically significant differences between Rac1TRac2−/− and WT mice are indicated (*P < .005; ***P < .001).
Aberrant survival of intracellular TCRβ− DN4 thymocytes in the absence of Rac1 and Rac2
To investigate this defect further, we measured expression of the pre-TCR components. We found that although DN3 and DN4 cells from Rac1TRac2−/− mice express normal amounts of intracellular CD3 complex and normal or increased levels of preTα (Figures 4C, 7A), they had anomalous expression of intracellular TCRβ. Around 25% of DN3 cells from both WT and Rac1TRac2−/− mice express intracellular TCRβ (icTCRβ+), suggesting that TCRβ rearrangement proceeds normally in the absence of Rac1 and Rac2. However, whereas in the WT DN4 subset 85% to 90% of cells are icTCRβ+, reflecting successful β-selection, only 39% of Rac1TRac2−/− DN4 cells are icTCRβ+ (Figure 4D). This results in a significantly decreased number of icTCRβ+ DN4 cells and an increased number of icTCRβ− DN4 cells in Rac1TRac2−/− mice (Figure 4E, and data not shown). These cells appear to be genuine DN4 T-lineage cells because they express Thy1, CD2, and CD5 (Figure 4F). In addition, they are not γδ T-cell precursors as more than 75% did not express intracellular TCRγδ (not shown). Analysis of competitive radiation chimeras showed that this decrease in icTCRβ+ DN4 cells and accumulation of icTCRβ− DN4 cells was due to an intrinsic requirement for Rac1 and Rac2 in the T-lineage itself (Figure 2B). Taken together, these results show that Rac1 and Rac2 are required for efficient pre-TCR–driven proliferation and β-selection, and that in their absence there is an aberrant accumulation of icTCRβ− DN4 cells.
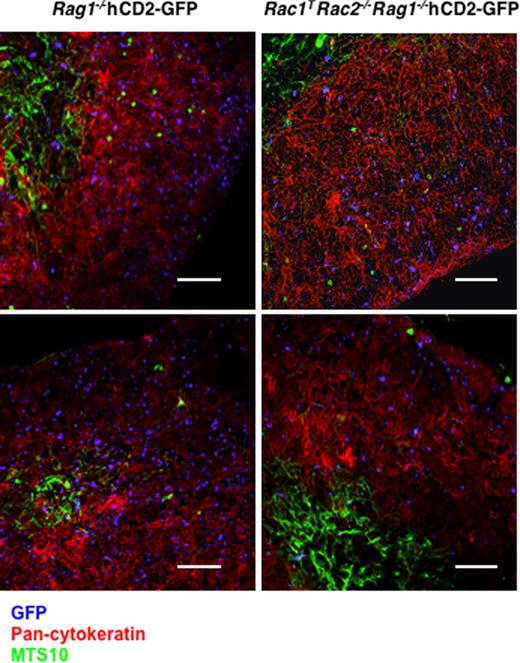
The Rac GTPases have been extensively documented to play critical roles in cell migration and adhesion.6 Thus we considered the possibility that the DN-DP developmental block in Rac1TRac2−/− mice, and the accumulation of icTCRβ− DN4 cells, may be caused by defects in migration. Hematopoietic progenitors arrive in the thymus at the corticomedullary junction, and then migrate to the outer zone of the cortex where β-selection allows DN3 cells to mature into DN4 and DP thymocytes.25 To evaluate the effect of Rac1 and Rac2 deficiency on this migratory program, we bred Rag1−/− and Rac1TRac2−/−Rag1−/− mice to hCD2-GFP transgenics, which express GFP from the earliest DN1 subset of the T lineage.20 In both of these strains development is completely arrested at the double-negative stage. To permit the evaluation of cell migration in the context of a normal thymic architecture, we generated mixed radiation chimeras reconstituted with bone marrow from Rag1−/−hCD2-GFP or Rac1TRac2−/−Rag1−/−hCD2-GFP mixed with wild-type marrow. Immunohistology showed that double-negative cells from both Rag1−/−hCD2-GFP or Rac1TRac2−/−Rag1−/−hCD2-GFP mice had a normal distribution throughout the thymic cortex, suggesting that, despite the absence of Rac1 and Rac2, migration is not impaired (Figure 5).
Absence of Rac1 and Rac2 does not impair localization of DN thymocytes. Fluorescence images of thymic sections from mixed radiation chimeras reconstituted with bone marrow from either Rac1+/+Rac2+/+Rag1−/−hCD2-iCre/hCD2-GFP (Rag1−/−hCD2-GFP) or Rac1flox/floxRac2−/−Rag1−/−hCD2-iCre/hCD2-GFP (Rac1TRac2−/−Rag1−/−hCD2-GFP) mice and bone marrow from C57BL/6 mice. Two images are shown for each genotype. Sections were stained with anti-GFP (blue) to identify the DN2/3 thymocytes from the hCD2-GFP–containing strains, anti–pan-cytokeratin (red) to recognize cortical epithelial cells, and MTS10 (green) to identify medullary epithelial cells. Note that GFP+ cells are distributed throughout the cortex in both strains. Bar indicates 145 μm.
Absence of Rac1 and Rac2 does not impair localization of DN thymocytes. Fluorescence images of thymic sections from mixed radiation chimeras reconstituted with bone marrow from either Rac1+/+Rac2+/+Rag1−/−hCD2-iCre/hCD2-GFP (Rag1−/−hCD2-GFP) or Rac1flox/floxRac2−/−Rag1−/−hCD2-iCre/hCD2-GFP (Rac1TRac2−/−Rag1−/−hCD2-GFP) mice and bone marrow from C57BL/6 mice. Two images are shown for each genotype. Sections were stained with anti-GFP (blue) to identify the DN2/3 thymocytes from the hCD2-GFP–containing strains, anti–pan-cytokeratin (red) to recognize cortical epithelial cells, and MTS10 (green) to identify medullary epithelial cells. Note that GFP+ cells are distributed throughout the cortex in both strains. Bar indicates 145 μm.
DN4 thymocytes that do not express TCRβ are usually eliminated by apoptosis.26 Interestingly, measurement of apoptosis showed that significantly fewer icTCRβ− DN4 cells in Rac1TRac2−/− mice were undergoing apoptosis (Figure 6A). Furthermore, these cells also had a significantly increased ratio of the antiapoptotic Bcl2 protein relative to the proapoptotic Bax protein (Figure 6B). Taken together these results suggest that the anomalous accumulation of these cells is caused by increased cell survival.
Increased survival of Rac1TRac2−/− icTCRβ− DN4 thymocytes. Analysis of DN3, icTCRβ+ DN4, and icTCRβ− DN4 thymocytes from mice of the indicated genotypes, showing (A) percentage of TUNEL+ cells as a measure of apoptosis, (B) ratio of Bcl2/Bax expression, and (C) surface levels of IL7Rα as judged by mean fluorescence intensity (MFI) of antibody staining. Statistically significant differences between Rac1TRac2−/− and WT mice are indicated (*P < .005; **P < .001).
Increased survival of Rac1TRac2−/− icTCRβ− DN4 thymocytes. Analysis of DN3, icTCRβ+ DN4, and icTCRβ− DN4 thymocytes from mice of the indicated genotypes, showing (A) percentage of TUNEL+ cells as a measure of apoptosis, (B) ratio of Bcl2/Bax expression, and (C) surface levels of IL7Rα as judged by mean fluorescence intensity (MFI) of antibody staining. Statistically significant differences between Rac1TRac2−/− and WT mice are indicated (*P < .005; **P < .001).
IL7 and Notch signaling in thymocytes deficient in Rac1 and Rac2
Cell survival at the DN stage of thymic development is under the control of IL7 receptor and Notch receptor signaling.1 As thymocytes transit from the DN3 to DN4 stage, expression of IL7 receptor α chain (IL7Rα) declines.27 Although this decrease was seen in wild-type, Rac1T, and Rac2−/− mice, icTCRβ− DN4 cells in Rac1TRac2−/− mice expressed elevated surface levels of IL7Rα, similar to those seen in DN3 thymocytes (Figure 6C). This result is consistent with the possibility that increased IL7 signaling may contribute to the increased cell survival. We note that IL7 signaling has been shown to lead to increased levels of Bcl2,28 potentially explaining the higher Bcl2/Bax ratio seen in Rac1TRac2−/− icTCRβ− DN4 cells.
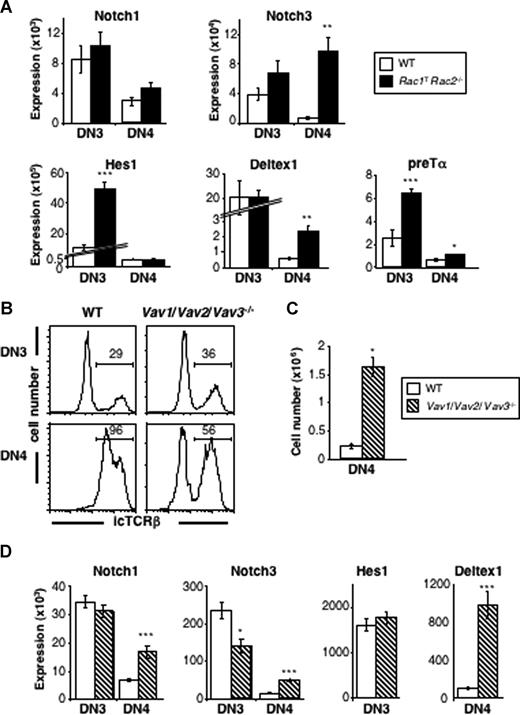
To evaluate Notch signaling in Rac1TRac2−/− thymocytes, we measured levels of mRNA for genes whose expression is known to be activated by Notch signaling. Although levels of mRNA for Notch1 itself were unaltered, we found significantly increased levels of mRNA for Notch3, Hes1, Deltex1, and preTα in DN3 and/or DN4 Rac1TRac2−/− thymocytes (Figure 7A). All 4 of these latter genes have been reported to be targets of Notch signaling,29,30 suggesting that there is hyperactive Notch signaling in Rac1TRac2−/− thymocytes. Because the IL7Rα gene has been proposed to be a Notch target,31 the elevated levels of IL7Rα may also reflect increased Notch signaling.
Increased Notch-regulated transcripts in the absence of Rac1 and Rac2 and Vav-family GEFs. (A) Graphs show mean (± SEM) mRNA expression of Notch1, Notch3, Hes1, Deltex1, and preTα in DN3 and DN4 thymocytes from WT and Rac1TRac2−/− mice. Expression of targets is relative to Hprt1 expression in the same samples. (B) Histograms showing expression of intracellular TCRβ (icTCRβ) in DN3 and DN4 thymocytes from WT or Vav1/Vav2/Vav3−/− mice. Markers indicate cells positive for icTCRβ. Numbers indicate percentage of cells falling into the markers. (C) Graph showing mean (± SEM) number of icTCRβ− DN4 thymocytes in mice of the indicated genotypes. (D) Graphs show mean (± SEM) mRNA expression of Notch1, Notch3, Hes1, and Deltex1 in DN3 and DN4 thymocytes from WT and Vav1/Vav2/Vav3−/− mice. Colors of bars as in panel C. Expression of targets is relative to Hprt1 expression in the same samples. Statistically significant differences between Rac1TRac2−/− or Vav1/Vav2/Vav3−/− mice and WT mice are indicated (*P < .005; **P < .0005; ***P < .0001).
Increased Notch-regulated transcripts in the absence of Rac1 and Rac2 and Vav-family GEFs. (A) Graphs show mean (± SEM) mRNA expression of Notch1, Notch3, Hes1, Deltex1, and preTα in DN3 and DN4 thymocytes from WT and Rac1TRac2−/− mice. Expression of targets is relative to Hprt1 expression in the same samples. (B) Histograms showing expression of intracellular TCRβ (icTCRβ) in DN3 and DN4 thymocytes from WT or Vav1/Vav2/Vav3−/− mice. Markers indicate cells positive for icTCRβ. Numbers indicate percentage of cells falling into the markers. (C) Graph showing mean (± SEM) number of icTCRβ− DN4 thymocytes in mice of the indicated genotypes. (D) Graphs show mean (± SEM) mRNA expression of Notch1, Notch3, Hes1, and Deltex1 in DN3 and DN4 thymocytes from WT and Vav1/Vav2/Vav3−/− mice. Colors of bars as in panel C. Expression of targets is relative to Hprt1 expression in the same samples. Statistically significant differences between Rac1TRac2−/− or Vav1/Vav2/Vav3−/− mice and WT mice are indicated (*P < .005; **P < .0005; ***P < .0001).
Vav-family GEFs may inhibit Notch signaling in the thymus
Previous studies had shown that deficiency of Vav-family proteins, GEFs for Rac GTPases, also causes developmental arrest at the pre-TCR checkpoint.3,4 In view of this, we investigated whether the absence of Vav proteins could also lead to an aberrant accumulation of icTCRβ− DN4 thymocytes and to an up-regulation of Notch target genes. We found that whereas in Vav1−/− mice there was no change in the numbers of icTCRβ− DN4 thymocytes (Figure 4E), in mice deficient in all 3 Vav proteins (Vav1−/−Vav2−/−Vav3−/−) there were significantly more icTCRβ− DN4 cells (Figure 7B,C). Furthermore, these Vav-deficient thymocytes showed increased levels of mRNA for the Notch targets Notch3 and Deltex1 though not Hes1, suggesting that they also may have hyperactive Notch signaling (Figure 7D). Taken together, these results show that absence of the Vav-family GEFs for Rac GTPases results in a phenotype similar to that seen in the absence of Rac1 and Rac2—a block at the pre-TCR checkpoint, accumulation of icTCRβ− DN4 thymocytes and apparent increased Notch signaling.
Discussion
Our work has shown that Rac1 and Rac2 are critical for the efficient transition of thymocytes from the DN to DP compartments, reflecting a key role in β-selection. Furthermore, Rac1 and Rac2 were required for positive selection of a class I–restricted TCR into the CD8+ lineage. Although we have not tested a class II–restricted TCR in the same assay, it seems likely that positive selection of the CD4+ lineage is also affected, as judged by the block in 4SP development. Mutation of the Vav-family of Rac-specific GEFs results in similar blocks in β-selection and positive selection,2 which have been ascribed to critical roles for Vav proteins in transducing pre-TCR and TCR signals, respectively. Thus it is likely that the phenotypes seen in the absence of Rac1 and Rac2 reflect important signal transducing functions for these GTPases downstream of the same receptors. Studies of Vav1-deficient T cells have shown that Vav1 plays an important signaling function proximal to the TCR, and is required for TCR-induced calcium flux, ERK activation, PI3 kinase activation, and inside-out activation of the integrin LFA-1.4,32-38 Thus in the absence of Rac1 and Rac2, similar pathways may be perturbed. However it remains unknown which of these pathways is dependent on the GEF activity of Vav1, since it has been suggested that Vav1 may also have GEF-independent activities, for example as an adapter protein.39,40
A recent study, published during the preparation of this paper, used a Cre transgene under the control of the Lck promoter (Lck-Cre) to delete the Rac1 gene.41 In agreement with our findings, the authors of this study found that deletion of Rac1 alone had no discernible effect on T-cell development or on the number of peripheral T cells. Furthermore, similarly to our results, this study showed reduced numbers of SP thymocytes and peripheral T cells in mice deficient in both Rac1 and Rac2, and concluded that the 2 GTPases play redundant roles in the positive selection of DP thymocytes. However, in contrast to our own work, the reported block in positive selection was not as strong, and this study did not find a block at the pre-TCR checkpoint. These differences are most likely due to incomplete deletion of Rac1 by the Lck-Cre transgene, because we found very variable efficiency of Rac1 deletion by this transgene (not shown). In the studies reported here, we have used the CD2-Cre transgene, which caused much more complete and earlier deletion of Rac1 (Figure 1), and hence resulted in a strong block at both the pre-TCR and TCR checkpoints.
An unexpected result from our studies was the accumulation of icTCRβ− DN4 thymocytes in the absence of Rac1 and Rac2. These cells accumulated most likely because of increased cell survival, as judged by decreased TUNEL staining and increased Bcl2/Bax ratio. This survival may, in turn be caused by increased IL7R or Notch signaling. Because the IL7Rα gene may be a target of Notch signaling,31 it is possible that increased Notch signaling results in more IL7Rα expression that leads to part of the increased cell survival. Consistent with this, forced expression of the constitutively active intracellular domain of Notch3 leads to aberrant survival of immature thymocytes, and, ultimately, to lymphoma.42 Intriguingly, an accumulation of icTCRβ− DN4 thymocytes has also been described in mice with a conditional thymic deletion of Notch1 or the RBP-J transcription factor that mediates Notch signaling,43,44 suggesting that both gain and loss of Notch signaling can promote aberrant DN4 maturation.
The presence of a similar accumulation of icTCRβ− DN4 thymocytes in Vav-deficient mice suggests that there may be a Vav/Rac pathway, which negatively regulates Notch signaling. We note that the pre-TCR controlled transition of DN3 into DN4 cells coincides with a large drop in expression of Notch targets and presumably Notch signaling (Figure 7A),45 consistent with the possibility that pre-TCR signaling represses Notch function. Thus, one possible explanation of our results is that the Rac and Vav proteins transduce pre-TCR signals that not only contribute to β-selection, but are also required to inhibit Notch signaling. An accumulation of icTCRβ− DN4 thymocytes has also been reported in mice deficient in the Erk1 and Erk2 MAP kinases.46 In this case, it is not known whether the phenotype is accompanied by increased Notch signaling, as this was not examined. Because Vav1 transduces TCR signals to the activation of the Erk kinases,36 it is possible that Vav proteins also activate Erk proteins downstream of the pre-TCR. Taking these observations together, we hypothesize that pre-TCR signals transduced by Vav and Rac proteins via the Erk kinases are required to inhibit Notch signaling in double-negative thymocytes.
Interestingly, observations on the regulation of Notch by Vav- and Rho-family proteins have previously been made in 2 very different systems. In Drosophila melanogaster, Drosophila Cdc42, a Rho-family GTPase, was shown to be a negative regulator of Notch signaling during wing growth and development.47 Secondly, during the specification of the secondary vulval precursor fate in Caenorhabditis elegans, down-regulation of Vav1 results in increased LIN-12/Notch signaling.48 Our data now extend these observations to a mammalian system, where we find that both Vav and Rac proteins may negatively regulate Notch signaling during early thymic development. The mechanism of this broadly conserved negative regulation of Notch by Vav and Rac will be an interesting area for future study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Alexander Saveliev, Robert Henderson, Edina Schweighoffer, Farnaz Fallah-Arani, Lesley Vanes, and Owen Williams for help and advice; Ben Seddon and Steve Ley for critical reading of the paper; Peter Fletcher for peptide synthesis; David Baltimore for Rag1−/− mice; and Iva Greenwald for helpful discussions.
This work was supported by the Medical Research Council (United Kingdom).
Authorship
Contribution: C.D. designed and performed research, analyzed and interpreted data, performed statistical analysis, and wrote the paper; A.C.-T. and S.R. performed research and analyzed and interpreted data; J.d.B., A.W., M.T., and D.K. contributed vital new reagents; and V.L.J.T. designed research, analyzed and interpreted data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Victor L. J. Tybulewicz, National Institute for Medical Research, The Ridgeway, Mill Hill, London, NW7 1AA, United Kingdom; e-mail: vtybule@nimr.mrc.ac.uk.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal