Abstract
The small molecule inhibitor of the MDM2/p53 interaction Nutlin-3 significantly up-regulated the steady-state mRNA and protein levels of Notch1 in TP53wild-type (OCI, SKW6.4) but not in TP53deleted (HL-60) or TP53mutated (BJAB) leukemic cell lines. A direct demonstration that NOTCH1 was a transcriptional target of p53 in leukemic cells was obtained in experiments carried out with siRNA for p53. Moreover, inhibition of Notch1 expression using Notch1-specific siRNA significantly increased cytotoxicity in TP53wild-type leukemic cells. Of note, Nutlin-3 up-regulated Notch1 expression also in primary TP53wild-type B-chronic lymphocytic leukemia (B-CLL) cells and the combined use of Nutlin-3 plus pharmacological γ-secretase inhibitors of the Notch signaling showed a synergistic cytotoxicity in both TP53wild-type leukemic cell lines and primary B-CLL cells. A potential drawback of γ-secretase inhibitors was their ability to enhance osteoclastic maturation of normal circulating preosteoclasts induced by RANKL + M-CSF. Notwithstanding, Nutlin-3 completely suppressed osteoclastogenesis irrespective of the presence of γ-secretase inhibitors. Taken together, these data indicate that the p53-dependent up-regulation of Notch1 in response to Nutlin-3 represents an antiapoptotic feedback mechanism able to restrain the potential therapeutic efficacy of Nutlin-3 in hematologic malignancies. Therefore, therapeutic combinations of Nutlin-3 + γ-secretase inhibitors might potentiate the cytotoxicity of Nutlin-3 in p53wild-type leukemic cells.
Introduction
The activation of p53 is tightly regulated by the human homolog of murine double minute 2 (MDM2) gene,1 which is an E3 ubiquitin ligase for p53 and itself and controls p53 half-life mainly via ubiquitin-dependent degradation. In response to a variety of stimuli, such as cellular stress, the p53-MDM2 interaction is disrupted and p53 rapidly accumulates within the cell.1 Potent and selective small molecule inhibitors of the p53-MDM2 interaction, the Nutlins, have been recently reported.2,3 These compounds bind MDM2 in the p53 binding pocket with high selectivity and can release p53 from negative control, leading to effective stabilization of p53 and activation of the p53 pathway.2,3 It has been demonstrated that treatment with the active enantiomer Nutlin-3a results in rising levels of p53 protein and subsequent induction of cell cycle arrest and apoptosis in a variety of tumor cells.2 In contrast to most solid tumors, TP53 is mutated in approximately 10% to 15% of both myeloid and lymphoid leukemias at diagnosis,4 and several recent studies have demonstrated that Nutlin-3 induces ex vivo cytotoxic cell death of most TP53wild-type primary hematologic malignancies, including acute myeloid leukemias, multiple myeloma, B-chronic lymphocytic leukemias (B-CLL), and B-cell lymphomas.5-15
Although it is well established that p53 mediates a variety of cellular functions, such as cell cycle arrest, cellular senescence, and, only as ultimate choice, apoptosis,16,17 some experimental evidence suggests that inhibition of the transcriptional activity of p53 might paradoxically result in an increase of the p53-mediated proapoptotic activity,18 also in B-CLL cells.19 Therefore, the aim of this study was to further investigate the relationship between the transcriptional and cytotoxic activities induced by Nutlin-3 and to elucidate whether Nutlin-3 promotes the transcription of antiapoptotic genes in leukemic cells, which might hamper and/or reduce its potential therapeutic efficacy. In this respect, it has recently emerged that a potential target gene of the p53 pathway is NOTCH1,20 which belongs to an evolutionarily conserved pathway that profoundly impacts mammalian development. Of note, a recent study published while this article was under preparation has demonstrated that circulating B-CLL cells overexpress Notch1 and Notch2 family members with respect to circulating normal B lymphocytes.21 The same study suggested that constitutively activated Notch signaling might be involved in survival and apoptosis resistance of B-CLL cells.21 For the purpose of our present study, it is also of particular interest that Notch family members contribute to the maintenance, renewal, and maturation of normal hematopoietic system,22 and have been implicated in the promotion of different types of cancer, including hematologic malignancies.23-27
On these bases, the aim of the present study was to evaluate the effect of Nutlin-3 treatment on Notch1 expression in both myeloid and lymphoid leukemic cell lines as well as in primary B-CLL cells and the role of Notch signaling in modulating Nutlin-3 cytotoxicity.
Methods
Leukemic cell lines and primary B-CLL cells
The myeloid TP53deleted HL-60 and TP53wild-type OCI and the lymphoid TP53wild-type SKW6.4 and TP53mutated BJAB leukemic cell lines were either purchased from the American Type Culture Collection (ATCC, Manassas, VA) or obtained from DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany). Cells were cultured in RPMI-1640 containing 10% FBS (both from Gibco BRL, Grand Island, NY).
For experiments with primary cells, peripheral blood samples were collected in heparin-coated tubes from 27 B-CLL patients (Table 1) and from healthy blood donors following informed consent, in accordance with the Declaration of Helsinki and in agreement with the institutional guidelines of the University Hospital of Udine. The diagnosis of B-CLL was made by peripheral blood morphology and immunophenotyping. T lymphocytes, natural killer (NK) lymphocytes, granulocytes, and monocytes were negatively depleted from peripheral blood leucocytes (PBLs) with immunomagnetic microbeads (MACS microbeads; Miltenyi Biotech, Auburn, CA), with a purity more than 95% of resulting CD19+ B-CLL population, as assessed by flow cytometry.
Clinical and laboratory features of patients with CLL
| Patient no. . | Sex . | Age, y . | Rai stage . | ZAP70* . | FISH† . | TP53 . |
|---|---|---|---|---|---|---|
| 1 | F | 82 | 1 | Absent/low | 13q− | Mut (exon V) |
| 2 | M | 66 | 4 | Interm/high | ND | WT |
| 3 | M | 65 | 1 | Absent/low | Nor | WT |
| 4 | M | 68 | 4 | Interm/high | 12+ | WT |
| 5 | F | 62 | 0 | Absent/low | Nor | WT |
| 6 | M | 64 | 1 | Absent/low | 13q−/11q− | WT |
| 7 | M | 67 | 2 | Interm/high | 17p− | WT |
| 8 | F | 78 | 2 | Absent/low | 11q− | WT |
| 9 | M | 51 | 0 | Absent/low | Nor | WT |
| 10 | F | 59 | 0 | Absent/low | Nor | WT |
| 11 | M | 69 | 2 | Absent/low | 13q−/17p− | WT |
| 12 | M | 40 | 4 | Interm/high | 11q− | WT |
| 13 | M | 74 | 2 | Interm/high | 13q− | WT |
| 14 | M | 62 | 4 | Interm/high | 17p− | WT |
| 15 | M | 62 | 0 | Absent/low | 11q− | WT |
| 16 | M | 69 | 2 | Absent/low | 17p− | Mut (exon VII) |
| 17 | F | 79 | 3 | Absent/low | 12+ | WT |
| 18 | M | 61 | 2 | Interm/high | 17p− | WT |
| 19 | M | 63 | 4 | Absent/low | 13q− | WT |
| 20 | M | 60 | 4 | Absent/low | 17p− | WT |
| 21 | M | 59 | 2 | Absent/low | 13q− | WT |
| 22 | M | 64 | 1 | Interm/high | 13q− | WT |
| 23 | M | 57 | 1 | Absent/low | 12+ | WT |
| 24 | M | 67 | 4 | ND | ND | WT |
| 25 | F | 68 | 2 | Interm/high | 17p− | Mut (exon V) |
| 26 | F | 55 | 0 | Absent/low | ND | WT |
| 27 | M | 61 | 2 | Absent/low | 17p− | Mut (exon VI) |
| Patient no. . | Sex . | Age, y . | Rai stage . | ZAP70* . | FISH† . | TP53 . |
|---|---|---|---|---|---|---|
| 1 | F | 82 | 1 | Absent/low | 13q− | Mut (exon V) |
| 2 | M | 66 | 4 | Interm/high | ND | WT |
| 3 | M | 65 | 1 | Absent/low | Nor | WT |
| 4 | M | 68 | 4 | Interm/high | 12+ | WT |
| 5 | F | 62 | 0 | Absent/low | Nor | WT |
| 6 | M | 64 | 1 | Absent/low | 13q−/11q− | WT |
| 7 | M | 67 | 2 | Interm/high | 17p− | WT |
| 8 | F | 78 | 2 | Absent/low | 11q− | WT |
| 9 | M | 51 | 0 | Absent/low | Nor | WT |
| 10 | F | 59 | 0 | Absent/low | Nor | WT |
| 11 | M | 69 | 2 | Absent/low | 13q−/17p− | WT |
| 12 | M | 40 | 4 | Interm/high | 11q− | WT |
| 13 | M | 74 | 2 | Interm/high | 13q− | WT |
| 14 | M | 62 | 4 | Interm/high | 17p− | WT |
| 15 | M | 62 | 0 | Absent/low | 11q− | WT |
| 16 | M | 69 | 2 | Absent/low | 17p− | Mut (exon VII) |
| 17 | F | 79 | 3 | Absent/low | 12+ | WT |
| 18 | M | 61 | 2 | Interm/high | 17p− | WT |
| 19 | M | 63 | 4 | Absent/low | 13q− | WT |
| 20 | M | 60 | 4 | Absent/low | 17p− | WT |
| 21 | M | 59 | 2 | Absent/low | 13q− | WT |
| 22 | M | 64 | 1 | Interm/high | 13q− | WT |
| 23 | M | 57 | 1 | Absent/low | 12+ | WT |
| 24 | M | 67 | 4 | ND | ND | WT |
| 25 | F | 68 | 2 | Interm/high | 17p− | Mut (exon V) |
| 26 | F | 55 | 0 | Absent/low | ND | WT |
| 27 | M | 61 | 2 | Absent/low | 17p− | Mut (exon VI) |
Negative (−) indicates deletion; Mut, mutated; ND, not done; Nor, normal cytogenetics; and positive (+), trisomy.
ZAP-70 expression was determined by Western blot analysis.
FISH defects were found using a B-CLL FISH panel.
Culture treatments and assessment of cell viability and apoptosis
Both leukemic cell lines and primary B-CLL cells were seeded at a density of 1 × 106 cells/mL before treatment with Nutlin-3 (Cayman Chemical, Ann Arbor, MI), pifithrin-α (PFT-α; Calbiochem, Nottingham, United Kingdom), and the γ-secretase inhibitors L-685,458 and DAPT (both from Sigma-Aldrich, St Louis, MO), used either alone or in various combinations as indicated.
Cell viability was examined at different time points after treatment by trypan blue dye exclusion and confirmed by MTT assay, whereas the degree of apoptosis was quantified by annexin V-FITC/propidium iodide (PI) staining (Immunotech, Marseille, France) followed by flow cytometry analysis, as previously detailed.28
RNA analyses
Total RNA was extracted using the RNeasy Plus Mini Kit (QIAGEN, Hilden, Germany) that also removes chromatin DNA. Total RNA was retrotranscribed into cDNA, using the AccuScript high fidelity 1st strand cDNA synthesis kit (Stratagene, La Jolla, CA). To ascertain the TP53 status, mutation analysis for cDNA of TP53 was performed analyzing 3 overlapping fragments spanning the TP53 coding region of exons II-X, which were amplified by polymerase chain reaction (PCR), purified, and sequenced. The primer sequences were as follows: no. 1, AGT CAG ATC CTA GCG TCG AG (14-33); no. 2, GGA GTA CGT GCA AGT CAC AG (381-362); no. 3, TGC ACC AGC AGC TCC TAC AC (225-244); no. 4, ACA GTC AGA GCC AAC CTC AG (687-668); no. 5, TTC GAC ATA GTG TGG TGG TG (635-654); no. 6, GAA TGT CAG TCT GAG TCA GG (1187-1168). Numbers refer to the human TP53 cDNA sequence.
Modulation of the NOTCH1 gene expression upon Nutlin-3 treatment was assessed with the real-time thermal analyzer Rotor-Gene 6000 (Corbett, Cambridge, United Kingdom) using SYBR Green based-technology and the quantitative PCR primer set for human Notch1 cDNA (SABioscience, Frederick, MD).
Western blot analyses
Cells were harvested in lysis buffer containing 1%Triton X-100, Pefablock (1 mM), aprotinin (10 μg/mL), pepstatin (1 μg/mL), leupeptin (10 μg/mL), NaF (10 mM), and Na3VO4 (1 mM), as described.29 Protein determination was performed by Bradford assay (Bio-Rad, Richmond, CA). Equal amounts of protein for each sample were migrated in acrylamide gels and blotted onto nitrocellulose filters.
The following antibodies were used in our experiments: MoAb anti-p53 (DO-1) and MoAb anti-Notch1 (nN1A; both purchased from Santa Cruz Biotechnology, Santa Cruz, CA), and MoAb antitubulin (Sigma-Aldrich). After incubation with peroxidase-conjugated anti–mouse IgG, specific reactions were revealed with the enhanced chemiluminescence (ECL) detection kit (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom). Densitometry values were estimated by the ImageQuant TL software (Amersham Pharmacia Biotech). Multiple film exposures were used to verify the linearity of the samples analyzed and to avoid saturation of the film.
Transfection experiments
Cells (1.25 × 106) were resuspended into 0.1 mL Nucleofector solution V of human nucleofector kit V (Amaxa, Cologne, Germany). Plasmid DNA (2 μg; GFP-construct) or 1 μg siRNA was mixed with the 0.1 mL of cell suspension, transferred into a 2.0-mm electroporation cuvette, and nucleofected using an Amaxa Nucleofector II apparatus, following the manufacturer's guidelines. After transfection, cells were immediately transferred into complete medium and cultured in 6-well plates at 37°C. Transfection efficiency was estimated in each experiment by scoring the number of GFP-positive cells by flow cytometry analysis. SiRNAs were designed and manufactured by Ambion (Austin, TX) according to the current guidelines for effective gene knockdown by this method. Based on preliminary validation experiments, the following siRNAs were selected: target no. 1 for TP53, 5′-GGGAGUUGUCAAGUCUUGCtt-3′ (sense) and 5′-GCAAGACUUGACAACUCCCtc-3′ (antisense); target no. 2 for TP53, 5′-GGGUUAGUUUACAAUCAGCtt-3′ (sense) and 5′-GCUGAUUGUAAACUAACCCtt-3′ (antisense); target no. 1 for NOTCH1, 5′-GGUGUGCACUGUGAGAUCAtt-3′ (sense) and 5′-UGAUCUCACAGUGCACACCct-3′ (antisense); target no. 2 for NOTCH1, 5′-GGACUGUGCGGAGCAUGUAtt-3′ (sense) and 5′-UACAUGCUCCGCACAGUCCag-3′ (antisense); target no. 3 for NOTCH1, 5′-UCGUCUACCUGGAGAUUGAtt-3′ (sense) and 5′-UCAAUCUCCAGGUAGACGAtg-3′ (antisense). A cocktail of 3 different negative control siRNAs, each composed of a 19 bp-scrambled sequence with 3′ dT overhangs (Ambion's Silencer negative control siRNA), was used to demonstrate that the transfection did not induce nonspecific effects on gene expression. The Ambion's Silencer negative control siRNA sequences have no significant homology to any known gene sequences from humans, and they have been previously tested for the lack of nonspecific effects on gene expression (Ambion).
Evaluation of osteoclastic differentiation
As model system of human osteoclastogenesis, we have used peripheral blood mononuclear cells (PBMCs), obtained from healthy blood donors, as previously described.30,31 Briefly, adherent PBMCs were cultured in RPMI medium containing 10% FBS and treated with 50 ng/mL human macrophage-colony stimulating factor (M-CSF; PeproTech, London, United Kingdom) alone for 6 days, followed by M-CSF (50 ng/mL) plus RANKL (50 ng/mL; Alexis Biochemicals, Lausen, Switzerland) for additional 12 days, replacing the medium containing fresh cytokines every 3 days. Nutlin-3 and L-685,458, either alone or in combination, were added to the cultures simultaneously with RANKL + M-CSF and at every medium replacement.
At the end of the culture time, the degree of osteoclastic differentiation was investigated by staining adherent cells for TRAP, using the leukocyte acid phosphatase kit (387-A; Sigma-Aldrich) according to the manufacturer's instructions, and for 4,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich), as previously described.30,31 After the stainings, cells were photographed under a light (for TRAP; Eclipse TE200 Inverted microscope; Nikon, Tokyo, Japan) or fluorescence (for DAPI; Axiophot 100; Zeiss, Jena, Germany) microscope and scoring of TRAP-positive and multinucleated cells was performed by examination of at least 8 independent fields.
In parallel, the culture supernatants were analyzed for the levels of osteoclast-derived tartrate-resistant acid phosphatase form 5b, a product of osteoclast activity. The assay used (TRACP 5b; Immunodiagnostic systems, Fountain Hills, AZ) detects only the active TRACP molecules, giving an accurate estimation of bone resorption activity. Results were read at an optical density of 450 nm using an Anthos 2010 enzyme-linked immunosorbent assay (ELISA) reader (Anthos Labtec Instruments, Wals Salzburg, Austria). Measurements were done in duplicate and corrected for the dilution factors.
Statistical analysis and assessment of the effect of combination treatment
The results were evaluated using analysis of variance with subsequent comparisons by Student t test and with the Mann-Whitney rank-sum test. Statistical significance was defined as P value less than .05. To investigate the effect of Nutlin-3 plus L-685,458 combination, leukemic cells were then treated with serial 2-fold dilutions of Nutlin-3 (from 10 to 1.25 μM) or L-685,458 (from 20 to 2.5 μM), individually or in combination using a constant ratio (Nutlin-3/L-685,458) of 1:2, for 24 hours. Dilutions of each compound were performed starting from the maximal concentrations of Nutlin-3 (10 μM) and L-685,458 (20 μM), which avoid aspecific cytotoxicity, as determined in preliminary experiments. Results were analyzed using the CalcuSyn software program (Biosoft, Cambridge, United Kingdom), which uses the method of Chou and Talalay,32 to determine whether combined treatment yields greater effects than expected from summation alone. A combination index (CI) of 1 indicates an additive effect, whereas a CI below 1 indicates synergism.
Results
Nutlin-3 selectively up-regulates Notch1 in p53wild-type leukemic cell lines
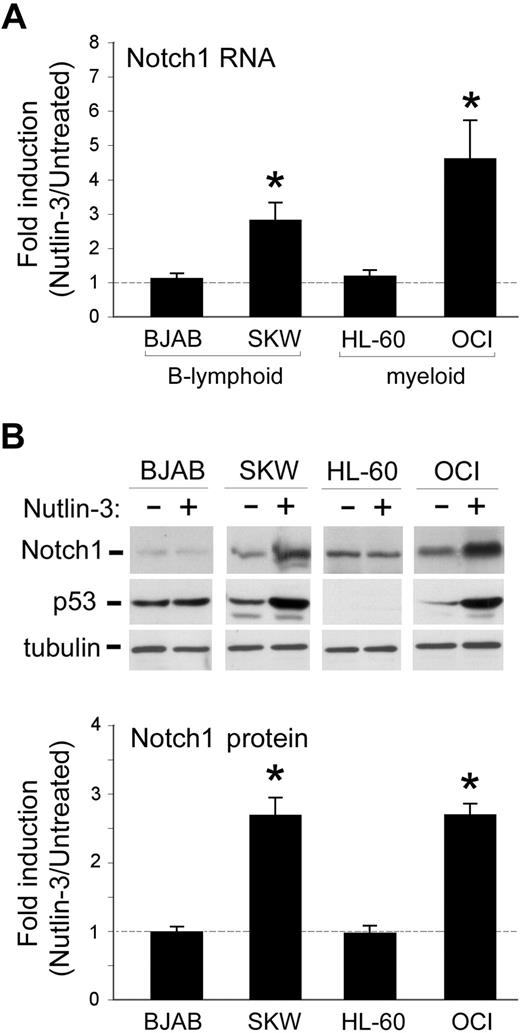
In the first set of experiments, we sought to investigate whether Nutlin-3 was able to affect the steady-state mRNA levels of Notch1 in B lymphoblastoid (SKW6.4 and BJAB) and myeloid (OCI, HL-60) cell lines. Quantitative reverse-transcription (RT)–PCR analysis demonstrated that the exposure for 24 hours to 10 μM Nutlin-3 selectively (P < .01) up-regulated the mRNA levels of Notch1 in SKW6.4 and OCI but not in BJAB and HL-60 cell lines (Figure 1A). Western blot analysis confirmed that the up-regulation of Notch1 mRNA was accompanied by a significant (P < .01) accumulation of the transmembrane/cytoplasmic (120-kDa) portion of Notch1 protein in response to Nutlin-3 in SKW6.4 and OCI leukemic cells (Figure 1B). Because the best characterized biologic activity of Nutlin-3 is to disrupt the interaction between MDM2 and p53 proteins and to prevent p53 from ubiquitination,1 changes in p53 protein were also investigated in the different leukemic cell lines before and after treatment with Nutlin-3 (Figure 1B). The amount of p53 in cell lysates obtained from untreated TP53wild-type SKW6.4 and OCI cells markedly increased after exposure to 10 μM Nutlin-3 for 24 hours. On the other hand, HL-60 cells carrying TP53deleted or BJAB carrying TP53mutated exhibited absent and high basal levels of p53, respectively (Figure 1B). As expected on the basis of previous findings obtained on a variety of hematologic malignancies,5-15 exposure to Nutlin-3 did not modulate p53 levels in either TP53deleted HL-60 or TP53mutated BJAB cell lines (Figure 1B).
Effect of Nutlin-3 on Notch1 mRNA expression levels in leukemic cell lines. B-lymphoid (BJAB and SKW6.4) and myeloid (HL-60 and OCI) leukemic cells were either left untreated or exposed for 24 hours to Nutlin-3 (10 μM). Levels of Notch1 mRNA and protein were analyzed by quantitative RT-PCR (A) and Western blot analysis (B), respectively. (A) For each cell line, after normalization to the level of GAPDH mRNA, results were expressed as fold of Notch1 mRNA induction in Nutlin-3–treated cultures with respect to the control untreated cultures. Data are reported as means ± SD of results from 3 to 5 independent experiments each performed in duplicate. (B) The levels of Notch1 and of p53 proteins were assessed by Western blot analysis in cell lysates. Tubulin staining is shown as a loading control. Representative examples of Western blot results of 5 independent experiments are shown. After densitometric analyses, results were expressed as fold of Notch1 protein induction in Nutlin-3–treated cultures with respect to the control untreated cultures. Data are reported as means plus or minus SD of results from 5 independent experiments. *P < .05 with respect to untreated cells.
Effect of Nutlin-3 on Notch1 mRNA expression levels in leukemic cell lines. B-lymphoid (BJAB and SKW6.4) and myeloid (HL-60 and OCI) leukemic cells were either left untreated or exposed for 24 hours to Nutlin-3 (10 μM). Levels of Notch1 mRNA and protein were analyzed by quantitative RT-PCR (A) and Western blot analysis (B), respectively. (A) For each cell line, after normalization to the level of GAPDH mRNA, results were expressed as fold of Notch1 mRNA induction in Nutlin-3–treated cultures with respect to the control untreated cultures. Data are reported as means ± SD of results from 3 to 5 independent experiments each performed in duplicate. (B) The levels of Notch1 and of p53 proteins were assessed by Western blot analysis in cell lysates. Tubulin staining is shown as a loading control. Representative examples of Western blot results of 5 independent experiments are shown. After densitometric analyses, results were expressed as fold of Notch1 protein induction in Nutlin-3–treated cultures with respect to the control untreated cultures. Data are reported as means plus or minus SD of results from 5 independent experiments. *P < .05 with respect to untreated cells.
Silencing of TP53 counteracts the ability of Nutlin-3 to up-regulate Notch1
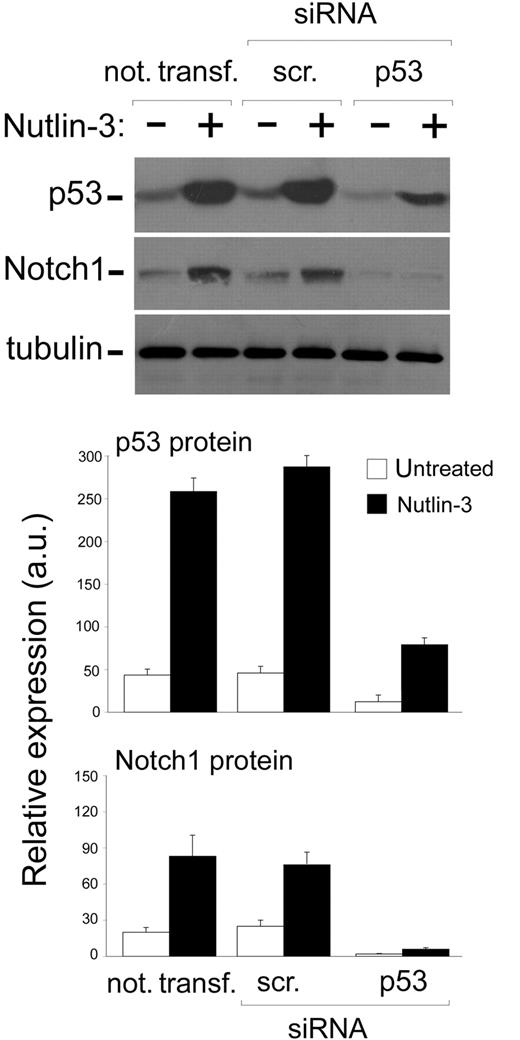
Since it has been shown that p53 might be involved in the up-regulation of Notch1 in human prostate and breast cancer cell lines,20 next, experiments were carried out to ascertain whether Nutlin-3 up-regulated Notch1 expression in leukemic cells via p53. For this purpose, we have used predetermined optimal experimental conditions for siRNA transfection to specifically attenuate p53 gene expression. The efficiency and specificity of the TP53 knockdown was documented at the protein level by a significant decrease of p53 accumulation upon treatment with Nutlin-3 in cells transfected with siRNA for p53 but not in cells transfected with control scrambled siRNA (Figure 2). Of note, TP53 knockdown specifically abrogated the ability of Nutlin-3 to induce the accumulation not only of p53 but also of Notch1 protein in cell lysates (Figure 2), strongly suggesting that the induction of Notch1 by Nutlin-3 was mediated by p53 also in leukemic cells.
Effect of TP53 silencing on the ability of Nutlin-3 to up-regulate Notch1 in leukemic cell lines. OCI cells were either not transfected or transfected with control scrambled (scr) siRNA or p53 siRNA before treatment with Nutlin-3 (10 μM), as indicated. Levels of p53 and Notch1 proteins were assessed by Western blot analysis of cell lysates; tubulin staining is shown as loading control. Representative examples of Western blot results of 3 independent experiments are shown. After densitometric analyses, p53 and Notch1 protein levels were expressed as arbitrary units (au). Data are reported as means ± SD of results from 3 independent experiments.
Effect of TP53 silencing on the ability of Nutlin-3 to up-regulate Notch1 in leukemic cell lines. OCI cells were either not transfected or transfected with control scrambled (scr) siRNA or p53 siRNA before treatment with Nutlin-3 (10 μM), as indicated. Levels of p53 and Notch1 proteins were assessed by Western blot analysis of cell lysates; tubulin staining is shown as loading control. Representative examples of Western blot results of 3 independent experiments are shown. After densitometric analyses, p53 and Notch1 protein levels were expressed as arbitrary units (au). Data are reported as means ± SD of results from 3 independent experiments.
Silencing of Notch1 increases the Nutlin-3–mediated cytotoxicity in leukemic cell lines
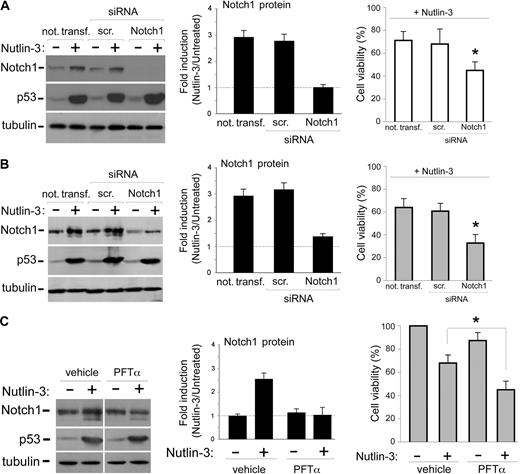
Notch1 has been shown to play a significant role in promoting leukemic development mainly through inhibition of apoptosis.21-25 Therefore, the data illustrated in the previous paragraphs suggested that the Notch1 up-regulation might represent a regulatory feedback antiapoptotic mechanism restraining the cytotoxic effects of p53 activation induced by Nutlin-3. Therefore, to investigate whether the inhibition of Notch1 might potentiate the Nutlin-3–mediated apoptosis, we have used siRNAs to attenuate Notch1 expression. Knockdown of Notch1 expression was demonstrated by Western blot, documenting a significant reduction of Notch1 protein both at the basal level as well as in response to Nutlin-3 in both OCI (Figure 3A) and SKW6.4 (Figure 3B) cells, transfected with a cocktail of Notch1-specific siRNAs. On the other hand, the ability of Nutlin-3 to induce the accumulation of p53 in OCI and SKW6.4 cells was not affected by Notch1 siRNAs (Figure 3A,B). Of note, in OCI and SKW6.4 samples in which Notch1 expression was knocked down by transfection with Notch1-specific siRNAs, the cytotoxicity of Nutlin-3 was significantly (P < .05) increased with respect to either cells transfected with a scrambled control siRNA or cells not transfected (Figure 3A,B).
Effects of Notch1 silencing and of inhibition of p53 transcriptional activity on Nutlin-3–mediated cytotoxicity in leukemic cell lines. (A,B) OCI and SKW6.4 cells were either not transfected or transfected with control scrambled (scr) siRNA or Notch1 siRNA before treatment with Nutlin-3 (10 μM), as indicated. (C) SKW6.4 cells were preincubated either with control vehicle or with PFTα (25 μM) before treatment with Nutlin-3 (10 μM). Levels of Notch1 and p53 proteins were assessed by Western blot analysis of cell lysates; tubulin staining is shown as loading control. Representative examples of Western blot results of 3 independent experiments for each cell line are shown. After densitometric analyses, for each of the indicated experimental conditions, results were expressed as fold of Notch1 protein induction in Nutlin-3–treated cultures with respect to the untreated cultures. Data are reported as means ± SD of results from 3 independent experiments. In parallel, cell viability in Nutlin-3–treated cells was calculated as percentage with respect to the untreated cultures in each of the indicated experimental conditions. Data are reported as means ± SD of results from 3 independent experiments. A,B: *P < .05 with respect to either cells transfected with a scrambled (scr) control siRNA or cells not transfected. C: *P < .05 with respect to Nutlin-3–treated cells.
Effects of Notch1 silencing and of inhibition of p53 transcriptional activity on Nutlin-3–mediated cytotoxicity in leukemic cell lines. (A,B) OCI and SKW6.4 cells were either not transfected or transfected with control scrambled (scr) siRNA or Notch1 siRNA before treatment with Nutlin-3 (10 μM), as indicated. (C) SKW6.4 cells were preincubated either with control vehicle or with PFTα (25 μM) before treatment with Nutlin-3 (10 μM). Levels of Notch1 and p53 proteins were assessed by Western blot analysis of cell lysates; tubulin staining is shown as loading control. Representative examples of Western blot results of 3 independent experiments for each cell line are shown. After densitometric analyses, for each of the indicated experimental conditions, results were expressed as fold of Notch1 protein induction in Nutlin-3–treated cultures with respect to the untreated cultures. Data are reported as means ± SD of results from 3 independent experiments. In parallel, cell viability in Nutlin-3–treated cells was calculated as percentage with respect to the untreated cultures in each of the indicated experimental conditions. Data are reported as means ± SD of results from 3 independent experiments. A,B: *P < .05 with respect to either cells transfected with a scrambled (scr) control siRNA or cells not transfected. C: *P < .05 with respect to Nutlin-3–treated cells.
In additional experiments, SKW6.4 cells were preincubated with PFTα (25 μM), a pharmacological inhibitor that has been shown to block p53-mediated transcription in B-CLL cells.19 Preincubation with PFTα abrogated Notch1 induction by Nutlin-3 without affecting the levels of p53 (Figure 3C) and resulted in a significant (P < .05) increase of Nutlin-3 cytotoxicity. Overall, these experiments strongly suggest that NOTCH1 was a downstream transcriptional target of p53.
Nutlin-3 up-regulates Notch1 also in primary B-CLL cells
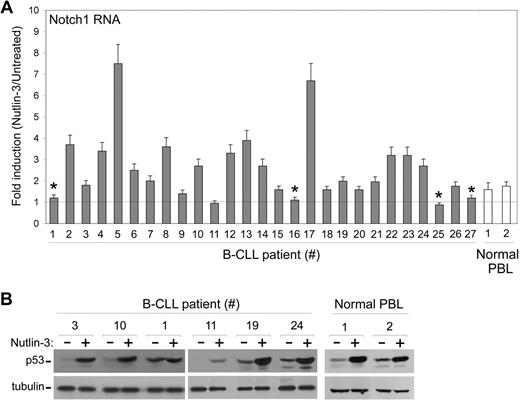
To exclude the possibility that the data illustrated above were confined to leukemic cell lines, next experiments were carried out using a series of primary B-CLL samples, obtained from 27 patients all characterized for Rai stage, ZAP70 levels, fluorescent in situ hybridization (FISH) analysis, and TP53 mutational status (Table 1). Nutlin-3 variably up-regulated the steady-state mRNA levels of Notch1 also in the majority of primary B-CLL samples examined (Figure 4A). Of note, all B-CLL samples characterized for having a mutated p53 (samples nos. 1, 16, 25, and 27 of Table 1 and Figure 4A) showed an absent induction of Notch1 mRNA in response to Nutlin-3. Consistently with the data obtained in leukemic cell lines, although TP53wild-type B-CLL samples displayed a significant accumulation of p53 in response to Nutlin-3 (exemplified by samples nos. 3, 10, 19, and 24 of Table 1 and Figure 4B), TP53mutated B-CLL samples displayed high basal levels of p53, which were not modified by Nutlin-3 (exemplified by sample no. 1 of Table 1 and Figure 4B). Moreover, in samples characterized by a high percentage of 17p− deletion at FISH analysis (exemplified by sample no. 11 of Table 1), Nutlin-3 poorly induced p53 accumulation (Figure 4B) and Notch1 mRNA expression (Figure 4A). Taken together, these data strongly suggest that Nutlin-3 up-regulates Notch1 in a p53-dependent manner also in primary B-CLL cells.
Modulation of Notch1 mRNA by Nutlin-3 in primary B-CLL patient samples. After exposure to Nutlin-3 for 24 hours, samples from B-CLL and from normal PBLs were analyzed for Notch1 mRNA levels (A) and for p53 accumulation (B). (A) Levels of Notch1 mRNA were analyzed by quantitative RT-PCR. After normalization to the level of GAPDH mRNA, results were expressed as fold of Notch1 mRNA induction in Nutlin-3–treated cultures with respect to the control untreated cultures. Data are reported as means ± SD of results from experiments each performed in triplicate. *TP53mutated B-CLL samples. (B) p53 protein levels, analyzed by Western blot, are shown for representative B-CLL and normal PBL samples. Tubulin staining is shown as loading control.
Modulation of Notch1 mRNA by Nutlin-3 in primary B-CLL patient samples. After exposure to Nutlin-3 for 24 hours, samples from B-CLL and from normal PBLs were analyzed for Notch1 mRNA levels (A) and for p53 accumulation (B). (A) Levels of Notch1 mRNA were analyzed by quantitative RT-PCR. After normalization to the level of GAPDH mRNA, results were expressed as fold of Notch1 mRNA induction in Nutlin-3–treated cultures with respect to the control untreated cultures. Data are reported as means ± SD of results from experiments each performed in triplicate. *TP53mutated B-CLL samples. (B) p53 protein levels, analyzed by Western blot, are shown for representative B-CLL and normal PBL samples. Tubulin staining is shown as loading control.
The combination of Nutlin-3 plus γ-secretase inhibitors promotes synergistic cytotoxicity in both leukemic cell lines and primary B-CLL cells
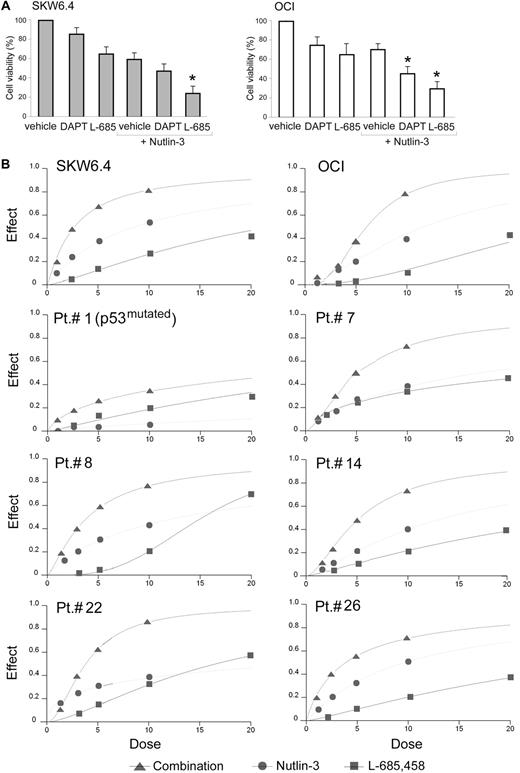
To analyze the potential role of Notch signaling in modulating the survival/apoptotic response of leukemic cells to Nutlin-3, we used a pharmacological approach using the γ-secretase inhibitors DAPT and L-685,458, which block the activation of Notch receptors, and induce phenotypes similar to NOTCH-loss-of-function mutations.33-35 Predetermined optimal concentrations of DAPT and L-685,458 in SKW6.4 and OCI cells induced a variable decrease of leukemic viability and enhanced Nutlin-3 cytotoxicity (Figure 5A).Among the 2 γ-secretase inhibitors used, L-685,458 exhibited more powerful cytotoxicity both alone and in combination with Nutlin-3.
Evaluation of cytotoxicity by Nutlin-3 and γ-secretase inhibitors used alone or in combination in leukemic cells. OCI and SKW6.4 cells were exposed for 24 hours to Nutlin-3 (10 μM), DAPT (20 μM), or L-685,458 (20 μM), used either alone or in combination, as indicated. Cell viability was calculated as percentage with respect to the control (vehicle) cultures. Data are reported as means ± SD of results from 3 independent experiments. *P < .05 with respect to cultures treated with vehicle + Nutlin-3. (B) Leukemic cells were exposed to serial doses of Nutlin-3 or L-685,458, used either alone or in combination, with a fixed ratio, for 24 hours. Dose-effect plots to determine drug efficacy are shown for SKW6.4, OCI, representative TP53wild-type B-CLL samples, and for one TP53mutated B-CLL (patient no. 1). The decrease of cell viability, labeled “effect” on the y-axis, was determined in assays done at least twice in duplicate.
Evaluation of cytotoxicity by Nutlin-3 and γ-secretase inhibitors used alone or in combination in leukemic cells. OCI and SKW6.4 cells were exposed for 24 hours to Nutlin-3 (10 μM), DAPT (20 μM), or L-685,458 (20 μM), used either alone or in combination, as indicated. Cell viability was calculated as percentage with respect to the control (vehicle) cultures. Data are reported as means ± SD of results from 3 independent experiments. *P < .05 with respect to cultures treated with vehicle + Nutlin-3. (B) Leukemic cells were exposed to serial doses of Nutlin-3 or L-685,458, used either alone or in combination, with a fixed ratio, for 24 hours. Dose-effect plots to determine drug efficacy are shown for SKW6.4, OCI, representative TP53wild-type B-CLL samples, and for one TP53mutated B-CLL (patient no. 1). The decrease of cell viability, labeled “effect” on the y-axis, was determined in assays done at least twice in duplicate.
To better define the nature of the pharmacological interaction between Nutlin-3 and L-685,458 on leukemic cell viability, a fixed ratio of these molecules was analyzed, as exemplified in Figure 5B. Based on the availability of B-CLL samples, the Chou and Talalay median-effect method was used to determine drug efficacy and the nature of the drug interaction for 16 of 27 B-CLL samples, as well as for SKW6.4 and OCI cell lines (Table 2). The interactions were judged to be synergistic, with CI less than 1, for SKW6.4 and OCI cell lines as well as for all TP53wild-type B-CLL samples (Table 2). Interestingly, the synergistic cytotoxicity was observed also in B-CLL samples carrying unfavorable cytogenetic abnormalities (samples nos. 6 and 8 carrying 11q− and samples nos. 7, 14, and 18 carrying 17p−). Of note, B-CLL sample no. 11 with a CI of 0.98, more additive than synergistic (Table 2), was characterized by a double cytogenetic abnormality (13q−/17p−, Table 1), and a poor accumulation of p53 protein (Figure 4B) as well as absent induction of Notch1 mRNA (Figure 4A) in response to Nutlin-3. The only TP53mutated B-CLL included in this analysis was sample no. 1, which showed a CI more than 1 (Table 2).
Combination index values for effects of Nutlin-3 and γ-secretase inhibitor L-685,458 on cell viability
| Sample . | ED50 . | ED75 . | ED90 . | Average CI* . |
|---|---|---|---|---|
| SKW6.4 | 0.22 | 0.35 | 0.76 | 0.44 |
| OCI | 0.74 | 0.60 | 0.48 | 0.61 |
| Patient no. 1 | 0.73 | 1.31 | 2.36 | 1.47 |
| Patient no. 2 | 0.42 | 0.81 | 1.59 | 0.94 |
| Patient no. 3 | 0.52 | 0.50 | 0.54 | 0.52 |
| Patient no. 6 | 0.35 | 0.38 | 0.52 | 0.42 |
| Patient no. 7 | 0.24 | 0.38 | 0.62 | 0.30 |
| Patient no. 8 | 0.56 | 0.63 | 0.84 | 0.68 |
| Patient no. 9 | 0.64 | 0.82 | 1.05 | 0.84 |
| Patient no. 11 | 0.40 | 0.93 | 1.61 | 0.98 |
| Patient no. 13 | 0.21 | 0.45 | 0.96 | 0.54 |
| Patient no. 14 | 0.56 | 0.46 | 0.38 | 0.47 |
| Patient no. 17 | 0.41 | 0.47 | 0.53 | 0.47 |
| Patient no. 18 | 0.70 | 0.75 | 0.95 | 0.80 |
| Patient no. 22 | 0.37 | 0.23 | 0.17 | 0.26 |
| Patient no. 23 | 0.24 | 0.38 | 0.62 | 0.41 |
| Patient no. 24 | 0.26 | 0.80 | 0.51 | 0.86 |
| Patient no. 26 | 0.52 | 0.60 | 0.70 | 0.61 |
| Sample . | ED50 . | ED75 . | ED90 . | Average CI* . |
|---|---|---|---|---|
| SKW6.4 | 0.22 | 0.35 | 0.76 | 0.44 |
| OCI | 0.74 | 0.60 | 0.48 | 0.61 |
| Patient no. 1 | 0.73 | 1.31 | 2.36 | 1.47 |
| Patient no. 2 | 0.42 | 0.81 | 1.59 | 0.94 |
| Patient no. 3 | 0.52 | 0.50 | 0.54 | 0.52 |
| Patient no. 6 | 0.35 | 0.38 | 0.52 | 0.42 |
| Patient no. 7 | 0.24 | 0.38 | 0.62 | 0.30 |
| Patient no. 8 | 0.56 | 0.63 | 0.84 | 0.68 |
| Patient no. 9 | 0.64 | 0.82 | 1.05 | 0.84 |
| Patient no. 11 | 0.40 | 0.93 | 1.61 | 0.98 |
| Patient no. 13 | 0.21 | 0.45 | 0.96 | 0.54 |
| Patient no. 14 | 0.56 | 0.46 | 0.38 | 0.47 |
| Patient no. 17 | 0.41 | 0.47 | 0.53 | 0.47 |
| Patient no. 18 | 0.70 | 0.75 | 0.95 | 0.80 |
| Patient no. 22 | 0.37 | 0.23 | 0.17 | 0.26 |
| Patient no. 23 | 0.24 | 0.38 | 0.62 | 0.41 |
| Patient no. 24 | 0.26 | 0.80 | 0.51 | 0.86 |
| Patient no. 26 | 0.52 | 0.60 | 0.70 | 0.61 |
ED indicates effect dose.
The averaged combination index (CI) values were calculated from ED50, ED75, and ED90.
Nutlin-3 efficiently counteracts the pro-osteoclastic activity of L-685,458
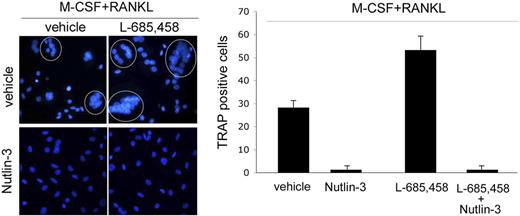
A potential concern in considering the therapeutic use of γ-secretase inhibitors in the antileukemic therapy is the reported ability of Notch1 signaling pathway to suppress osteoclastogenesis.36,37 Consistently with these previously published data,36,37 we found that L-685,458 significantly (P < .05) increased the number of mature osteoclasts, when added to peripheral blood adherent mononuclear cells simultaneously with M-CSF + RANKL (Figure 6). As previously observed in leukemic cells, exposure to Nutlin-3 induced Notch1 expression in normal preosteoclasts (data not shown). However, Nutlin-3 either added alone or in combination with L-685,458 completely suppressed the formation of multinucleated (Figure 6) giant osteoclasts in cultures supplemented by M-CSF + RANKL.
Effect of Nutlin-3 and L-685,458, used alone or in combination, on osteoclastic differentiation. Adherent PBMCs were cultured in presence of M-CSF + RANKL for 12 days, in the absence or presence of Nutlin-3 and/or L-685,458. After 12 days, cultures were analyzed for osteoclastic differentiation. Representative fluorescence microscopy fields of osteoclastic cultures, treated as indicated, are shown in the left panel. DAPI-stained nuclei of polynucleated osteoclasts are circled. Original magnification: ×400; 40×/0.75 NA objective. In the right panel, osteoclastic differentiation was quantified by scoring the number of TRAP-positive multinucleated cells, containing 3 or more nuclei. Data represent the means ± SD of 4 different experiments.
Effect of Nutlin-3 and L-685,458, used alone or in combination, on osteoclastic differentiation. Adherent PBMCs were cultured in presence of M-CSF + RANKL for 12 days, in the absence or presence of Nutlin-3 and/or L-685,458. After 12 days, cultures were analyzed for osteoclastic differentiation. Representative fluorescence microscopy fields of osteoclastic cultures, treated as indicated, are shown in the left panel. DAPI-stained nuclei of polynucleated osteoclasts are circled. Original magnification: ×400; 40×/0.75 NA objective. In the right panel, osteoclastic differentiation was quantified by scoring the number of TRAP-positive multinucleated cells, containing 3 or more nuclei. Data represent the means ± SD of 4 different experiments.
Discussion
The Notch receptors and ligands are single-pass transmembrane proteins expressed on the surface of adjacent cells, and, therefore, activation of Notch signaling usually requires cell/cell contact and the cleavage of the amino terminal region of Notch family members by γ-secretase.38 This cleavage releases the intracellular domain of Notch, which translocates to the nucleus and induces the transcription of Notch target genes. Aberrant NOTCH activation has been linked to cancer since 1991 when mammalian Notch1 was first identified as part of the translocation t(7;9) in a subset of human T-cell acute lymphoblastic leukemias (T-ALLs).27,38 Recent findings indicate an important role of Notch also in the pathogenesis of human T and B cell–derived lymphomas.38,39 In this context, the interplay between Notch1 and p53 signaling pathways appears rather complex. In fact, previous data obtained in primary keratinocytes40 have demonstrated that a p53-responsive element could be identified in the NOTCH1 promoter, and therefore NOTCH1 gene represents a novel target of p53 and plays a key role of keratinocyte differentiation. Conversely, a couple of recent studies41,42 have demonstrated that cells expressing the intracellular active domain of human Notch1 are chemoresistant in a wild-type p53-dependent manner. In keeping with an antiapoptotic role of Notch family members, a recent study21 has demonstrated that B-CLL shows a constitutive high expression of both Notch1 and Notch2 as well as their ligands Jagged1 and Jadded2 with respect to normal lymphocytes and both Notch family members appear to contribute to apoptosis resistance of B-CLL. Other studies24,25 have associated Notch2 overexpression in B-CLL with high levels of surface CD23 antigen.
In this study, we have demonstrated for the first time that the nongenotoxic activator of p53 pathway Nutlin-3 significantly up-regulated Notch1 at both the mRNA and protein levels in both myeloid and lymphoblastoid TP53wild-type cell lines as well as in the majority of primary TP53wild-type B-CLL patient cells. Moreover, down-regulation of Notch1 expression/function using either siRNA specific for Notch1 or 2 different types of γ-secretase inhibitors significantly potentiated the cytotoxic activity of Nutlin-3 toward TP53wild-type leukemic cells. In this respect, it is noteworthy that also p21WAF/CIP1, a major p53 transcriptional target potently up-regulated by Nutlin-3,15 besides inducing cell cycle arrest also exhibits antiapoptotic activity.43-45 Thus, our present study suggests that, similarly to p21WAF/CIP1, the activation of NOTCH1 by p53 represents a negative feedback loop, able to restrain the induction of apoptosis mediated by p53. On the other hand, since several studies have shown that the basal levels of p53 expression are very low in B-CLL cells,7-10 transcription factors other than p53 likely account for the high basal expression of Notch1 in B-CLL cells.21
Experiments performed with PFT-α, a pharmacologic inhibitor of the transcriptional activity of p53, demonstrated that PFT-α was able to enhance the cytotoxic activity of Nutlin-3, confirming recent data of Steele et al,19 who demonstrated that activation of p53 by Nutlin-3 induced apoptosis of B-CLL mainly through the nontranscriptional association of p53 to the mitochondria. These authors rather showed that the transcriptional activity of p53 counteracts the apoptosis-inducing activity of both Nutlin-3 and chemotherapeutic drugs.19 Since we have demonstrated that PFT-α significantly counteracted the transcriptional induction of NOTCH1, these data further indicate that NOTCH1 is one of the transcriptional targets of p53 with antiapoptotic activity.
The synergistic cytotoxic activity of Nutlin-3 and γ-secretase inhibitors are particularly noteworthy since clinical trials with γ-secretase inhibitors have commenced for refractory T-ALL,but it has been shown that γ-secretase inhibitors may be useful for the treatment of hematologic malignancies other than T-ALL.46 Although the mechanism behind the cytotoxic effects of γ-secretase inhibitors remains to be clarified, it has been proposed that Notch1 signaling confers chemoresistance by inhibiting p53 pathway through mTOR-dependent PI3K-Akt/PKB pathway.41
In considering the potential cytotoxic activity of the combination of Nutlin-3 plus γ-secretase inhibitors toward normal tissues, previous studies have shown that Nutlin-3 is much less toxic against normal tissues with respect to neoplastic cells,2 and might even protect normal cells from apoptosis induced by mitotic inhibitors.47 Moreover, although Notch1 mediates cell survival signal in leukemic cells, in vitro and in vivo studies strongly support a role for Notch signaling in the regulation of stem cell renewal and hematopoiesis.48,49 In normal hematopoiesis, proliferation is tightly linked to differentiation in ways that involve cell-cell interaction with stromal elements in the bone marrow stem cell niches, and it has been shown that activation of Notch signaling in hemangioblasts dramatically reduces their survival and proliferative capacity and lowers the levels of hematopoietic stem cell markers CD34 and c-Kit.48 In keeping with a key role of Notch in hematopoiesis, it has also been shown that the down-regulation of Notch1 in osteoclastic precursors enhances osteoclastogenesis,36,37 identifying osteoporosis as a potential complication of therapeutic inhibition of Notch activity in humans. Consistently, we also found that the addition of the γ-secretase inhibitor L-685,458 in vitro significantly increased the number of mature osteoclasts differentiating from peripheral blood adherent mononuclear cells in the presence of M-CSF + RANKL. On the other hand, recently we have demonstrated that Nutlin-3 suppresses osteoclastic maturation.50 In this context, an additional important finding of our study was that although Nutlin-3 potently induced Notch1 accumulation in preosteclasts, the simultaneous addition of Nutlin-3 + L-685,458 to normal preosteclasts completely suppressed osteoclastic maturation, driven by M-CSF + RANKL.
In conclusion, we have demonstrated for the first time that Nutlin-3 induces the transcriptional activation of NOTCH1 in TP53wild-type leukemic cell lines and primary B-CLL cells and that the simultaneous treatment with Nutlin-3 and γ-secretase inhibitors enhances the potential therapeutic efficacy of Nutlin-3. Moreover, the combined used of Nutlin-3 plus γ-secretase inhibitors might be safely used also in hematologic malignancies characterized by hyperactivation of osteoclastogenesis.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by grants from the Italian Association for Cancer Research (AIRC, Milan, Italy; G.Z.) and from the CariFe Foundation (Ferrara, Italy; P.S.).
Authorship
Contribution: P.S. conceived and designed the study, provided financial support, assembled, analyzed, and interpreted the data, and wrote the paper; E.M., M.G.d.I., E.R., and F.C. carried out the experiments, and collected, assembled, analyzed, and interpreted data; M.T. and V.G. were responsible for provision of study material, and analyzed and interpreted data; and G.Z. conceived and designed the study, provided financial support, wrote the paper, and was responsible for final approval of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Paola Secchiero, Department of Morphology and Embryology, University of Ferrara, Via Fossato di Mortara 66, 44100 Ferrara, Italy; e-mail: paola.secchiero@unife.it.