Abstract
There exists a unique group of persons who are able to durably control HIV in the absence of therapy. The mechanisms of control in these persons remain poorly defined. In this study, we examined CD8+ T-cell responses in blood and rectal mucosa from 17 “elite controllers” (viral load < 75 copies/mL), 11 “viremic controllers” (75-2000 copies/mL), 14 noncontrollers (> 10 000 copies/mL), and 10 antiretroviral-treated persons (< 75 copies/mL). Production of interferon-γ, interleukin-2, tumor necrosis factor-α, macrophage inflammatory protein-1β, and CD107a by CD8+ T cells in response to HIV-1 Gag stimulation was measured using flow cytometry. Our hypothesis was that “polyfunctional” T cells producing multiple antiviral factors would be most abundant in mucosal tissues of HIV controllers. Mucosal CD8+ T-cell responses were significantly stronger and more complex in controllers than in antiretroviral-suppressed persons (P = .0004). The frequency of 4-function responses in rectal mucosa was higher in controllers than in noncontrollers and patients on therapy (P < .0001). Mucosal responses in controllers were frequently stronger and more complex than blood responses. These findings demonstrate that many controllers mount strong, complex HIV-specific T-cell responses in rectal mucosa. These responses may play an important role in mucosal immune surveillance, as suggested by their relative enrichment among persons who control HIV in the absence of therapy.
Introduction
There are approximately 33 million people infected with HIV worldwide.1 Many believe that the only way to effectively prevent the ongoing spread of HIV is to develop a protective vaccine. Consequently, it has become increasingly important to understand how HIV interacts with its host and under what conditions the immune system is able to effectively control viral replication and limit disease progression.
Crucial to understanding immune control over viral replication in humans is the study of those rare persons who remain healthy in the absence of antiretroviral therapy. Initial studies focused on persons who maintained normal CD4+ T-cell counts for more than 10 years (long-term nonprogressors [LTNPs])2 ; such persons comprise between 5% and 15% of the chronically infected population. Plasma HIV RNA levels were often low in these persons, but there were exceptions. Because many factors may prevent disease progression, we and others have increasingly defined nonprogressors based on plasma HIV RNA levels. Two distinct groups have been identified: those who maintain undetectable plasma HIV RNA levels (“elite controllers”) and those who have persistently detectable but low plasma HIV RNA levels (“viremic controllers”). Elite controllers represent less than 1% of the HIV-infected population.2
Cytotoxic T lymphocytes (CTLs) are important in the initial decline of peak viremia during acute HIV infection and the determination of viral set point during chronic infection, a measure that often predicts progression rates to AIDS.3-5 Despite a strong initial response from HIV-specific CTLs during primary infection, these cells are unable to completely clear the virus. As chronic infection ensues, HIV-specific CTLs begin to lose function.6-8 However, several studies have shown that persons with better control over viral replication retain circulating HIV-specific CTLs with higher capacities for cytokine production,9-13 perforin expression,14 and proliferation.14,15 CTLs from controllers also appear to have an altered activation phenotype.16 Thus, immunologic features of circulating HIV-specific CTLs may contribute to the apparent control of viral replication.
Several host genetic factors have been associated with viral control and protection against disease progression. Most frequently cited is the enrichment of certain class I human leukocyte antigen (HLA) alleles, in particular HLA-B27 and B57, among HIV controllers.17-19 Much less is known about the potential role of class II HLA molecules in HIV infection; however, a few reports have suggested an association of HLA-DRB1*13 and /or DQB1*06 with the LTNP phenotype.20-22
Cell-mediated immune responses are typically studied in peripheral blood because of the ease of obtaining samples from this compartment. However, much of the virus, and presumably much of the host response, resides in lymphoid and mucosal tissues. HIV replicates to substantial levels in the gut,23,24 where a large proportion of the body's lymphocytes reside,25 particularly activated memory CD4+ T cells expressing CXCR4 and CCR5.26,27 These cells are highly susceptible to HIV infection and replication. Consequently, CD4+ T cells in this compartment are depleted,28-30 and there is rapid emergence of mucosal CTLs.27,31 Because of the differing microenvironments in peripheral blood and gut,32 it is important to understand the differences in immune pressures imposed on HIV in these compartments. To date, very few studies have addressed the role of mucosal T-cell function in HIV infection, and there have been no comprehensive studies of mucosal T-cell function in a large, well-defined group of HIV controllers.
To better understand how immune responses in the gastrointestinal tract relate to viral replication and disease progression, we analyzed HIV-specific T-cell function in paired peripheral blood and rectal biopsy samples from a well-characterized cohort of elite and viremic controllers and compared them with noncontrollers and patients on highly active antiretroviral therapy (HAART). Although these correlative studies cannot definitively establish a causal role for mucosal CD8+ T cells in protection from disease progression, our results do reveal that strong, polyfunctional CD8+ T-cell responses in gut mucosa are associated with the ability of some persons to control viral replication and delay disease progression in the absence of antiretroviral therapy. Accordingly, mucosal T-cell responses may be one of several critical immunologic, virologic, and genetic factors that contribute to the “HIV controller” phenotype.
Methods
Subjects and tissue collection
Most subjects were enrolled through an ongoing study of chronic HIV infection, based at San Francisco General Hospital. Some subjects were recruited through the Center for AIDS Research, Education and Services Clinic (Sacramento, CA). Five unique groups were recruited. Elite controllers were defined as antiretroviral untreated persons with plasma HIV RNA less than 75 copies/mL on at least 3 occasions. Viremic controllers were defined as antiretroviral untreated persons with plasma HIV RNA between 75 and 2000 copies/mL on at least 3 occasions. Noncontrollers had viral loads consistently more than 10 000 copies/mL in the absence of antiretroviral therapy, and the HAART-suppressed group included persons with plasma viremia artificially suppressed below the limit of detection (< 75 copies/mL) by HAART. HIV-uninfected controls were also included. Written informed consent for phlebotomy and flexible sigmoidoscopy was obtained from all persons in accordance with the Declaration of Helsinki, and with study protocols approved by the Institutional Review Board, University of California–Davis, and the Committee on Human Subjects Research, University of California–San Francisco.
Approximately 20 mL of blood was collected by sterile venipuncture into tubes containing ethylenediaminetetraacetic acid. Blood was processed on the day of collection. Rectal biopsy tissue was obtained at approximately 10 cm from the anal verge by flexible sigmoidoscopy.33 This approach is well documented, involves minimal discomfort, and provides sufficient lymphoid cells for cellular immunology assays.27,33-35 The sigmoidoscope was equipped with a biopsy channel, and tissues were procured with single-use biopsy forceps (Radial Jaw 3; Boston Scientific, Natick, MA). At each procedure, 20 to 25 tissue pieces were collected and placed in RPMI 1640 medium supplemented with 15% fetal calf serum, penicillin (100 U/mL), streptomycin (100 μg/mL), and l-glutamine (2 mM). This medium is hereafter referred to as R15. Specimens were immediately transported to the laboratory at the University of California–Davis for processing and analysis.
Peripheral blood and rectal biopsy tissue processing
Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Hypaque (Pfizer, New York, NY) density gradient centrifugation, washed in phosphate-buffered saline, and allowed to rest overnight in R15. Rectal mononuclear cells (RMCs) were isolated from biopsy specimens following a published protocol that was optimized for high yield and viability of mucosal lymphocytes without compromising the detection of most surface antigens.33,36 Briefly, biopsy pieces underwent 3 rounds of digestion in 0.5 mg/mL collagenase type II (Sigma-Aldrich, St Louis, MO). Each digestion was followed by disruption of the tissue using a syringe with a 16-gauge blunt end needle and subsequent passage through a 70-μm cell strainer. Strained and washed cells were pooled and centrifuged on a 35%/65% Percoll gradient (Sigma-Aldrich). RMCs were collected from both interfaces to maximize cell yield. RMCs were allowed to rest overnight in R15 containing 0.5 mg/mL piperacillin-tazobactam (Zosyn; Wyeth-Ayerst, Princeton, NJ) to discourage bacterial growth. Yields ranged from 4 to 21 × 106 RMCs from 20 to 25 biopsy pieces (mean, 10 × 106 RMCs).
HLA class I and II typing
DNA was isolated from approximately 10 × 106 PBMCs using the QIAamp DNA blood mini kit (QIAGEN, Valencia, CA) and quantified on a spectrophotometer. Typing was performed by polymerase chain reaction with sequence-specific primers (SSP-PCR) using commercial kits (Invitrogen, Carlsbad, CA). Low-resolution HLA-A, -B, and -C typing was performed with ABC SSP Unitray kits; low-resolution HLA-DR, DQ typing was performed with DR/DQ 2T Locus SSP Unitray kits; and high-resolution HLA-A and B typing was performed using direct to high-resolution SSP-PCR kits. PCR products were resolved by electrophoresis on a 2% agarose gel, photographed, and the patterns were analyzed with UniMatch Plus software (Invitrogen).
Antibodies and peptide pools
Fluorochrome-labeled monoclonal antibodies to CD3 (clone UCHT-1), CD107a (clone H4A3), interferon-γ (IFN-γ; clone B27), macrophage inflammatory protein-1β (MIP-1β; clone D21-1351), tumor necrosis factor-α (TNF-α; clone MAb11), interleukin-2 (IL-2; clone 5344.111), and unlabeled antibodies to CD28 (clone L293) and CD49d (clone L25) were purchased from BD Biosciences (San Jose, CA). Fluorochrome-labeled antibodies to CD4 (clone SFCI12T4D11) were purchased from Beckman Coulter (Fullerton, CA) and CD8 (clone SK1) from Invitrogen. All antibodies were titrated to determine optimum concentrations for assay conditions (data not shown). HIV Gag (p55, HXB2 sequence) peptide pools containing 15-mer peptides overlapping by 11 amino acids were purchased from BD Biosciences.
Intracellular cytokine staining and flow cytometry
Intracellular cytokine staining was performed using previously published protocols with minor modifications.10,36,37 All intracellular cytokine staining assays were performed on freshly isolated PBMCs and RMCs that had been rested overnight at 37°C and 5% CO2. Cells treated with HIV Gag peptide pool (3.5 μg/mL) served to measure the HIV-specific response; dimethyl sulfoxide (peptide vehicle) served as a negative control, and Staphylococcus enterotoxin B (5 μg/mL) served as a positive control. During the 5-hour stimulation, which included antibodies to CD28 and CD49d for costimulation, cells were stained for CD107a. Cells were then stained with antibodies to surface markers, CD4 and CD8, as well as 7-amino actinomycin D (7-AAD; BD Biosciences) for dead cell discrimination.38,39 Nonfluorescent actinomycin D (2 μg/mL) was included in reagents after the 7-AAD staining step to saturate sites that could bind residual 7-AAD. Cells were fixed in 4% paraformaldehyde and permeablized using FACS Perm 2 (BD Biosciences). This was followed by intracellular staining for CD3, IFN-γ, IL-2, TNF-α, and MIP-1β. Within 24 hours, samples were read on an LSRII flow cytometer using FACSDiva software (BD Biosciences).
Data analysis
Flow cytometric data were analyzed with FlowJo software (TreeStar, Ashland, OR). Boolean gate analysis was applied to separate cells into functional categories. Data were then evaluated and graphed in SPICE software (version 4.1.5; Mario Roederer, Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD). Bivariate plots were also constructed to better visualize raw data and confirm the placement of Boolean gates.
Statistical analysis
To analyze response data, we first determined whether antigen-specific responses differed significantly from the negative control using a previously described formula that assumes a Poisson distribution for both conditions and takes into account the actual numbers of gated events rather than the percentages within the gates.37 Net antigen-specific responses were then calculated by subtracting the negative control values from antigen-specific responses. This step was important, particularly for analyzing mucosal cells, because mucosal lymphocyte yield is generally small and these partially activated cells have been shown to produce low levels of cytokines in the absence of antigen-specific stimulation.32,36
Comparisons between 2 groups (tissues or patient groups) were done using a 2-tailed Mann-Whitney test in GraphPad Prism software, version 5 (GraphPad Software, San Diego, CA). Correlations between variables were done using the Spearman correlation, and linear regression was used to graph a best-fit line to the data (GraphPad Prism). Statistical analysis of Boolean-gated data (polyfunctional responses) was performed in SPICE software using the Wilcoxon rank test to compare individual polyfunctional response categories at a significance level of P less than .01 and a permutation test, based on χ2 statistics, to compare pie charts.
Results
Baseline characteristics
This study included 52 HIV-infected subjects and 8 uninfected, healthy controls. Each of the 52 HIV-infected subjects was assigned to 1 of 4 categories based on viral load and treatment status: elite controllers (no antiretroviral therapy, plasma HIV RNA < 75 copies/mL, n = 17), viremic controllers (no antiretroviral therapy, plasma HIV RNA 75-2000 copies/mL, n = 11), noncontrollers (no antiretroviral therapy, plasma HIV RNA > 10 000 copies/mL, n = 14), and HAART-suppressed (on treatment, plasma HIV RNA < 75 copies/mL, n = 10). The median blood CD4+ T-cell counts at the time of the analysis were 765, 475, 510, 488, and 692 for the elite controllers, viremic controllers, noncontrollers, HAART-suppressed subjects, and HIV-uninfected controls, respectively (Table 1).
Patient characteristics
| Patient ID . | Sex . | Race . | Plasma viral load, RNA copies/mL . | CD4 count, cells/mm3 . |
|---|---|---|---|---|
| Elite controllers | ||||
| 85 | M | W | < 75 | 883 |
| 1099 | M | B | < 75 | 139 |
| 1122 | M | W | < 75 | 884 |
| 1125 | F | B | < 75 | 765 |
| 1139 | M | NAm | < 75 | 536 |
| 1155 | M | B | < 75 | 225 |
| 1161 | M | W | < 75 | 912 |
| 1170 | M | As | < 75 | 535 |
| 1198 | M | W | < 75 | 1208 |
| 1203 | M | Mixed race | < 75 | 575 |
| 1511 | F | B | < 75 | 455 |
| 1517 | M | W | < 75 | 1537 |
| 1518 | T | B | < 75 | 1224 |
| 1529 | M | W | < 75 | 727 |
| 1545 | M | B | < 75 | 725 |
| 1552 | T | B | < 75 | 894 |
| 1555 | F | B | < 75 | 1115 |
| Group median | < 75 | 765 | ||
| Viremic controllers | ||||
| 50 | F | B | 140 | 619 |
| 1116 | M | W | 94 | 381 |
| 1119 | F | W | 209 | 1131 |
| 1127 | F | W | 1499 | 555 |
| 1128 | M | B | 546 | 398 |
| 1143 | M | W | 104 | 533 |
| 1148 | M | NAm | 426 | 389 |
| 1164 | M | Mixed race | 311 | 475 |
| 1181 | F | W | 2451 | 420 |
| 1516 | M | B | 104 | 657 |
| 4008 | M | W | 260 | 404 |
| Group median | 260 | 475 | ||
| Noncontrollers | ||||
| 56 | M | W | 38 441 | 304 |
| 58 | M | W | 44 956 | 538 |
| 65 | M | W | 31 975 | 719 |
| 66 | M | W | 42 049 | 481 |
| 67 | M | W | 13 582 | 715 |
| 98 | F | W | 34 882 | 608 |
| 118 | M | W | 36 286 | 289 |
| 1141 | M | H | 19 997 | 1123 |
| 1237 | M | W | > 50 118 | 564 |
| 1254 | F | B | > 50 118 | 242 |
| 1259 | M | B | > 50 118 | 275 |
| 1265 | F | Mixed race | 20 420 | 692 |
| 1275 | F | NAm | 23 205 | 347 |
| 1562 | T | B | 1108-25 115* | 474 |
| Group median | 35 584 | 510 | ||
| HAART-suppressed | ||||
| 54 | T | W | < 75 | 512 |
| 57 | M | W | < 75 | 442 |
| 63 | F | W | < 75 | 464 |
| 64 | M | W | < 75 | 851 |
| 77 | M | B | < 75 | 408 |
| 80 | M | W | < 75 | 927 |
| 83 | M | B | < 75 | 287 |
| 88 | M | W | < 75 | 316 |
| 92 | M | W | < 75 | 545 |
| 93 | F | W | < 75 | 809 |
| Group median | < 75 | 488 | ||
| Seronegatives | ||||
| 27 | M | B | < 75 | 837 |
| 74 | M | B | < 75 | 322 |
| 86 | F | B | < 75 | 670 |
| 87 | F | W | < 75 | 742 |
| 91 | F | W | < 75 | 714 |
| 94 | F | As | < 75 | 228 |
| 96 | M | B | < 75 | 1396 |
| 100 | M | Mixed race | < 75 | 609 |
| Group median | < 75 | 692 |
| Patient ID . | Sex . | Race . | Plasma viral load, RNA copies/mL . | CD4 count, cells/mm3 . |
|---|---|---|---|---|
| Elite controllers | ||||
| 85 | M | W | < 75 | 883 |
| 1099 | M | B | < 75 | 139 |
| 1122 | M | W | < 75 | 884 |
| 1125 | F | B | < 75 | 765 |
| 1139 | M | NAm | < 75 | 536 |
| 1155 | M | B | < 75 | 225 |
| 1161 | M | W | < 75 | 912 |
| 1170 | M | As | < 75 | 535 |
| 1198 | M | W | < 75 | 1208 |
| 1203 | M | Mixed race | < 75 | 575 |
| 1511 | F | B | < 75 | 455 |
| 1517 | M | W | < 75 | 1537 |
| 1518 | T | B | < 75 | 1224 |
| 1529 | M | W | < 75 | 727 |
| 1545 | M | B | < 75 | 725 |
| 1552 | T | B | < 75 | 894 |
| 1555 | F | B | < 75 | 1115 |
| Group median | < 75 | 765 | ||
| Viremic controllers | ||||
| 50 | F | B | 140 | 619 |
| 1116 | M | W | 94 | 381 |
| 1119 | F | W | 209 | 1131 |
| 1127 | F | W | 1499 | 555 |
| 1128 | M | B | 546 | 398 |
| 1143 | M | W | 104 | 533 |
| 1148 | M | NAm | 426 | 389 |
| 1164 | M | Mixed race | 311 | 475 |
| 1181 | F | W | 2451 | 420 |
| 1516 | M | B | 104 | 657 |
| 4008 | M | W | 260 | 404 |
| Group median | 260 | 475 | ||
| Noncontrollers | ||||
| 56 | M | W | 38 441 | 304 |
| 58 | M | W | 44 956 | 538 |
| 65 | M | W | 31 975 | 719 |
| 66 | M | W | 42 049 | 481 |
| 67 | M | W | 13 582 | 715 |
| 98 | F | W | 34 882 | 608 |
| 118 | M | W | 36 286 | 289 |
| 1141 | M | H | 19 997 | 1123 |
| 1237 | M | W | > 50 118 | 564 |
| 1254 | F | B | > 50 118 | 242 |
| 1259 | M | B | > 50 118 | 275 |
| 1265 | F | Mixed race | 20 420 | 692 |
| 1275 | F | NAm | 23 205 | 347 |
| 1562 | T | B | 1108-25 115* | 474 |
| Group median | 35 584 | 510 | ||
| HAART-suppressed | ||||
| 54 | T | W | < 75 | 512 |
| 57 | M | W | < 75 | 442 |
| 63 | F | W | < 75 | 464 |
| 64 | M | W | < 75 | 851 |
| 77 | M | B | < 75 | 408 |
| 80 | M | W | < 75 | 927 |
| 83 | M | B | < 75 | 287 |
| 88 | M | W | < 75 | 316 |
| 92 | M | W | < 75 | 545 |
| 93 | F | W | < 75 | 809 |
| Group median | < 75 | 488 | ||
| Seronegatives | ||||
| 27 | M | B | < 75 | 837 |
| 74 | M | B | < 75 | 322 |
| 86 | F | B | < 75 | 670 |
| 87 | F | W | < 75 | 742 |
| 91 | F | W | < 75 | 714 |
| 94 | F | As | < 75 | 228 |
| 96 | M | B | < 75 | 1396 |
| 100 | M | Mixed race | < 75 | 609 |
| Group median | < 75 | 692 |
M indicates male; F, female; T, transgender; W, white; B, black; NAm, Native American; As, Asian; and H, Hispanic.
Patient 1562: plasma viral load fluctuates. For the last 2 years, more than 50% of viral loads have been more than 23 000 copies/mL.
Mucosal CD4+ T-cell levels are preserved in controllers compared with noncontrollers
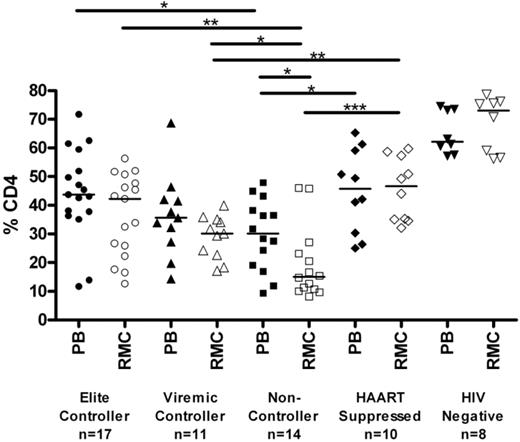
The relative percentage of CD4+ T cells in rectal mucosa was determined by flow cytometry (Figure 1). Elite and viremic controllers had significantly higher levels of CD4+ T cells in rectal mucosa than noncontrollers (42.2%, 30.1%, and 15.1% in elite controllers, viremic controllers, and noncontrollers, respectively; P < .05 for each pairwise comparison). Mucosal CD4+ T-cell percentages in HAART-suppressed patients were comparable with those observed in controllers (45.8%.) However, the percentage of mucosal CD4+ T cells was significantly higher in seronegatives (73%) compared with any of the HIV-seropositive groups, including controllers and HAART-suppressed patients (P < .05 for each pairwise comparison). The CD4+ T-cell percentages in peripheral blood were comparable with those in mucosa for each of the groups, with the weakest association observed among the noncontrollers (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Percentage of CD4+ T cells in blood and mucosa. Percentage of CD4+ T cells, shown as a subset of CD3+ T cells, as determined by flow cytometry in peripheral blood (PB, closed symbols) and rectal mucosa (RMC, open symbols) across patient groups. Horizontal bars represent the median of each group. *P < .05, **P < .01, ***P < .001. All HIV-infected patient groups had significantly lower percentages of CD4+ T cells than seronegatives (P < .01).
Percentage of CD4+ T cells in blood and mucosa. Percentage of CD4+ T cells, shown as a subset of CD3+ T cells, as determined by flow cytometry in peripheral blood (PB, closed symbols) and rectal mucosa (RMC, open symbols) across patient groups. Horizontal bars represent the median of each group. *P < .05, **P < .01, ***P < .001. All HIV-infected patient groups had significantly lower percentages of CD4+ T cells than seronegatives (P < .01).
Mucosal Gag-specific responses are strongest in controllers
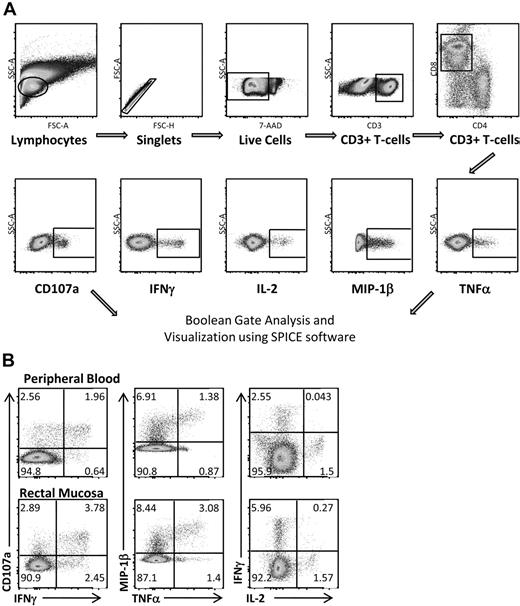
Multicolor flow cytometry was used to assess HIV Gag-specific T-cell responses in blood and rectal mucosa. A representative gating strategy for rectal mononuclear cells is shown in Figure 2A. Doublet and dead cell discrimination was used to reduce background noise. For CD8+ T cells, positive responses for each individual function (CD107a, IFN-γ, IL-2, MIP-1β, and TNF-α) were selected. Conventional bivariate plots were also created to confirm the placement of individual gates. A representative subject from the elite controller group is shown in Figure 2B. Boolean gates were created from the 5 individual gates to produce 31 distinct responding populations, encompassing all possible combinations of these 5 functions.
Measuring HIV Gag-specific immune responses: intracellular cytokine staining. (A) Flow cytometric gating strategy for RMCs. Doublet and dead cell discrimination is used to reduce background noise (second and third graphs). For CD8+ T cells, positive responses for each individual function (CD107a, IFN-γ, IL-2, MIP-1β, and TNF-α) are selected. Boolean gates are created from these 5 individual gates to divide responding cells into 31 distinct populations corresponding to all possible combinations of these functions. These are graphed using SPICE software to better visualize polyfunctional responses. (B) CD8+ T-cell responses in both peripheral blood and rectal mucosa from an elite controller using standard bivariate plots.
Measuring HIV Gag-specific immune responses: intracellular cytokine staining. (A) Flow cytometric gating strategy for RMCs. Doublet and dead cell discrimination is used to reduce background noise (second and third graphs). For CD8+ T cells, positive responses for each individual function (CD107a, IFN-γ, IL-2, MIP-1β, and TNF-α) are selected. Boolean gates are created from these 5 individual gates to divide responding cells into 31 distinct populations corresponding to all possible combinations of these functions. These are graphed using SPICE software to better visualize polyfunctional responses. (B) CD8+ T-cell responses in both peripheral blood and rectal mucosa from an elite controller using standard bivariate plots.
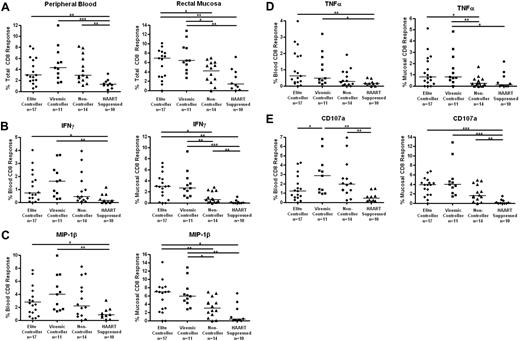
The total HIV-specific CD8+ T-cell response was determined as the percentage of cells capable of responding in any way to Gag stimulation after the subtraction of background (Figure 3A). Both elite and viremic controllers had significantly stronger mucosal CD8+ T-cell responses than either noncontrollers or HAART-suppressed subjects (medians, 6.9% elite controllers and 6.4% viremic controllers vs 4.1% noncontrollers and 1.4% HAART; P < .05 for elite controllers vs noncontrollers, P < .01 for other pairwise comparisons). The total Gag-specific CD8+ T-cell responses were more similar in peripheral blood (3.0%, 4.3%, and 2.9% for elite controllers, viremic controllers, and noncontrollers, respectively), but responses in all 3 off-treatment groups were stronger than responses in the HAART group (1.3%; P < .05 compared with viremic controllers, P < .01 compared with elite controllers and noncontrollers).
HIV Gag-specific CD8+ T-cell responses in blood and rectal mucosa. (A) Total percentage of CD8+ T cells from peripheral blood (left panels) or rectal mucosa (right panels) able to respond in any way (CD107a, IFN-γ, IL-2, MIP-1β, or TNF-α) to Gag stimulation. Percentage of CD8+ T cells capable of producing IFN-γ (B), MIP-1β (C), TNF-α (D), and CD107 (E) in response to Gag stimulation. Horizontal bars represent the median for each group. *P < .05, **P < .01, ***P < .001.
HIV Gag-specific CD8+ T-cell responses in blood and rectal mucosa. (A) Total percentage of CD8+ T cells from peripheral blood (left panels) or rectal mucosa (right panels) able to respond in any way (CD107a, IFN-γ, IL-2, MIP-1β, or TNF-α) to Gag stimulation. Percentage of CD8+ T cells capable of producing IFN-γ (B), MIP-1β (C), TNF-α (D), and CD107 (E) in response to Gag stimulation. Horizontal bars represent the median for each group. *P < .05, **P < .01, ***P < .001.
We also compared response magnitudes in blood versus mucosa for each patient group. Although we did not quantify individual memory/effector T-cell subsets in this study, it is well known that peripheral blood includes many naive T cells, whereas mucosal T cells are primarily effector memory cells.40 We would therefore expect higher magnitude responses in rectal mucosa compared with blood. Indeed, we found mucosal CD8+ T-cell responses to be consistently higher than responses in blood for most patients; however, this difference was significant only in the case of elite controllers (P = .034; Figure 3).
Mucosal IFN-γ, MIP-1β, and TNF-α responses in controllers are particularly strong
HIV Gag-specific CD8+ T-cell responses were further broken down into individual functions (IFN-γ, IL-2, MIP-1β, TNF-α, and CD107a; Figure 3B-E). Elite and viremic controllers had significantly stronger mucosal Gag-specific CD8+ T-cell responses than noncontrollers for IFN-γ, MIP-1β, and TNF-α (P < .05, Figure 3B-D right panels). Again, the differences in PBMCs were less robust, although elite and viremic controllers had higher responses than HAART-suppressed patients (Figures 3B-D left panels). IL-2 production by Gag-specific CD8+ T cells was generally very low, and no differences were found between patient or tissue groups (data not shown). Degranulation (CD107a) responses were generally higher for the elite controller, viremic controller, and noncontroller groups than for patients on HAART in either blood or mucosal tissue (P = .0532 for elite controllers vs HAART in blood, all others P < .01; Figure 3E).
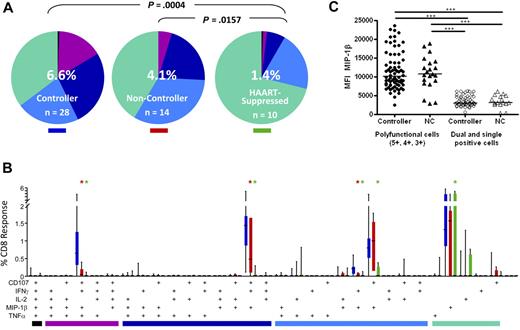
Higher frequencies of polyfunctional CD8+ T cells in rectal mucosa of controllers
We next analyzed the complexity of the CD8+ T-cell response by looking at all 5 functional parameters in various combinations. To do this, we used Boolean gate analysis, which expanded the 5 response parameters into 31 individual functional categories. The pie charts in Figure 4A show the overall pattern of functionality of HIV Gag-specific CD8+ T cells in rectal mucosa. Elite and viremic controllers were grouped together into a single controller category as they did not differ significantly from one another (data not shown). The pattern of Gag-specific CD8+ T-cell response categories, as revealed by pie charts, differed significantly between controllers and HAART-suppressed persons in both blood (P = .0034, data not shown) and rectal mucosa (P = .0004). As we reported earlier,36,37 mucosal Gag-specific CD8+ T-cell responses from noncontrollers were also significantly more complex than those from patients on HAART (P = .0157).
HIV Gag-specific polyfunctional CD8+ T-cell responses. (A) The overall polyfunctionality of the mucosal CD8+ T-cell response can be visualized with pie charts in which each slice represents a different functional category: black indicates 5 functions; purple, 4 functions; dark blue, 3 functions; light blue, 2 functions; green, 1 function. The number in the center of each pie represents the median total percentage of cells responding in any way to Gag stimulation. Elite and viremic controllers are combined into a single “controller” category. NC indicates noncontroller; HAART, HAART-suppressed. Statistical differences between groups are indicated above pie charts. (B) The bar chart shows the total Gag-specific response broken down into 31 individual response categories. Each bar represents the median and interquartile ranges for the frequency of responding cells for a different patient group as indicated by the colored rectangle beneath each pie (ie, blue indicates controllers; red, noncontrollers; green, HAART-suppressed). The colored bars below the x-axis relate to the pie slice colors in Figure 4A. Statistical differences between patient groups are marked by an asterisk (*) and color coded (blue indicates controllers; red, noncontrollers; green, HAART). All differences were significant at P < .01. (C) Expression of MIP-1β in mucosal CD8+ T cells as measured by MFI comparing polyfunctional cells to dual- and single-function cells in controllers and noncontrollers. Each data point represents the MFI from a separate population of cells (5+, 4+, 3+, 2+, or 1+). Horizontal bars represent the median for each group. ***P < .001.
HIV Gag-specific polyfunctional CD8+ T-cell responses. (A) The overall polyfunctionality of the mucosal CD8+ T-cell response can be visualized with pie charts in which each slice represents a different functional category: black indicates 5 functions; purple, 4 functions; dark blue, 3 functions; light blue, 2 functions; green, 1 function. The number in the center of each pie represents the median total percentage of cells responding in any way to Gag stimulation. Elite and viremic controllers are combined into a single “controller” category. NC indicates noncontroller; HAART, HAART-suppressed. Statistical differences between groups are indicated above pie charts. (B) The bar chart shows the total Gag-specific response broken down into 31 individual response categories. Each bar represents the median and interquartile ranges for the frequency of responding cells for a different patient group as indicated by the colored rectangle beneath each pie (ie, blue indicates controllers; red, noncontrollers; green, HAART-suppressed). The colored bars below the x-axis relate to the pie slice colors in Figure 4A. Statistical differences between patient groups are marked by an asterisk (*) and color coded (blue indicates controllers; red, noncontrollers; green, HAART). All differences were significant at P < .01. (C) Expression of MIP-1β in mucosal CD8+ T cells as measured by MFI comparing polyfunctional cells to dual- and single-function cells in controllers and noncontrollers. Each data point represents the MFI from a separate population of cells (5+, 4+, 3+, 2+, or 1+). Horizontal bars represent the median for each group. ***P < .001.
Comparing controllers and noncontrollers, there were significant differences between groups in the “polyfunctional” 3- and 4-function populations (purple and navy pie slices, Figure 4A bar graphs, Figure 4B). However, permutation tests did not reveal an overall difference between pie charts for these 2 patient groups, probably because the high magnitudes and large variance in monofunctional responses (green pie slices) masked the differences in polyfunctionality between patient groups.
Figure 4B shows the detailed breakdown of mucosal Gag-specific CD8+ T-cell responses in each of 31 functional categories. Although it is theoretically possible for antigen-specific cells to respond to stimulation by producing any of the 31 possible combinations of factors, 4 combinations predominated in both blood and mucosa (Figure 4B; and data not shown). The major 4-function category consisted of cells expressing CD107a, IFN-γ, MIP-1β, and TNF-α but not IL-2; the major 3-function category expressed CD107a, IFN-γ, and MIP-1β; and the major dual-function category included cells expressing CD107a and MIP-1β. Among single-function cells, the dominant response was production of MIP-1β. Very few CD8+ T cells produced all 5 analytes in any patient group, including controllers. In peripheral blood as in rectal mucosa, the major differences were observed between controllers and patients on HAART, with controllers having significantly stronger responses in each functional category (P < .01, data not shown).
Differences between controllers and noncontrollers were more apparent in rectal mucosa than in blood (Figure 4B; and data not shown). Notably, mucosal CD8+ T-cell responses in controllers were significantly more robust than mucosal responses in noncontrollers in the 4-function (CD107a, IFN-γ, MIP-1β, and TNF-α) and 3-function (CD107a, IFN-γ, and MIP-1β) categories (P < .01) as well as a minor dual-function category (IFN-γ, MIP-1β, P < .01). Thus, when functional populations were considered individually, HIV controllers had significantly higher frequencies of 4- and 3-function Gag-specific CD8+ T cells in rectal mucosa compared with noncontrollers.
Polyfunctional CD8+ T cells produce more MIP-1β on a per-cell basis
Previous studies have demonstrated that antigen-specific T cells that produce multiple cytokines/chemokines (ie, polyfunctional cells) also produce greater amounts of these analytes on a per-cell basis than monofunctional cells.9,37,41 We wished to determine whether polyfunctional mucosal CD8+ T cells (defined as those capable of 5, 4, or 3 functions) from controllers produced larger quantities of cytokines or chemokines than those from noncontrollers. We chose to examine the median fluorescence intensity (MFI) of MIP-1β as a representative effector molecule because it was the one factor common to all 5 major functional categories. As seen previously,37 CD8+ T cells producing 5, 4, or 3 factors produced larger quantities of MIP-1β than dual- and single-function cells (P < .001; Figure 4C). However, this effect was independent of both clinical status (controller or noncontroller) and tissue type (Figure 4C; and data not shown).
Association of HLA class I genotype with CD8+ T-cell response magnitude and polyfunctionality
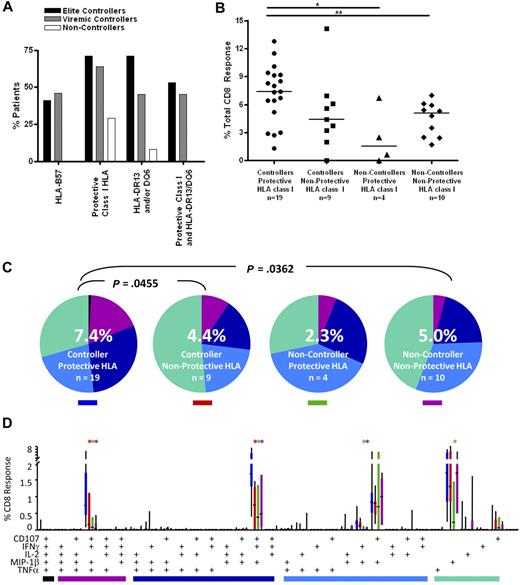
High-resolution HLA class I genotyping was performed on DNA from the patients in this study (Table S1). As observed in previous studies,12,13,18 HLA-B57 was highly enriched in both elite and viremic controllers (41% and 46%, respectively) compared with the noncontroller group (0%; Figure 5A). A recent study of the larger University of California–San Francisco controller cohort from which the current subjects were drawn also found a higher prevalence of HLA-B13, 27, 58, and 81 in controllers compared with noncontrollers.12 As expected, these alleles were also more prevalent in the controllers we studied here (71% and 64% in elite and viremic controllers, respectively) compared with noncontrollers (29%; Figure 5A).
CD8+ T-cell responses in controllers and noncontrollers with and without protective class I HLA alleles. (A) Patient immunogenetics. The bar graphs show the percentage of patients in this study with HLA-B57 (far left); any protective class I HLA (HLA-B13, B27, B57, B58, B81; second from left); HLA-DRB1*13 and/or DQB1*6 (third from left); and both protective class I and class II alleles (far right). (B) Total magnitude of the CD8+ T-cell response in rectal mucosa. Horizontal bars represent the median for each group. *P < .05, **P < .01, ***P < .001. (C) Overall polyfunctionality of the CD8+ T-cell response in rectal mucosa. Each pie slice represents a different functional category: black indicates 5 functions; purple, 4 functions; dark blue, 3 functions; light blue, 2 functions; green, 1 function. The number in the center of each pie represents the median percentage of responding cells responding in any way to Gag stimulation. Statistical differences between groups are written above pie charts. (D) Mucosal CD8+ T-cell polyfunctional responses broken down into 31 individual response categories. Each bar represents the median and interquartile ranges for the frequency of responding cells for a different patient group and class I HLA status as indicated by the colored rectangle beneath each pie (ie, blue indicates controllers/protective HLA). The colored bars below the x-axis relate to the pie slice colors in panel B. Statistical differences between controllers with protective class I alleles and other patient groups are marked by an asterisk (*) and color coded for each patient group (red indicates controller/nonprotective HLA; green, noncontroller/protective HLA; purple, noncontroller/nonprotective HLA). All differences are significant at P < .01.
CD8+ T-cell responses in controllers and noncontrollers with and without protective class I HLA alleles. (A) Patient immunogenetics. The bar graphs show the percentage of patients in this study with HLA-B57 (far left); any protective class I HLA (HLA-B13, B27, B57, B58, B81; second from left); HLA-DRB1*13 and/or DQB1*6 (third from left); and both protective class I and class II alleles (far right). (B) Total magnitude of the CD8+ T-cell response in rectal mucosa. Horizontal bars represent the median for each group. *P < .05, **P < .01, ***P < .001. (C) Overall polyfunctionality of the CD8+ T-cell response in rectal mucosa. Each pie slice represents a different functional category: black indicates 5 functions; purple, 4 functions; dark blue, 3 functions; light blue, 2 functions; green, 1 function. The number in the center of each pie represents the median percentage of responding cells responding in any way to Gag stimulation. Statistical differences between groups are written above pie charts. (D) Mucosal CD8+ T-cell polyfunctional responses broken down into 31 individual response categories. Each bar represents the median and interquartile ranges for the frequency of responding cells for a different patient group and class I HLA status as indicated by the colored rectangle beneath each pie (ie, blue indicates controllers/protective HLA). The colored bars below the x-axis relate to the pie slice colors in panel B. Statistical differences between controllers with protective class I alleles and other patient groups are marked by an asterisk (*) and color coded for each patient group (red indicates controller/nonprotective HLA; green, noncontroller/protective HLA; purple, noncontroller/nonprotective HLA). All differences are significant at P < .01.
We examined the functionality of CD8+ T cells from controllers and noncontrollers with and without protective HLA class I alleles. Controllers with protective alleles had a greater median response magnitude in rectal mucosa compared with peripheral blood (7.4% vs 3.0%, P < .001; Figure 5B; and data not shown). In all other groups, response magnitudes were similar in the 2 compartments. Controllers with protective alleles had more robust mucosal responses than noncontrollers with (7.4% vs 2.3%, P < .05) or without (7.4% vs 5.0%, P < .01) protective alleles. There was a trend toward higher responses in controllers with protective alleles compared with controllers lacking protective alleles (7.4% vs 4.4%, P = .09).
Similarly, in rectal mucosa, controllers with protective alleles displayed greater response complexity than either controllers (P = .0455) or noncontrollers (P = .0362) lacking protective alleles (Figure 5C). Controllers with protective alleles had higher frequencies of mucosal Gag-specific 4- and 3-function cells than any of the other patient groups (P < .01; Figure 5D).
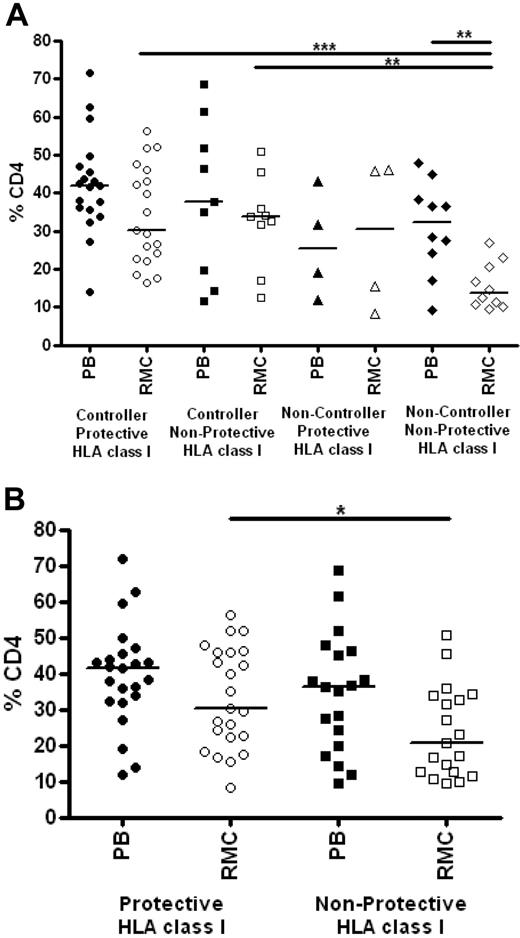
Persons with protective HLA class I genotypes have increased levels of CD4+ T cells in rectal mucosa
We also assessed the relationship between class I genotype and CD4+ T-cell percentages in peripheral blood and rectal mucosa. Persons with and without protective HLA class I genotypes had similar levels of CD4+ T cells in blood (Figure 6A). In rectal mucosa, noncontrollers lacking protective alleles had significantly lower CD4+ T-cell percentages compared with controllers with or without protective class I genotypes (medians 13.7% vs 30.1% and 33.9%, respectively; P < .01). There was no difference between the elite and viremic controller groups (Figure 6A). However, analysis of CD4+ T-cell percentages irrespective of clinical status revealed that those with protective class I alleles had higher percentages of CD4+ T cells in rectal mucosa compared with those who lacked protective alleles (30.1% vs 20.5%; P = .0298; Figure 6B).
Relationship between HLA class I genotype and CD4+ T-cell levels. (A) Percentage of CD4+ T cells as determined by flow cytometry in peripheral blood (PB, closed symbols) and rectal mucosa (RMC, open symbols) among controllers and noncontrollers with and without protective class I HLA alleles. (B) Percentages of CD4+ T cells in peripheral blood (PB, closed symbols) and rectal mucosa (RMC, open symbols) among persons with or without protective class I HLA alleles, defined as in Figure 5, irrespective of clinical status. Horizontal bars represent the median for each group. *P < .05, **P < .01, ***P < .001.
Relationship between HLA class I genotype and CD4+ T-cell levels. (A) Percentage of CD4+ T cells as determined by flow cytometry in peripheral blood (PB, closed symbols) and rectal mucosa (RMC, open symbols) among controllers and noncontrollers with and without protective class I HLA alleles. (B) Percentages of CD4+ T cells in peripheral blood (PB, closed symbols) and rectal mucosa (RMC, open symbols) among persons with or without protective class I HLA alleles, defined as in Figure 5, irrespective of clinical status. Horizontal bars represent the median for each group. *P < .05, **P < .01, ***P < .001.
Stronger mucosal CD8+ T-cell responses in persons with HLA-DRB1*13 and/or DQB1*06
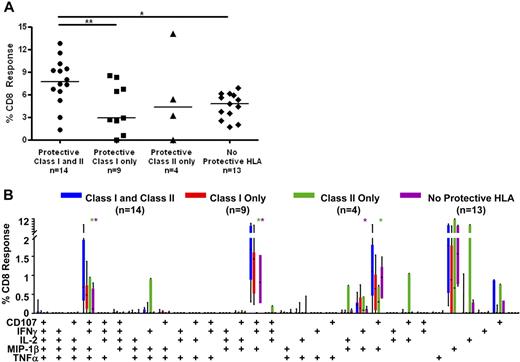
To determine whether certain major histocompatibility complex (MHC) class II alleles were associated with controller status, we also performed HLA-DR/DQ typing (Figure 5A; Table S1). We found that HLA-DRB1*13 and DQB1*06 were enriched among HIV controllers (70% elite controllers and 45% viremic controllers vs 8% noncontrollers; Figure 5A). These 2 alleles were previously reported to be associated with slow HIV disease progression in several cohorts.20,21,42 In addition, 53% of elite controllers and 45% of viremic controllers, but none of the noncontrollers, had one or both of these class II alleles in addition to at least one protective class I allele. We then compared Gag-specific responses in persons with and without these class II alleles (Figure 7). Persons who possessed one or both of these class II alleles as well as one or more protective class I allele had significantly higher median Gag-specific CD8+ T-cell responses (7.7%) in rectal mucosa compared with those with only protective class I HLA alleles (2.9%, P < .05) and those lacking any protective alleles (4.8%, P < .01; Figure 7A). There was also a trend toward persons with both class I and II “protective” alleles having higher magnitude mucosal CD8+ T-cell responses than those with only HLA-DRB1*13 and/or DQB1*06 (4.3%).
CD8+ T-cell responses in persons with protective class I and II HLA alleles. (A) Total magnitude of the CD8+ T-cell responses in rectal mucosa in persons with or without protective class I and/or class II HLA alleles. Horizontal bars represent the median for each group. *P < .05, **P < .01. (B) Mucosal CD8+ T-cell polyfunctional responses broken down into 31 individual response categories. Each bar represents the median and interquartile ranges for the frequency of responding CD8+ cells in persons with or without protective class I and/or class II HLA alleles. Statistical differences between persons with both protective class I and class II alleles and other groups are marked by an asterisk (*) and color coded for each patient group (green indicates class II only; purple, no protective HLA). All differences were significant at P < .01.
CD8+ T-cell responses in persons with protective class I and II HLA alleles. (A) Total magnitude of the CD8+ T-cell responses in rectal mucosa in persons with or without protective class I and/or class II HLA alleles. Horizontal bars represent the median for each group. *P < .05, **P < .01. (B) Mucosal CD8+ T-cell polyfunctional responses broken down into 31 individual response categories. Each bar represents the median and interquartile ranges for the frequency of responding CD8+ cells in persons with or without protective class I and/or class II HLA alleles. Statistical differences between persons with both protective class I and class II alleles and other groups are marked by an asterisk (*) and color coded for each patient group (green indicates class II only; purple, no protective HLA). All differences were significant at P < .01.
The presence of certain class I and class II alleles also appeared to confer an advantage in terms of mucosal CD8+ T-cell response complexity (Figure 7B). Persons with both protective class I alleles and HLA-DRB1*13 and/or DQB1*06 had significantly higher frequencies of 4- and 3-function mucosal Gag-specific CD8+ T cells than those with only protective class II alleles or those lacking protective HLA alleles (P < .01). However, they had only marginally higher frequencies of polyfunctional CD8+ T cells than patients with only protective class I alleles.
Discussion
Numerous studies have supported a relationship between blood CD8+ T-cell responses and HIV control.9,10,14,43 Here, we have expanded on these studies by examining for the first time HIV-specific CD8+ T-cell responses in gastrointestinal mucosal tissues from a large, well-characterized cohort of HIV controllers. The gastrointestinal mucosa is the largest lymphoid organ in the body, a major site of HIV replication,23-25 and houses a significant population of effector memory CD8+ T cells. Many of these cells are HIV-specific,27,31,36,40 yet their frequency and effector functions have not been thoroughly assessed.
In the patients studied here, CD8+ T-cell responses in rectal mucosa were more strongly associated with controller status than peripheral blood responses. Not only did controllers have higher magnitude Gag-specific mucosal CD8+ T-cell responses than noncontrollers and persons on HAART, but these responses were of greater complexity: that is, controllers had higher frequencies of mucosal CD8+ T cells capable of 3 or more simultaneous effector functions. It has become increasingly clear that T-cell “quality” rather than magnitude alone is important in defining an effective immune response to HIV. Several studies have demonstrated that the production of IFN-γ alone is insufficient to define a protective immune response.11,44-47 The term “polyfunctionality” has come to describe the ability of cells to produce multiple cytokines and chemokines as well as release cytolytic granules.10,44,45 Polyfunctional T cells are generated after exposure to influenza virus, Epstein-Barr virus, varicella zoster virus, and cytomegalovirus.45 In the mouse model of Leishmania major, mice were administered several vaccine formulations and subsequently challenged; mice that demonstrated the greatest protection were those that produced the highest frequencies of polyfunctional cells (IFN-γ+TNF-α+IL-2+).41 In previous studies, polyfunctional T cells were shown to be enriched in the blood of certain HIV-positive LTNP, and the presence of cells capable of 5 simultaneous effector functions negatively correlated with viral load.10 However, it is important to keep in mind that strong, polyfunctional T-cell responses may also be viewed as the product of a relatively intact immune system and that HIV controllers may be shielded from profound immunologic damage by mechanisms other than CD8+ T-cell activity. Our findings do not suggest that polyfunctional mucosal CD8+ T-cell responses are sufficient for immune control: some controllers in our cohort did not exhibit unusually robust CD8+ T-cell responses, and some with relatively strong mucosal CD8+ T-cell responses were indeed noncontrollers. Accordingly, there remains an urgent need to explore additional factors contributing to HIV nonprogression.
We observed a consistent association between certain class I HLA alleles and the CD8+ T-cell response, suggesting that MHC class I-restricted responses can contribute to immune control of HIV infection. Previous studies have shown certain class I alleles to be overrepresented in cohorts of HIV controllers, particularly HLA-B27 and B57.17-19 CD8+ T cells restricted by these alleles represent a large proportion of the total immune response in LTNP.48 In a recent study of persons from the University of California–San Francisco controller cohort, we observed that subjects with protective HLA alleles (HLA-B13, 27, 57, 58, and 81) had higher frequencies of HIV-specific IFN-γ+IL-2+ CD8+ T cells in peripheral blood compared with those without protective alleles.12 In this earlier study, we argued that HIV-specific T-cell responses probably contribute to immune control of HIV infection but that other, “non–T-cell” mechanisms also play a role. The findings of the present study confirm and extend our earlier work, and reveal that the gastrointestinal mucosa is a site of robust and polyfunctional CD8+ T-cell responses in many HIV controllers. Nevertheless, it remains apparent that CD8+ T-cell responses alone cannot fully explain the phenomenon of HIV control.
The preservation of a strong CD4+ T-cell population appears to be beneficial to the maintenance of CD8+ T-cell responses, and our findings suggest that certain class II alleles may be associated with polyfunctional CD8+ T-cell responses. The question remains as to why specific class I and II HLA alleles are associated with long-term viral control. First, T-cell responses restricted by these alleles may be targeting functionally important regions within the viral genome. Several studies have shown an association between Gag-specific CTL responses and low viral loads.49-54 Targeting of multiple Gag epitopes is associated with HIV control, whereas responses directed toward Env epitopes fail to limit viral replication.50 It has also been shown that HLA molecules associated with slower disease progression preferentially bind peptides from the p24 region of Gag.49,52,53 Gag is highly conserved, and mutations occurring in Gag often come with a high fitness cost to the virus.51,55 In particular, HLA-B57 targets a region of p24 involved in assembly,49,51 and CTL escape mutations in this region result in a significant fitness cost to the virus.51 In addition to polyfunctionality, CD8+ T cells restricted by certain MHC alleles may exhibit high functional avidity, as demonstrated for HIV-specific T cells restricted by HLA-B27,9 B57,56 and B81.57 Furthermore, HLA-B alleles containing the Bw4 epitope serve as ligands for killer immunoglobulin-like receptors that regulate natural killer cell function.58,59 Finally, protective HLA alleles may also be in linkage disequilibrium with other as yet unknown factors contributing to viral containment.
However, as recently demonstrated, it is now clear that particular MHC alleles are neither entirely necessary, nor are they sufficient, to determine controller status.12 HIV-specific T-cell responses clearly do not account for all cases of viral containment, as some controllers have weak HIV-specific CD8+ T-cell responses and/or lack “protective” HLA alleles. It is therefore probable that virologic and/or genetic factors account for some instances of elite control. Multiple varieties of HIV control may exist: controllers with “protective” MHC alleles may rely heavily on strong, polyfunctional CD8 and/or CD4+ T-cell responses to limit viral replication, whereas those lacking protective MHC alleles may rely on other mechanisms, such as low viral fitness, innate immune mechanisms, and/or host restriction factors.
It is also noteworthy that a small subset of HIV controllers shows increased markers of immune activation and a decline in CD4 counts despite low plasma viremia.60 Therefore, in future studies, it will be important to compare and contrast those controllers who progress with those who maintain controller status. It may also be of interest to consider persons who fit the classical definition of long-term nonprogression (ie, stable CD4+ T-cell counts over a period of 7 or more years and the absence of AIDS defining illnesses). A successful AIDS vaccine should be capable of both controlling viral replication and preventing CD4+ T-cell loss and disease progression.
Taken together, our findings suggest that multiple mechanisms contribute to the HIV controller phenotype, including but not limited to MHC class I/II genotype and mucosal HIV-specific T-cell responses. Additional studies will be required to address issues, such as (1) the role of innate immunity in HIV control; (2) the level of ongoing viral replication and the replication competence of virus isolated from blood and tissues of controllers; and (3) the impact of host genetics on T-cell infectability and viral replication kinetics.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Rebecca Hoh (University of California–San Francisco) and Melissa Schreiber (University of California–Davis) for assistance with patient recruitment and enrollment.
This study was supported by the National Institutes of Health (grant AI057020, B.L.S.; grants AI069994 and AI044595, S.G.D.; grant AI065244, P.W.H.; grant AI027763, the University of California–San Francisco Center for AIDS Research), the California HIV/AIDS Research Program (grant CH05-D-606; R.B.P.), the American Foundation for AIDS Research (106710-40-RGRL; S.G.D.), and the University of California–San Francisco Clinical and Translational Science Institute (UL1 RR024131). This investigation was conducted in a facility constructed with support from the Research Facilities Improvement Program (grant C06 RR-12 088-01) from the National Center for Research Resources, National Institutes of Health. The LSR-II violet laser was upgraded with funding from the James B. Pendleton Charitable Trust.
National Institutes of Health
Authorship
Contribution: A.L.F., P.W.H., S.G.D., and B.L.S. conceived and planned the study; A.L.F., J.W.C., D.H.Y., and M.M.M. performed the studies; A.L.F. analyzed the data; J.N.M. provided statistical guidance; J.C.G., R.B.P., and H.F.Y. provided clinical expertise, referred patients, and/or procured samples; and A.L.F. and B.L.S. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Barbara L. Shacklett, Department of Medical Microbiology and Immunology and Division of Infectious Diseases, Department of Internal Medicine, School of Medicine, University of California–Davis, Davis, CA 95616; e-mail: blshacklett@ucdavis.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal