Abstract
To further investigate potential mechanisms of resistance to FLT3 inhibitors, we developed a resistant cell line by long-term culture of MV4-11 cells with ABT-869, designated as MV4-11-R. Gene profiling reveals up-regulation of FLT3LG (FLT3 ligand) and BIRC5 (survivin), but down-regulation of SOCS1, SOCS2, and SOCS3 in MV4-11-R cells. Hypermethylation of these SOCS genes leads to their transcriptional silencing. Survivin is directly regulated by STAT3. Stimulation of the parental MV4-11 cells with FLT3 ligand increases the expression of survivin and phosphorylated protein STAT1, STAT3, STAT5. Targeting survivin by short-hairpin RNA (shRNA) in MV4-11-R cells induces apoptosis and augments ABT-869–mediated cytotoxicity. Overexpression of survivin protects MV4-11 from apoptosis. Subtoxic dose of indirubin derivative (IDR) E804 resensitizes MV4-11-R to ABT-869 treatment by inhibiting STAT signaling activity and abolishing survivin expression. Combining IDR E804 with ABT-869 shows potent in vivo efficacy in the MV4-11-R xenograft model. Taken together, these results demonstrate that enhanced activation of STAT pathways and overexpression of survivin are important mechanisms of resistance to ABT-869, suggesting that the STAT pathways and survivin could be potential targets for reducing resistance developed in patients receiving FLT3 inhibitors.
Introduction
Internal tandem duplication (ITD) in juxtamembrane (JM) and point mutation (PM) in the kinase domain (KD) of fms-like tyrosine kinase 3 (FLT3) are common genetic lesions in acute myeloid leukemia (AML).1-5 Recently, additional PMs were identified in the extracellular domain and JM domain by high-throughput DNA sequencing.6
FLT3 mutations induce receptor dimerization and autophosphorylation of the KD, in turn, resulting in constitutive activation of phosphoinositide 3-kinase (PI3K)–AKT, RAS-MEK–mitogen-activated protein kinase (MAPK), and signal transducers and activators of transcription (STAT) 5 pathways.1-5 On the biologic level, it leads to uncontrolled cell proliferation, blockage of differentiation, and cell survival. Therefore, FLT3 mutations play an important role in leukemogenesis and represent attractive therapeutic targets.1,2,4,5 Several small molecule tyrosine kinase inhibitors (TKIs) are currently undergoing different phases of clinical development.7 Although most FLT3 inhibitors show potent efficacy in vitro with the half maximal inhibitory concentration (IC50) values in the nanomole range, the majority of patients only have moderate and transient responses.8 Furthermore, under prolonged therapy with TKIs, leukemic cells could develop resistance to FLT3 inhibitors when used as monotherapy. This is exemplified by the resistance phenomenon to imatinib mesylate (Gleevec), the first small molecule kinase inhibitor for the treatment of chronic myeloid leukemia (CML) harboring the BCR-ABL fusion oncogene. The identification of PMs in the ATP binding site or gene amplification of BCR-ABL from imatinib-resistant CML patients9 promoted researchers to investigate the role of acquired mutations in resistance to FLT3 inhibitors.
Mutations in the ATP-binding pocket have been identified through polymerase chain reaction (PCR)–based mutagenesis screening in murine Ba/F3-FLT3-ITD cells and selected for growth in the presence of PKC412,10 or in a resistant Ba/F3-FLT3-ITD cell line developed by coculture with SU5416.11 Resistance to PKC412 resulting from the N676K PM in the FLT3 KD has been described in a clinical trial patient.12 Human leukemia cell lines are valuable disease models. Piloto et al used long-term exposure of human leukemia cell lines, including MOLM-14 (AML-M5, one allele wild-type and the other FLT3-ITD allele), Hb1119 (acute lymphoblastic leukemia [ALL], FLT3-D836H), and SEM-K2 (overexpression of wild-type FLT3), to FLT3 inhibitors, CEP-5214 and CEP-701, to generate 6 resistance human cell lines.13 Selection of activating Ras mutations has been found in 2 of 6 FLT3 inhibitor resistant cell lines, but no PM in the FLT3 KD was found in all 6 resistant cell lines.13
To further investigate other potential mechanisms of resistance to multitargeted TKIs, we developed 3 resistant cell lines (designated as MV4-11-R1, -R2, -R3) by long-term coculture of the human leukemia cell line, MV4-11 (AML, both allele FLT-ITD), with ABT-869, a multitargeted TKI with activity against FLT3.14 We also explored the combination of ABT-869 with other small molecule inhibitors to overcome resistance and thereby potentially provide novel treatments in vitro and in vivo.
Methods
Small molecular inhibitors and reagents
ABT-869, a multitargeted TKI with activity against FLT3, was kindly provided by Abbott Laboratories (Chicago, IL). For in vitro and in vivo experiments, the preparation for ABT-869 was previously published.15 Indirubin derivative (IDR) E804, Tyrene CR4, AG490, AG1296, JAK3 inhibitor II, NU6140, and FLT3 inhibitor III were purchased from Calbiochem (Gibbstown, NJ) and dissolved in dimethyl sulfoxide (DMSO) before use. SU5416 and 5-aza–deoxycytidine (5-aza) were purchased from Sigma-Aldrich (St Louis, MO). Human FLT3 ligand was obtained from PeproTech (Rocky Hill, NJ).
Cell lines and development of resistant cell lines
Human MV4-11 cells were cultured with RPMI 1640 (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; JRH Bioscience, Lenexa, KS) at a density of 2 to 10 × 105 cells/mL in a humid incubator with 5% CO2 at 37°C. Log phase growing MV4-11 cells were cocultured with increasing concentration of ABT-869 for 3 months. Three parallel experiments were performed in parallel for selection of resistant lines. These resistant lines were grown in normal medium without ABT-869 for at least 48 hours before experiments.
Cell viability assays
Leukemic cells were seeded in 96-well culture plates at a density of 2 × 104 viable cells/100 μL/well in triplicates and were treated with small molecular inhibitors. Colorimetric CellTiter 96 AQueous One Solution Cell Proliferation assay (MTS assay; Promega, Madison, WI) was used to determine the cytotoxicity as described previously.15 IC50 values were determined by MTS assay and calculated with CalcuSyn software (Biosoft, Cambridge, United Kingdom). Each experiment was in triplicate.
Flow cytometric analysis
For analysis of MRP1 and multidrug-resistance protein (MDR) expression, 2 million cells were fixed and stained according to the manufacturer's instruction and analyzed with a DakoCytomation Cyan LX (DakoCytomation Denmark, Glostrup, Denmark) flow cytometer, using Summit (version 4.3) software. For apoptosis assays, annexin V–FITC binding assay (BD Pharmingen, San Diego, CA) was used as recommended by the manufacturer. For cell-cycle analysis, 1 million cells were fixed, stained with propidium iodide (PI; BD Pharmingen) and analyzed by flow cytometry.
Western blot analysis
Preparation of the cell lysate and immunoblotting were performed as previously described.16 Antibodies used were as follows: anti-FLT3, anti–p-FLT3, anti–p-STAT1 (Tyr701), anti–p-STAT3 (Tyr705, clone 3E2), anti-p–STAT5 (Tyr694), anti-STAT1, anti-STAT3, anti-STAT5, anti-Survivin, anti–poly (ADP-ribose) polymerase (PARP), anti-cleaved PARP, from Cell Signaling Technology (CST; Danvers, MA) and anti-actin, anti–lung resistance protein (anti-LRP), anti-MRP1, anti-MDR, immunoglobulin G (IgG) isotype control from Santa Cruz Biotechnology (Santa Cruz, CA).
Low-density array
Gene expression profiling was investigated with custom real-time PCR-based analysis using TaqMan low-density arrays (LDA; Applied Biosystems, Foster City, CA) as described before.15 Briefly, RNA was extracted from cells using the Purescript RNA isolation kit (Genetra Systems, Minneapolis, MN). First strand cDNA was synthesized with SuperScript III First-Strand Synthesis SuperMix (Invitrogen). PCR amplification was performed in the 7900HT Fast Real-Time system (Applied Biosystems). Each sample was in triplicate. The target genes analyzed include anti- and proapoptotic genes, cell cycle–regulated genes, DNA damage genes, stress gene, PI3K/AKT pathway, MAPK pathway, JAK/STAT pathway, mTOR pathway, VEGF pathway, NOTCH pathway, WNT pathway, NFκB pathway, invasion- and metastasis-related genes, oncogenes, as well as housekeeping genes (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Sequence Detection System (SDS) 2.2.1 software (Applied Biosystems) was used to perform relative quantitation (RQ) of target genes using the comparative cycle threshold (CT; ΔΔCT) method.
RT-PCR and RQ-PCR
The primers and reverse transcription–polymerase chain reaction (RT-PCR) conditions for survivin analysis were adopted from Mahotka et al.17 Sequences of primers for survivin real-time quantitative (RQ)–PCR were described before.18 The sequences of primers of STAT3 for RQ-PCR were as follows: STAT3-RQ forward, 5′-CCTGAAGCTGACCCAGGTAGC-3′; STAT3-RQ reverse, 5′-CACCTTCACCATTATTTCCAAACTG-3′. Sequences of primers of suppressor of cytokine signaling (SOCS) family (SOCS1, SOCS2, and SOCS3) for RQ-PCR were published before.19 Power SYBR Green PCR Master Mix (Applied Biosystems) was used as recommendation by the manufacturer. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as internal control. SDS 2.2.1 software was used to perform RQ of target genes using the comparative CT (ΔΔCT) method.
Transfection
Human STAT3 cDNA was purchased from Open Biosystems (Huntsville, AL) and cloned into pEGFP vector (Clontech, Mountain View, CA). MV4-11 cells were transfected with pEGFP control vector and pEGFP-STAT3 separately, using Nucelofector device (Amaxa, Cologne, Germany) according to the manufacturer's protocol. Briefly, 3 × 106 cells were mixed with 2 μg vector and 100 μL Solution-L, transferred to a cuvet. The program Q-001 was used to transfect the cells in the Nucelofector device. After transfection, cells were immediately transferred into a 6-well plate containing prewarmed (37°C) complete medium. After 48 hours posttransfection, the cells were spun into pellets and followed by RNA extraction, cDNA synthesis, and RQ-PCR analysis for gene expression.
Human full-length of survivin cDNA was obtained from Open Biosystems and cloned into lentivirus pLVX-puro vector (Clontech) within EcoRI/BamHI site. The construct was validated by sequencing. The production and harvest of high titer lentivirus was performed using Lenti-X HT Packaging System (Clontech) as recommended by the manufacturer. MV4-11 cells were infected with pLVX-puro–Survivin lentivirus particulars and selected in culture medium containing gradually incrementally increased concentration of puromycin ranging from 400 ng/mL to 2 μg/mL for 3 weeks. The stable transfectant cell line was designated as MV4-11-Survivin.
shRNA studies
A pool of survivin (RHS4529-NM_001168) shRNA, as well as nonsilencing shRNA control (RHS1707) was purchased from Open Biosystems. RetroPack PT67 cells (Clontech) were seeded into a 6-well plate at 60%-80% confluence (4 × 105 cells/well) 24 hours before transfection, 5 μg of each short-hairpin RNA (shRNA) vector and 10 μL Lipofectamine 2000 (Invitrogen) were used for transfection. PT67 cells were diluted and plated after transfection for 24 hours in culture medium with 2 μg/mL puromycin (Clontech). After 1 week selection, the large, healthy colonies were isolated and transferred into individual plates. Filtered medium containing viral particles together with 6 μg/mL polybrene were used for infecting cells (2 × 106), respectively. Twenty-four hours after infection, cultures were replaced with fresh medium and subjected to immunoblot and cell viability assay.
ChIP assays
Chromatin immunoprecipitation (ChIP) assays were done by using CHIP-IT Express Kit from Active Motif (Carlsbad, CA). Briefly, log-phase growing MV4-11-R cells were incubated with 37% formaldehyde to cross-link protein-DNA complexes. The cross-linked chromatin was then extracted, diluted with lysis buffer, and sheared with Enzymatic Shearing Cocktail (Active Motif). Total sheared chromatin (10 μL) was used as positive control in PCR analysis. The remaining chromatin was divided into equal amount for immunoprecipitation with either anti-Stat3 or anti-IgG (negative control) polyclonal antibody (Santa Cruz Biotechnology) on magnetic beads. The immunoprecipitates were eluted, reversed cross-linked, and treated with proteinase K. Purified DNA was subjected to PCR with primers specific for a region (nucleotides 1821-2912) in human the Survivin promoter (GenBank accession no. U75285). The sequences of the PCR primers used are as follows: pSurvivin forward primer, 5′-CTGGCCATAGAACCAGAGAAGTGA-3′; pSurvivin reverse primer, 5′-CCACCTCTGCCAACGGGTCCCGCG-3′.
Xenograft mouse model
Female Balb/C nude mice (17-20 g, 4-6 weeks old) were purchased from Animal Resources Center (Canning Vale, Australia). Exponentially growing MV4-11-R cells (5 × 106) were subcutaneously injected into loose skin between the shoulder blades and left front leg of recipient mice. All treatments were started 10 days after the injection, when the mice had palpable tumors of an average size of approximately 200 mm3. ABT-869 was administrated at 15 mg/kg per day by oral gavage daily.15,20 IDR E804 was prepared and given the same as ABT-869, but at dose of 10 mg/kg per day. In the combination group, mice were treated with both compounds at the same dose as monotherapy. Treatments lasted for 14 days. Each group comprised 10 mice.
The length (L) and width (W) of the tumor were measured with callipers, and tumor volume (TV) was calculated as TV = (L × W2)/2. The protocol was reviewed and approved by Institutional Animal Care and Use Committee of the National University of Singapore in compliance to the guidelines on the care and use of animals for scientific purpose.
Immunohistochemistry
Tissue fixation followed by hematoxylin and eosin staining were done as described previously.16 Sources and incubation conditions for the primary antibodies were as follows: anti-survivin (clone 71G4; CST), anti–Ki-67 (Neomarkers, Fremont, CA), and anti–cleaved PARP (CST). The slides were counterstained in hematoxylin for 30 seconds and mounted with cover slides. The images were analyzed by a Zeiss Axioplan 2 imaging system with AxioVision 4 software (Zeiss, Germany).
Statistical analysis
Number of viable cells, tumor volume, and survival time were expressed in mean plus or minus standard deviation (SD). Tumor volume reduction of the treatment groups was compared with the untreated control group by Student t test, and P values of less than .05 were considered to be significant.
Results
Long-term coculture of MV4-11 cells with ABT-869 resulted in cross-resistance to other FLT3 inhibitors
Human leukemia MV4-11 cells with both alleles FLT3-ITD were cocultured with gradually increasing concentration of ABT-869 for 3 months. Three separate cultures were performed in parallel, resulting in 3 resistant lines, designated as MV4-11-R1, MV4-11-R2, and MV4-11-R3. In addition, MV4-11-R represents a pool of MV4-11-R1, -R2, and -R3.
The MTS assay and CalcuSyn software were used to determine the cytotoxic effects of ABT-869, FLT3 inhibitor III, AG1296, and SU5416 on resistant lines and the parental MV4-11 cell line. The IC50 values of ABT-869 on resistant lines was approximately 9 times higher than parental MV4-11 cells. Furthermore, the resistant lines were cross-resistance to structurally unrelated FLT3 inhibitors (Table 1). Similarly, annexin V binding assay revealed that the resistant lines were also resistant to ABT-869–, FLT3 inhibitor III-, AG1296-, and SU5416-induced apoptosis as compared with the parent MV4-11 cells (data not shown).
Comparison of the potency (IC50 values) of ABT-869 and other structurally unrelated FLT3 inhibitors for inhibiting the proliferation of MV4-11, MV4-11-R, MV4-11 + FLT3 ligand, and MV4-11-Survivin cells
| Drugs . | Structure . | IC50 (nM) . | |||
|---|---|---|---|---|---|
| MV4-11 . | MV4-11-R . | MV4-11 + FLT3 ligand* . | MV4-11-Survivin . | ||
| ABT-869 | 3-Aminoindazole | 6 | 52 | 40 | > 200 |
| FLT3 inhibitor III | 5-Phenyl-2-thiazolamine | 26 | 83 | 1300 | 713 |
| AG 1296 | Tyrphostin | 1657 | > 7000 | > 7000 | > 10 000 |
| SU5416 | 3-Substituted indolinone | 270 | 3039 | 3076 | > 10 000 |
| Drugs . | Structure . | IC50 (nM) . | |||
|---|---|---|---|---|---|
| MV4-11 . | MV4-11-R . | MV4-11 + FLT3 ligand* . | MV4-11-Survivin . | ||
| ABT-869 | 3-Aminoindazole | 6 | 52 | 40 | > 200 |
| FLT3 inhibitor III | 5-Phenyl-2-thiazolamine | 26 | 83 | 1300 | 713 |
| AG 1296 | Tyrphostin | 1657 | > 7000 | > 7000 | > 10 000 |
| SU5416 | 3-Substituted indolinone | 270 | 3039 | 3076 | > 10 000 |
Cells were seeded in 96-well culture plates at a density of 2 × 104 viable cells/100 μL/well in triplicates and were treated with each compound for 48 hours. Colorimetric MTS assay was used to determine the cytotoxicity. IC50 was determined by MTS assay and calculated with CalcuSyn software. Each experiment was performed in triplicate.
FLT3 ligand 50 ng/mL.
Overexpression of FLT3, p-FLT3 receptor or multidrug-resistant related proteins, or mutations in KD were not responsible for resistance to FLT3 inhibitors in MV4-11-R
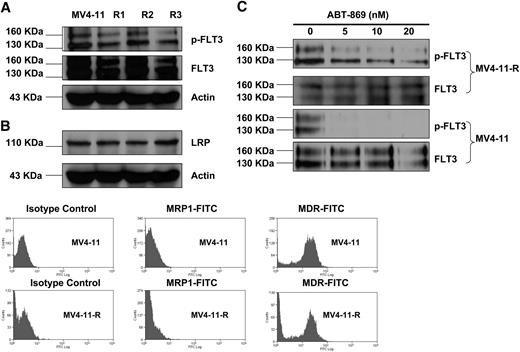
To investigate the mechanisms of drug resistance, immunoprecipitation, and Western blot analysis were performed to compare the expression of the wild-type FLT3 and p-FLT3 receptor in resistant lines with parental MV4-11 cells. This analysis demonstrated that their expression levels were similar (Figure 1A). Western blot and flow cytometric analysis were used to determine the expression of multidrug-resistant related proteins. LRP (Figure 1A) was not up-regulated in MV4-11-R cells. MDR (Figure 1B) was expressed 10-fold higher in both MV4-11-R and parental MV4-11 as compared with isotype controls. MDR-related protein (MRP1; Figure 1B) was not detected in MV4-11 and the resistant lines.
Comparison of the expression of phosphorylated FLT3 receptor, total FLT3 receptor, and multidrug-resistant related proteins (LRP, MRP1, and MDR) among the parental MV4-11 and resistant lines. R1, R2, and R3 indicate MV4-11-R1, MV4-11-R2, and MV4-11-R3, respectively. (A) Immunoprecipitation (IP) and immunoblot analysis reveals that there is no significant difference in the expression of p-FLT3 and FLT3 receptor among MV4-11 and MV4-11-R1, -R2, and -R3. IP was performed using anti-FLT3 antibody, followed by Western blot analysis with anti–p-Tyrosine antibody. The same blot was then stripped and reprobed with anti-FLT3 antibody. (B) Western blot analysis and fluorescence-activated cell sorting (FACS) analysis found the expression of LRP, MRP1, and MDR was not varied significantly among MV4-11 and MV4-11-R1, -R2, and -R3. (C) MV4-11 and MV4-11-R cells were treated with ABT-869 at a dose of 0, 5, 10, or 20 nM for 1 hour. IP and Western blot analysis were performed the same way as described in panel A.
Comparison of the expression of phosphorylated FLT3 receptor, total FLT3 receptor, and multidrug-resistant related proteins (LRP, MRP1, and MDR) among the parental MV4-11 and resistant lines. R1, R2, and R3 indicate MV4-11-R1, MV4-11-R2, and MV4-11-R3, respectively. (A) Immunoprecipitation (IP) and immunoblot analysis reveals that there is no significant difference in the expression of p-FLT3 and FLT3 receptor among MV4-11 and MV4-11-R1, -R2, and -R3. IP was performed using anti-FLT3 antibody, followed by Western blot analysis with anti–p-Tyrosine antibody. The same blot was then stripped and reprobed with anti-FLT3 antibody. (B) Western blot analysis and fluorescence-activated cell sorting (FACS) analysis found the expression of LRP, MRP1, and MDR was not varied significantly among MV4-11 and MV4-11-R1, -R2, and -R3. (C) MV4-11 and MV4-11-R cells were treated with ABT-869 at a dose of 0, 5, 10, or 20 nM for 1 hour. IP and Western blot analysis were performed the same way as described in panel A.
No PM in KD of FLT3 receptor was found in 3 resistant lines by sequencing analysis (data not shown). Treatment of MV4-11-R cells with ABT-869 still lead to inhibition of FLT3 phosphorylation (Figure 1C), but it was not completely abolished as in MV4-11 parental cells under the same treatment condition.
Identification of enhanced activation of STAT pathways and overexpression of survivin in the resistant lines
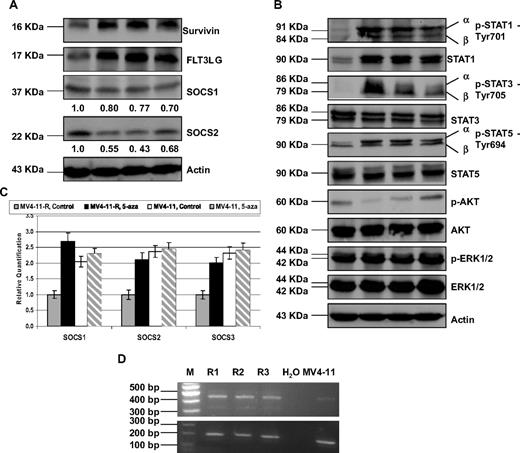
To explore possible novel mechanisms of resistance, we used a real-time PCR-based approach to profile and compare the gene expression among MV4-11 cells and the 3 resistant lines. The list of all differentially expressed genes more than 2-fold among them was shown in Table S2. Based upon low-density array analysis, FLT3 ligand (FLT3LG) and BIRC5 (Survivin) were up-regulated approximately 2-fold, while SOCS family (SOCS1, 2, and 3) were down-regulated 2-fold (Table 2). Consistent with the transcriptional changes, FLT3LG and survivin also were elevated, and SOCS1 and SOCS2 were reduced at the protein level by Western blot analysis (Figure 2A). The level of decrease reduction in SOCS1 and SOCS2 expression was quantified by densitometry analysis. The increment of FLT3LG was not due to gene amplification, because RQ-PCR analysis showed same level of DNA expression (data not shown).
LDA analysis revealed that the expression of FLT3LG and BIRC5 was up-regulated, while the expression of SOCS family 1, 2, and 3 was down-regulated in the MV4-11-R1, -R2, and -R3 cells as compared to MV4-11 cells
| Gene ID* . | RefSeq . | Fold change . | SD . |
|---|---|---|---|
| Up-regulation list | |||
| BIRC5-Hs00 1533 53_m1 | NM_00 10122 71.1 | 2.05 | 0.14 |
| FLT3LG-Hs0018 1740_m1 | NM_00 1459.2 | 2.38 | 0.19 |
| Down-regulation list | |||
| SOCS1-Hs00 7051 64_s1 | NM_00 3745.1 | −2.01 | 0.23 |
| SOCS2-Hs00 3744 16_m1 | NM_00 3877.3 | −2.43 | 0.18 |
| SOCS3-Hs00 2695 75_s1 | NM_00 3955.3 | −2.38 | 0.14 |
| Gene ID* . | RefSeq . | Fold change . | SD . |
|---|---|---|---|
| Up-regulation list | |||
| BIRC5-Hs00 1533 53_m1 | NM_00 10122 71.1 | 2.05 | 0.14 |
| FLT3LG-Hs0018 1740_m1 | NM_00 1459.2 | 2.38 | 0.19 |
| Down-regulation list | |||
| SOCS1-Hs00 7051 64_s1 | NM_00 3745.1 | −2.01 | 0.23 |
| SOCS2-Hs00 3744 16_m1 | NM_00 3877.3 | −2.43 | 0.18 |
| SOCS3-Hs00 2695 75_s1 | NM_00 3955.3 | −2.38 | 0.14 |
These results represent triplicated experiments.
SOCS indicates suppressor of cytokine signaling.
ID denotes the TaqMan Gene Expression assays.
Validation of FLT3LG, survivin, and SOCS1 and SOCS2 expression, and STAT pathway overactivation at the translational level, RQ-PCR quantification of SOCS gene family and confirmation of normal transcript of Survivin in MV4-11-R cells. MV4-11 and MV4-11-R cells were washed, then lysed and subjected to 10% to 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Western blot analyses were detected with the indicated antibodies for the assessment of expression level changes in (A) FLT3LG, survivin, SOCS1, and SOCS2. Densitometric analysis was performed using Amersham Image Scanner with LabScan ImageQuant TL Software (Amersham Biosciences, Piscataway, NJ). The protein levels of SOCS1 and SOCS2 were normalized with each respective actin level. (B) Western blot analyses of STAT, AKT, and MAPK pathway molecules. (C) MV4-11 parental and MV4-11-R cells were seeded at densities of 2 × 105/mL in 10 mL culture medium and treated with PBS control and 3 μM (final concentration) of 5-aza. Fresh medium was changed, and new drug was added every day. After 3 days, cells were harvested, washed with 1× PBS twice. Then the pellets were lysed, followed by RNA extraction, and RQ-PCR. (D) RT-PCR confirmed the overexpression of Survivin transcripts in resistant lines. The size of normal transcript is 431 bp, and 2 other transcript variants, Survivin-2B and Survivin-ΔEx3, are 500 and 329 bp, respectively (top panel). GAPDH was used as internal control (bottom panel).
Validation of FLT3LG, survivin, and SOCS1 and SOCS2 expression, and STAT pathway overactivation at the translational level, RQ-PCR quantification of SOCS gene family and confirmation of normal transcript of Survivin in MV4-11-R cells. MV4-11 and MV4-11-R cells were washed, then lysed and subjected to 10% to 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Western blot analyses were detected with the indicated antibodies for the assessment of expression level changes in (A) FLT3LG, survivin, SOCS1, and SOCS2. Densitometric analysis was performed using Amersham Image Scanner with LabScan ImageQuant TL Software (Amersham Biosciences, Piscataway, NJ). The protein levels of SOCS1 and SOCS2 were normalized with each respective actin level. (B) Western blot analyses of STAT, AKT, and MAPK pathway molecules. (C) MV4-11 parental and MV4-11-R cells were seeded at densities of 2 × 105/mL in 10 mL culture medium and treated with PBS control and 3 μM (final concentration) of 5-aza. Fresh medium was changed, and new drug was added every day. After 3 days, cells were harvested, washed with 1× PBS twice. Then the pellets were lysed, followed by RNA extraction, and RQ-PCR. (D) RT-PCR confirmed the overexpression of Survivin transcripts in resistant lines. The size of normal transcript is 431 bp, and 2 other transcript variants, Survivin-2B and Survivin-ΔEx3, are 500 and 329 bp, respectively (top panel). GAPDH was used as internal control (bottom panel).
Because the SOCS family is a negative regulator of STAT pathway,21 we hypothesize that that STAT pathways would be up-activated in the resistant lines. Indeed, Western blot analysis confirmed the overexpression of p-STAT1, p-STAT3, and p-STAT5 in the resistant lines compared with the parental MV4-11 (Figure 2B), which suggest that STAT activity is constitutively enhanced in the resistant lines. It is interesting to note that wild-type STAT1, but not wild-type STAT3 and STAT5, was also increased in the resistant lines, which likely resulted from intensified STAT1 activity (p-STAT1), because STAT1 itself has been identified as one of the STAT1 target genes (Figure 2B). In addition to the STAT pathways, PI3I/AKT and MAPK signaling pathways also play an important role in promoting cell survival and proliferation; however, p-AKT and p-ERK1/ERK2 were not overexpressed in the resistant lines (Figure 2B).
Aberrant methylation of SOCS genes have been reported in AML and solid tumors,19,22 so we further determined whether this epigenetic changes caused down-regulation of SOCS genes in MV4-11-R cells. The expression of SOCS1, 2, and 3 genes was restored by the demethylating agent 5-aza treatment in MV4-11-R cells, but essentially not changed in MV4-11 parental cells, suggesting SOCS promoters in MV4-11 parental cells are not sensitive to demethylating therapy (Figure 2C).
We have looked at the 3 most widely studied survivin splice variants,17 and RT-PCR analysis showed that all 3 transcripts appeared to be up-regulated with the normal transcript (431 bp) as the dominant transcript in the resistant lines (Figure 2D); however, the expression of other variants is unknown in our resistant lines.23
Up-regulation of survivin in MV4-11-R cells resulted in changes in cell cycle and apoptosis
Survivin has dual roles in suppressing apoptosis and modulating cell cycle.24 We sought to investigate the influence of up-regulated survivin on cell cycle and apoptosis in MV4-11-R cells. After serum deprivation for 48 hours, MV4-11 parental cells and MV4-11-R cells were transferred into complete medium for an additional 24 hours. Flow cytometric analysis revealed that MV4-11 parental cells had a significantly decreased S-phase population (6.5% vs 17.8%, P < .01), but a dramatically increased G2/M phase population (49.6% vs 20.3%, P < .01) as compared with MV4-11-R cells.
Furthermore, there were 4.5 times more dead cells in MV4-11 cells than in MV4-11-R cells as determined by the trypan blue dye exclusion method at the end of serum depletion for 48 hours. Taken together, these results suggest that overexpression of survivin in MV4-11-R cells leads to accelerated S-phase shift and resistance to apoptosis.
FLT3 ligand–mediated STAT activities and survivin expression
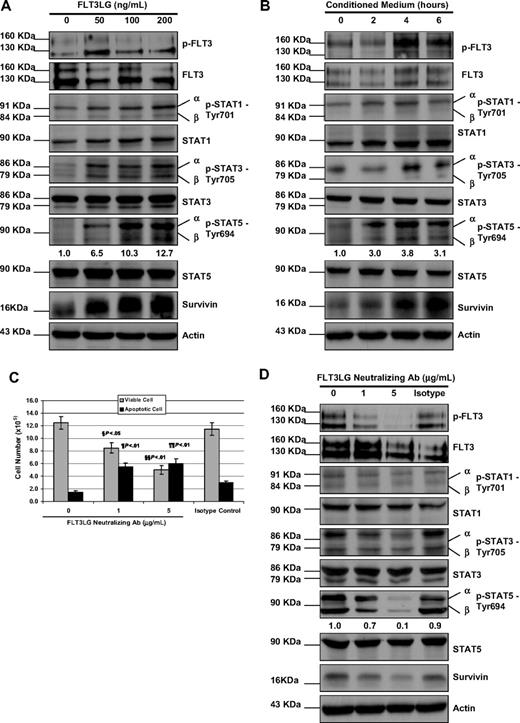
To mimic the overexpression of FLT3LG in the resistant cells, we cultured the parental MV4-11 cells with increasing concentration of FLT3 ligand in the cell culture for 48 hours. Additional FLT3 ligand stimulation fairly elevated the expression level of p-STAT1, p-STAT3, and p-STAT5 (Figure 3A). The expression of survivin was also increased in a concentration-dependent manner in response to FLT3 ligand stimulation (Figure 3A). To test whether leukemia cells can be protected by FLT3 ligand, we treated MV4-11 cells with the same panel of FLT3 inhibitors in the presence of 50 ng/mL FLT3 ligand in culture medium for 48 hours. Adding FLT3 ligand rendered MV4-11 cells resistance to all the FLT3 inhibitors tested, although the degree of IC50 increment varied (Table 1).
The effect of FLT3LG on activity of STAT signaling pathway and the expression of survivin. (A) MV4-11 cells were cultured with FLT3 ligand at concentrations of 0, 50, 100, and 200 ng/mL for 48 hours; then they were washed, lysed, and subjected to either IP of p-FLT3 receptor as described in Figure 1 or 10% to 12% SDS-PAGE. Western blot analyses were detected with the indicated antibodies for the assessment of expression level changes in STAT pathway molecules and survivin. β-Actin was used as a loading control. (B) MV4-11 cells were cultured in conditioned medium for 0, 2, 4, and 6 hours. Cells were then washed, lysed, and followed by IP and immunoblot analysis. (C) MV4-11-R cells were treated with FLT3LG neutralizing antibody at concentrations of 0, 1, 5 μg/mL, and istotype control antibody for 48 hours. Viable cells and apoptotic cells were counted by the trypan blue dye exclusion method. The experiments were triplicated. Bars represent SD. § and ¶ refer to P values of comparison of the viable and apoptotic cell numbers in 1 μg FLT3LG antibody-treated MV4-11-R samples with those in isotype control. §§ and ¶¶ refer to P values of comparison of the viable and apoptotic cell numbers in 5 μg FLT3LG antibody-treated MV4-11-R samples with those in isotype control. (D) After counting, cells were then washed, lysed, and followed by IP and immunoblot analysis. Densitometric analysis was performed for p-STAT5 using Amersham Image Scanner with LabScan ImageQuant TL software.
The effect of FLT3LG on activity of STAT signaling pathway and the expression of survivin. (A) MV4-11 cells were cultured with FLT3 ligand at concentrations of 0, 50, 100, and 200 ng/mL for 48 hours; then they were washed, lysed, and subjected to either IP of p-FLT3 receptor as described in Figure 1 or 10% to 12% SDS-PAGE. Western blot analyses were detected with the indicated antibodies for the assessment of expression level changes in STAT pathway molecules and survivin. β-Actin was used as a loading control. (B) MV4-11 cells were cultured in conditioned medium for 0, 2, 4, and 6 hours. Cells were then washed, lysed, and followed by IP and immunoblot analysis. (C) MV4-11-R cells were treated with FLT3LG neutralizing antibody at concentrations of 0, 1, 5 μg/mL, and istotype control antibody for 48 hours. Viable cells and apoptotic cells were counted by the trypan blue dye exclusion method. The experiments were triplicated. Bars represent SD. § and ¶ refer to P values of comparison of the viable and apoptotic cell numbers in 1 μg FLT3LG antibody-treated MV4-11-R samples with those in isotype control. §§ and ¶¶ refer to P values of comparison of the viable and apoptotic cell numbers in 5 μg FLT3LG antibody-treated MV4-11-R samples with those in isotype control. (D) After counting, cells were then washed, lysed, and followed by IP and immunoblot analysis. Densitometric analysis was performed for p-STAT5 using Amersham Image Scanner with LabScan ImageQuant TL software.
The FLT3 ligand exists in membrane-bound and soluble forms, which are both biologically active. To test whether the secreted soluble form of FLT3 ligand by MV4-11-R cells contributes to resistance, we first harvested conditioned medium from MV4-11-R cells incubated in complete medium for 12 hours. Then, MV4-11 cells were washed twice with 1× phosphate-buffered saline (PBS) and cultured in conditioned medium for 2, 4, and 6 hours, followed by Western blot analysis. As shown in Figure 3B, incubation in conditioned medium resulted in elevated expression of p-FLT3, p-STATs, and survivin. We noticed that the expression of p-STAT5 slightly dropped at 6 hours, while survivin continued to increase.
To investigate the effect of down-regulation of FLT3 ligand, MV4-11-R cells were treated with a FLT3 ligand neutralizing antibody for 48 hours, and cell viability was analyzed. Figure 3C showed the viable cell number was significantly decreased and apoptotic cell number was significantly increased in FLT3 ligand neutralizing antibody-treated samples as compared with untreated or isotype control–treated samples. As expected, in neutralizing antibody-treated samples, the expression of p-FLT3, p-STATs, and survivin was reduced (Figure 3D). These data suggest that FLT3 ligand plays an important role in mediating the resistance to FLT3 inhibitors.
Modulation of survivin expression influenced drug sensitivity
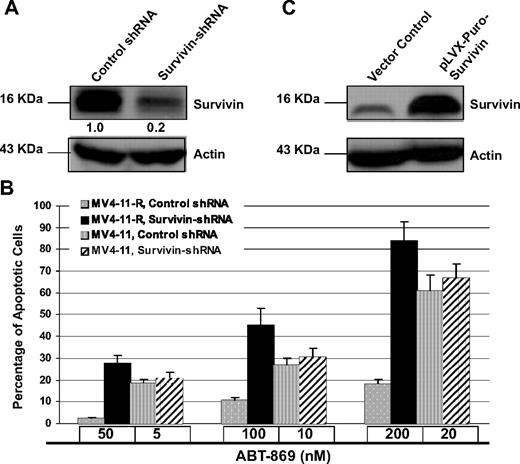
To demonstrate the critical role of survivin in the regulation of resistance, we used a pool of shRNA to specially target survivin. Western blot analysis confirmed specific inhibition of survivin by approximately 80% with the pool of survivin-shRNAs (Figure 4A). Silencing survivin remarkably potentiated ABT-869–induced apoptosis in MV4-11-R cells compared with control shRNA treatment (P < .001). On the contrary, MV4-11 parental cells, in the presence of IC50 dose of ABT-869, are not sensitive to Survivin-shRNA (P > .05; Figure 4B).
Knockdown of Survivin potentiated ABT-869–induced apoptosis in MV4-11-R cells. (A) MV4-11-R cells were treated with nontarget control shRNA or Survivin shRNA pools for 48 hours and then harvested for Western blot analysis. Actin level served as loading controls. Densitometric analysis was performed using Amersham Image Scanner with LabScan ImageQuant TL software. The level of survivin was normalized with each actin level. (B) After knockdown, MV4-11-R cells were treated with ABT-869 at doses of 50, 100, and 200 nM, and MV4-11 parental cells were treated with ABT-869 at doses of 5, 10, and 20 nM for 48 hours. As residual expression of survivin persists after treatment of survivin shRNA, it may provide some level of protection from a full-scale apoptosis. Apoptosis was measured by annexin V–FITC binding assay. P values demonstrate the comparison between survivin shRNA-treated and control shRNA-treated group. All P values of MV4-11-R samples are < .001. All P values of MV4-11 samples are greater than 0.05. Means are for 3 replicated experiments; bars represent SD. (C) Immunoblot analysis of the survivin protein level in MV4-11-Survivin and MV4-11 vector control cells.
Knockdown of Survivin potentiated ABT-869–induced apoptosis in MV4-11-R cells. (A) MV4-11-R cells were treated with nontarget control shRNA or Survivin shRNA pools for 48 hours and then harvested for Western blot analysis. Actin level served as loading controls. Densitometric analysis was performed using Amersham Image Scanner with LabScan ImageQuant TL software. The level of survivin was normalized with each actin level. (B) After knockdown, MV4-11-R cells were treated with ABT-869 at doses of 50, 100, and 200 nM, and MV4-11 parental cells were treated with ABT-869 at doses of 5, 10, and 20 nM for 48 hours. As residual expression of survivin persists after treatment of survivin shRNA, it may provide some level of protection from a full-scale apoptosis. Apoptosis was measured by annexin V–FITC binding assay. P values demonstrate the comparison between survivin shRNA-treated and control shRNA-treated group. All P values of MV4-11-R samples are < .001. All P values of MV4-11 samples are greater than 0.05. Means are for 3 replicated experiments; bars represent SD. (C) Immunoblot analysis of the survivin protein level in MV4-11-Survivin and MV4-11 vector control cells.
To further confirm the role of survivin in drug resistance, we evaluated the effect of overexpression of survivin in transfected MV4-11 parental cells. The stable transfectants (MV4-11-Survivin) showed overexpression of survivin protein (Figure 4C). MTS assays revealed an exceptional increase in resistance to the panel of FLT3 inhibitors in MV4-11-Survivin cells (Table 1).
Taken together, these data unequivocally demonstrated that survivin is crucial in mediating resistance to FLT3 inhibitors.
IDR E804 induced apoptosis through inhibition of STAT pathway and survivin and sensitized MV4-11-R to ABT-869
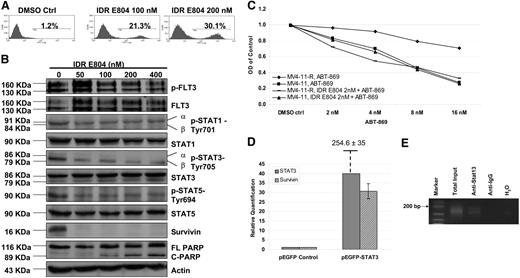
Next, we screened a panel of small molecule inhibitors of CDKs, SRC, BCR-ABL, and JAKs including IDR E804, Tyrene CR4, AG490, JAK3 inhibitor II, and NU6140. We found that MV4-11-R cells are most sensitive to IDR E804, an inhibitor of SRC-STAT3 pathway, using MTS assay (data not shown). IDR E804 treatment dose-dependently induced MV4-11-R cells to undergo apoptosis (Figure 5A). Western blot analysis also showed that IDR E804 inhibited the expression of p-STAT1, p-STAT3, p-STAT5, and completely blocked survivin (Figure 5B). It is worthy to note that IDR E804 completely inhibits survivin in the absence of complete inhibition of p-STATs. This apparently incongruous inhibition could be because survivin expression is regulated in a cell cycle–dependent manner and rapidly decline in G1/G0 phase and IDR E804 significantly arrested MV4-11-R cell in G1/G0 phase (P < .01; Figure S1). Furthermore, cleaved PARP, a hallmark of apoptosis, was detected at concentrations of 100 nM and higher (Figure 5B). Notably, IDR E804 did not inhibit FLT3-ITD kinase activity (Figure 5B), so its cytotoxicity to MV4-11-R cells was derived specifically from targeting STAT pathway and survivin. The IC50 value of ABT-869 in MV4-11-R decreased from 52 to 6 nM calculated by CalcuSyn software in the presence of a subtherapeutic concentration (2 nM) of IDR E804, suggesting a synergistic effect (Figure 5C; P < .01). Whereas the same combination treatment did not augment the inhibition effect in MV4-11 parental cells as compared with ATB-869 alone (Figure 5C; P > .05). These results are in accordance with the data obtained by shRNA study as above. To confirm the molecular mechanism of synergism via targeting STAT-Survivin pathway, we further tested the effect of lower doses of IDR E804 on MV4-11-R cells. IDR E804 from 2 to 20 nM inhibited the STAT activities and the expression of survivin in a dose-dependent fashion (Figure S2). Approximately 23% reduction of survivin was observed at 2 nM IDR E806 as compared with the control treatment.
IDR E804-induced apoptosis and sensitized MV4-11-R to ABT-869. (A) Two million cells of MV4-11-R were treated with either DMSO control or IDR E804 at concentrations of 100 and 200 nM for 48 hours. Cells were then washed and stained with annexin V–FITC for apoptosis assay. The shown graphs represent 3 independent experiments. (B) MV4-11-R cells (10 × 106) were cultured with DMSO control or IDR E804 at concentrations of 50, 100, 200, and 400 nM for 48 hours. The IP of p-FLT3 receptor was performed as in Figure 1. Cells were washed, lysed, and subjected to 10% to 12% SDS-PAGE. Western blot analyses were detected with the indicated antibodies for the assessment of the expression level changes in STAT pathway molecules and Survivin, PARP, and cleaved PARP. Actin was used as a loading control. (C) MV4-11-R and MV4-11 cells were treated with various concentrations of ABT-869 alone or together with 2 nM IDR E804 for 48 hours. MTS assay was used to determine the viable cell number. Means are shown for 3 replicated experiments. (D) After parental MV4-11 cells were transiently transfected with pEGFP empty vector or pEGFP-STAT3 for 48 hours, RNA was extracted, followed by cDNA synthesis and relative quantification by RQ-PCR. The baseline expression of STAT3 and survivin in MV4-11 cells transfected with pEGFP vector was set as 1.0. The relative quantification of STAT3 in MV4-11 cells transfected with pEGFP-STAT3 was 354.6 ± 35 from 3 independent experiments. (E) ChIP assays were done using anti-STAT3 antibody or control anti-IgG antibody. PCR primers for the survivin gene promoter were applied to detect promoter fragment in immunoprecipitates. PCR controls included total sheared chromatin (total input), DNA isolated through the negative control IgG-ChIP, and no DNA at all (H2O).
IDR E804-induced apoptosis and sensitized MV4-11-R to ABT-869. (A) Two million cells of MV4-11-R were treated with either DMSO control or IDR E804 at concentrations of 100 and 200 nM for 48 hours. Cells were then washed and stained with annexin V–FITC for apoptosis assay. The shown graphs represent 3 independent experiments. (B) MV4-11-R cells (10 × 106) were cultured with DMSO control or IDR E804 at concentrations of 50, 100, 200, and 400 nM for 48 hours. The IP of p-FLT3 receptor was performed as in Figure 1. Cells were washed, lysed, and subjected to 10% to 12% SDS-PAGE. Western blot analyses were detected with the indicated antibodies for the assessment of the expression level changes in STAT pathway molecules and Survivin, PARP, and cleaved PARP. Actin was used as a loading control. (C) MV4-11-R and MV4-11 cells were treated with various concentrations of ABT-869 alone or together with 2 nM IDR E804 for 48 hours. MTS assay was used to determine the viable cell number. Means are shown for 3 replicated experiments. (D) After parental MV4-11 cells were transiently transfected with pEGFP empty vector or pEGFP-STAT3 for 48 hours, RNA was extracted, followed by cDNA synthesis and relative quantification by RQ-PCR. The baseline expression of STAT3 and survivin in MV4-11 cells transfected with pEGFP vector was set as 1.0. The relative quantification of STAT3 in MV4-11 cells transfected with pEGFP-STAT3 was 354.6 ± 35 from 3 independent experiments. (E) ChIP assays were done using anti-STAT3 antibody or control anti-IgG antibody. PCR primers for the survivin gene promoter were applied to detect promoter fragment in immunoprecipitates. PCR controls included total sheared chromatin (total input), DNA isolated through the negative control IgG-ChIP, and no DNA at all (H2O).
Survivin was a direct target of STAT3
We examined whether STAT3 directly regulated survivin. In transient transfection studies with pEGFP-STAT3, we showed that forced expression of STAT3 in MV4-11 cells induced expression of survivin approximately 30-fold calculated by relative quantification RQ-PCR, as compared with pEGFP vector (Figure 5D). To test whether STAT3 could bind the survivin promoter, we performed ChIP assays in MV4-11-R cells. The amplified survivin promoter DNA was present in chromatin immunoprecipitate with an anti-STAT3 antibody (Figure 5E).
In vivo efficacy of IDR E804 in combination with ABT-869 for treatment of MV4-11-R mouse xenografts
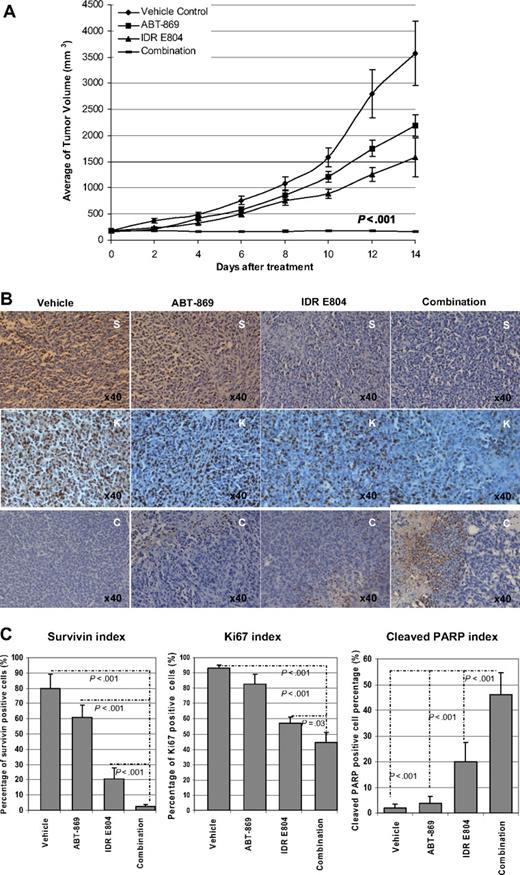
Based on the in vitro results that IDR E804 could sensitize the resistant line to ABT-869, we tested the combination of IDR E804 and ABT-869 in a subcutaneous mouse xenograft model in vivo. MV4-11-R tumors in mice treated with vehicle control developed rapidly up to 3569 plus or minus 619 mm3 after 2 weeks. Growth of tumors in mice treated with a single agent (ABT-869 or IDR E804) was reduced to 2189 plus or minus 211 mm3 and 1588 plus or minus 368 mm3, respectively (Figure 6A). However, in the combination group, tumors size did not increase and was kept at 158 plus or minus 16 mm3 throughout the course of treatment. The antitumor effects of the combination were significantly better compared with single agent or control (all P < .001).
In vivo effect of combination therapy on the MV4-11-R tumor xenograft model. (A) Combination of ABT-869 with IDR E804 achieved impressive regression of tumor growth compared with either vehicle control or single treatment (ABT-869 or IDR E804) alone (all P < .001). (B) Excised tumor pieces from each group were embedded in paraffin and stained with anti-survivin (S), anti-Ki67 (K), and anti–cleaved PARP (C). Photographs are representative of similar observations in 3 different mice receiving same treatment. (C) Quantitative analysis of the expressions of survivin, ki67, and cleaved PARP in IHC sections from each group shown in panel B. The survivin index, ki67 index, and cleaved PARP were calculated as the percentage of positive staining cells of total nucleated cells in a ×400 field. A total of 10 fields for each index were counted. Bars indicate SD. Statistical comparison and associated P values are indicated by the broken lines in each photograph.
In vivo effect of combination therapy on the MV4-11-R tumor xenograft model. (A) Combination of ABT-869 with IDR E804 achieved impressive regression of tumor growth compared with either vehicle control or single treatment (ABT-869 or IDR E804) alone (all P < .001). (B) Excised tumor pieces from each group were embedded in paraffin and stained with anti-survivin (S), anti-Ki67 (K), and anti–cleaved PARP (C). Photographs are representative of similar observations in 3 different mice receiving same treatment. (C) Quantitative analysis of the expressions of survivin, ki67, and cleaved PARP in IHC sections from each group shown in panel B. The survivin index, ki67 index, and cleaved PARP were calculated as the percentage of positive staining cells of total nucleated cells in a ×400 field. A total of 10 fields for each index were counted. Bars indicate SD. Statistical comparison and associated P values are indicated by the broken lines in each photograph.
In addition to reducing TV by approximately 22-fold compared with vehicle control, combination therapy demonstrated significant biochemical effects on MV4-11-R xenografts tumor. Histologic examination of tumor specimens showed that ABT-869 alone had minimal impact on the expression of survivin (Figure 6B top panel), whereas IDR E804 alone triggered a modest decrease in survivin-positive cells (brown color) compared with tumors from vehicle control. However, the combination therapy markedly inhibited the number of survivin-positive cells compared with either single-agent treatment (Figure 6B top panel, 6C left panel; all P < .001). In agreement with these data, a significant decrease in expression of Ki67 (Figure 6B,C middle panels) and an increase in the number of cleaved PARP-positive cells (Figure 6B bottom panel, 6C right panel) were observed in tumor sections from ABT-869 plus IDR E804–treated mice compared with tumors from mice receiving either treatment alone. Together, these data demonstrate a potent in vivo antileukemic effect of ABT-869 in combination with IDR E804 and support the potential clinical utility of combing ABT-869 with inhibitors of the STAT-signaling pathway in the treatment of TKI-resistant AML.
Discussion
FLT3 mutations represent one of the most common genetic lesions in AML. FLT3 inhibitors, like CEP-701, PKC412, MLN518, SU11248, or ABT-869 are in different phases of clinical development as monotherapy or in combination studies.1,2,4,5,7,8,15 It is predictable that patients could develop resistance to receptor tyrosine kinase (RTK) inhibitors after a long period of monotherapy as suggested by the clinical use of Gleevec. Several PMs in the KD were identified in murine Ba-F3-FLT3-ITD cells, which led to resistance to these agents.10,25 It is also found that overexpression of FLT3-ITD proteins in one resistant subline of Ba-F3-ITD lead to resistance to PKC412.26
However, so far, acquired PMs are a rare event in patient samples in FLT3 inhibitor clinical trials.12 Here, for the first time, we report the enhanced activation of STAT pathway and overexpression of survivin as a novel mechanism of resistance to ABT-869 and other FLT3 inhibitors. The resistance can be overcome by inhibition of the STAT pathway or by targeting survivin, thereby inducing MV4-11-R cells to undergo apoptosis and resensitizing them to ABT-869 in vitro and in vivo.
We first excluded the overexpression of multidrug resistant-related efflux proteins such as MDR, MRP1, by flow cytometric analysis, and LRP by Western blot analysis in our MV4-11-R1, -R2, and -R3 cell lines. We also did not find PMs in the FLT3 KD by sequencing analysis. In addition, overexpression of total FLT3 receptors was not evident in the resistant lines. These results are consistent with the findings from Piloto et al in which 3 different human leukemia cell lines and various FLT3 inhibitors were used.13
STAT pathways have been intensively investigated in cancer biology, because they regulate an array of fundamental cell functions such as survival, proliferation, differentiation, apoptosis, and immunity.27 Aberrant activation of STAT pathways, particularly STAT3, STAT5, and less frequency STAT1, has been found in the majority of solid tumors and hematologic malignancies, including AML.28,29 We demonstrated hypermethylation of SOCS genes correlating lower expression status and restored expression by 5-aza treatment in MV4-11-R cells, indicating the epigenetically regulated, transcriptional silencing plays an important role in the development of resistance. SOCS proteins are the part of key pathways that negatively regulate STAT signaling.21 SOCS inhibits STAT pathways either by directly competing for binding with STAT proteins to receptor complex, or by degradation of upstream JAK kinase or competing binding with JAK protein.30 So overactivation of STAT pathways in MV4-11-R cells results from, at least in part, decreasing expression of SOCS molecules as revealed by LDA analysis and rendering their resistance to FLT3 inhibitors. The observation that the activity of PI3K/AKT and MAPK pathways are not enhanced in the resistant lines relative to the parental MV4-11 cells further supports the importance of STAT-mediated resistance in MV4-11-R cells.
Both soluble and membrane-bound FLT3 ligand isoforms are biologically active. FLT3 ligand in conjunction with other cytokine growth factors, like granulocyte colony-stimulating factor (G-CSF), granulocyte macrophage-CSF (GM-CSF), and thrombopoietin (TPO), stimulates survival, proliferation, and differentiation of hematopoietic stem and progenitor cells (HSPC).31 Specifically, FLT3 ligand has potent direct-acting stimulating/costimulating activities on myeloid stem/progenitor cells.32 Compelling evidence shows that the existence of an autocrine FLT3LG/FLT3 loop promotes proliferation and prevents apoptosis of primary AML blasts and AML cell lines.33-35 In MV4-11-R cells, up-regulation of FLT3 ligand triggers a stronger autocrine reaction, thus further enhancing STAT pathway activity and survivin expression. This is also supported by observations of elevated phosphorylated proteins and survivin in the parental MV4-11 cells stimulated with FLT3LG in a cell culture system.
Survivin (encoded by BIRC5), the smallest member of inhibitor of apoptosis protein (IAP) family,36 has been identified as the fourth most highly expressed transcript in cancer37 and is one of the most cancer-specific molecules. Survivin is detected in a broad spectrum of different types of tumors, but is undetectable in most terminally differentiated normal tissues,24 except several normal tissues, particularly those high proliferative and self-renewal rates, ie, hematopoietic cells,38 neuronal stem cells,39 keratinocyte,40 and mucosal epithelial cells.41 Survivin antagonizes apoptosis through stabilization of X-linked IAP (XIAP), another member of IAP family, against proteasomal degradation.24 Overall, strong survivin expression has been associated with shorter disease-free or overall survival in the majority of patients with hematologic malignancies and solid tumors.18,24,42,43 Moreover, survivin proves to be a direct downstream target gene in BCR-ABL positive cells.44,45 Several studies indicate survivin plays an important role in resistance to (1) paclitaxel in ovarian cancer,46 (2) antiandrogen therapy in prostate cancer,47 and (3) doxorubicin in thyroid cancer.48 Here we demonstrate that increased expression of survivin contributes to acquired resistance to a molecularly targeted therapy (an FLT3 inhibitor), expanding its role in mediating resistance to conventional chemotherapy. Survivin has been identified as a direct target of the STAT3 transcription factor in primary effusion lymphoma,49 breast cancer,50 and endothelial cells stimulated with interleukin-11 (IL-11).51 Now we confirm this relationship in AML and provide further understanding that STAT3 directly binds and regulates the survivin promoter. The continuous activation of STAT3 signaling in the FLT3 inhibitor-resistant AML cells enhances the expression of survivin and grants resistance to apoptosis.
STAT pathways and survivin play a pivotal role in oncogenesis and have been validated as targets for cancer therapy.52,53 Targeting survivin by shRNA induced apoptosis and augmented ABT-869–mediated toxicity in MV4-11-R cells. On the contrary, overexpression of survivin in MV4-11 cells leads to remarkable resistance to the panel of FLT3 inhibitors. These results are consistent with the previous finding that silencing survivin by RNA interference (RNAi) restores sensitivity to doxorubicin in resistant thyroid cancer cells.48 IDR E804 has been shown to inhibit the SRC-STAT3 pathway and to down-regulate survivin in breast cancer cells.54 In our study, treatment with IDR E804 prompts MV4-11-R cells to undergo apoptosis as demonstrated by an increase in annexin V–binding assay and in the 89-kDa fragment of PARP, which is responsible for DNA breakage. The inhibitory effect of IDR E804 is not only on STAT3 activity, but it also abolishes STAT1 and STAT5 activity, which could possibly reinforce its cytotoxicity to MV4-11-R cells. A subtherapeutic concentration of IDR E804 signifcantly resensitizes MV4-11-R cells to ABT-869 treatment. This synergism is not evident in the parental MV4-11 cells. The animal experiments provide further evidence to support the therapeutic benefit of targeting STAT pathways and survivin. The dramatic inhibition of tumor growth in mice treated with the combination therapy is correlated with almost complete disappearance of survivin expression and signifcantly increased expression of cleaved PARP, as well as a decrease in the number of Ki67-positive (an indictor of proliferation) cells in tumor specimens from the combination therapy group compared with either single agent treatment alone.
The in vitro coculture resistance model mimics the clinical practice of targeted agents given on a chronic dosing schedule. It recapitulates to a certain extent the clonal heterogeneity in clinical tumors where resistant clones emerge as oligoclonal population and eventually expand, and therefore may reflect the natural course of many cancers, which later relapse after initial therapy. However, it may also signify underlying clonal heterogeneity and other potential resistance mechanism(s) are yet to be identified.
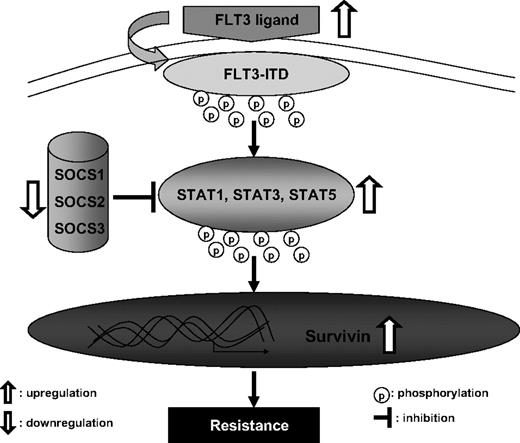
In conclusion, our results suggest a novel mechanism of resistance to the FLT3 inhibitor ABT-869. In this model, depicted in Figure 7, up-regulation of FLT3 ligand and methylation silencing of the SOCS family integrate to enhance STAT signaling activity and overexpression of survivin, in turn suppressing apoptosis and promoting survival, which leads to a resistant phenotype. Understanding the mechanism of resistance to FLT3 inhibitors could help develop new antileukemic agents or uncover compelling combinations. Our data strongly support the combination of FLT3 inhibitors with agents targeting STAT pathway or survivin, such as small molecular inhibitors or shRNA. It may represent a novel strategy to minimize resistance or resensitize resistant cells to FLT3 inhibitors in AML patients with FLT3-ITD mutation.
A model of enhanced STAT activation and overexpression of survivin leading to resistant phenotype in MV4-11-R cells.
A model of enhanced STAT activation and overexpression of survivin leading to resistant phenotype in MV4-11-R cells.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
This work was supported by Singapore Cancer Syndicate grant TN0031, AN0038 (C.-S.C.).
Authorship
Contribution: J.Z. and C.-S.C. conceptualized the original idea and designed the experiments; J.Z. performed the experiments and wrote the paper; J.Z., C.B., K.-G.T., and L.-F.P. contributed to the in vitro experiments; J.Z., S.-C.L., and J.V.J. carried out the animal experiments; J.Z., C.-S.C., C.B., Z.X., S.P., H.Y., and W.-J.C. were involved in discussion and data analysis; and K.B.G., D.H.A., and S.K.D. provided ABT-869.
Conflict-of-interest disclosure: K.B.G., D.H.A., and S.K.D. are employees of Abbott Laboratories, whose potential product was studied in the present work. The remaining authors declare no competing financial interests.
Correspondence: Chien-Shing Chen, Department of Medicine, Yong Loo Lin School of Medicine, National University of Singapore, 5 Lower Kent Ridge Rd, Singapore 119074; e-mail: mdcccs@nus.edu.sg and cschen@llu.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal