Abstract
Hematopoiesis is a carefully controlled process that is regulated by complex networks of transcription factors that are, in part, controlled by signals resulting from ligand binding to cell-surface receptors. To further understand hematopoiesis, we have compared gene expression profiles of human erythroblasts, megakaryocytes, B cells, cytotoxic and helper T cells, natural killer cells, granulocytes, and monocytes using whole genome microarrays. A bioinformatics analysis of these data was performed focusing on transcription factors, immunoglobulin superfamily members, and lineage-specific transcripts. We observed that the numbers of lineage-specific genes varies by 2 orders of magnitude, ranging from 5 for cytotoxic T cells to 878 for granulocytes. In addition, we have identified novel coexpression patterns for key transcription factors involved in hematopoiesis (eg, GATA3-GFI1 and GATA2-KLF1). This study represents the most comprehensive analysis of gene expression in hematopoietic cells to date and has identified genes that play key roles in lineage commitment and cell function. The data, which are freely accessible, will be invaluable for future studies on hematopoiesis and the role of specific genes and will also aid the understanding of the recent genome-wide association studies.
Introduction
The hematopoietic system represents one of the best-studied cellular differentiation processes in mammals. The differentiation of the hematopoietic stem cell (HSC) into the blood cell lineages, which is depicted as a stepwise process, generates diverse types of cells that perform many different functions. Historical observations of the blood, made in the late 18th century using some of the first microscopes, revealed that blood is composed of a heterogeneous population of cells that are distinct in number, morphology, and function. Since these early studies, the application of both technologic and methodologic advances to the investigation of blood has led to an ever-increasing understanding of the nature and function of the different types of blood cells. For example, the use of monoclonal antibodies (mAbs) and the designation of the cluster of differentiation (CD) markers, of which there are now more than 300,1 allows hematologists to assign detailed phenotypes to malignant blood cells, which form the basis of decisions on therapeutic intervention.
The value of the current understanding of the hematopoietic system to patient care is perhaps best illustrated in the field of malignancy where gene and protein expression profiles permit rapid and routine patient stratification. It is now possible to stratify patients with leukemia and lymphoma with unprecedented accuracy using gene expression profiles. Signature gene expression profiles may be used for diagnosis and predicting disease prognosis. In addition to studies in patients, gene expression profiles are available for a wide range of healthy tissue types. However, many of these resources, although broad in tissue coverage, are limited in the number of samples analyzed for each tissue type (eg, Symatlas).2 Consequently, the false-positive and false-negative discovery rates are high, and limited reliable information is available regarding variation in gene expression profiles between healthy persons. Similarly, platform differences between studies do not facilitate rapid comparison between datasets.
We set out to generate a focused gene expression atlas for cells of the hematopoietic system from healthy persons, a so-called Hematology Expression Atlas (HaemAtlas). We have taken advantage of recent advances in cell purification, RNA amplification, and microarray technologies that allow the study of gene expression of purified subsets of cells on a genome-wide scale. Using whole-genome expression arrays, we have compared the gene expression profiles of the precursors of erythrocytes and platelets (erythroblasts [EBs], megakaryocytes [MKs]) and of B cells, cytotoxic T cells (Tc), helper T cells (Th), natural killer (NK) cells, granulocytes, and monocytes. In total, 50 expression profiles were obtained using the Illumina HumanWG-6 version 2 Expression BeadChip (Illumina, San Diego, CA), which have more than 48 000 probes, targeting genes and known alternative splice variants from the RefSeq database release 17 and UniGene build 188.
The data described represent an extremely useful resource for the clinical hemato-oncologist and for the research community as a whole. In addition, we demonstrated the utility of this dataset by performing a focused bioinformatic analysis of transcription factor and immunoglobulin superfamily (IgSF) member gene expression. The dataset has already been used in conjunction with genome-wide association studies and in the characterization of tetraspanins.3,4 Finally, by comparing expression profiles between cell types, we have identified sets of transcripts that are lineage specific and show, in an accompanying manuscript, the expression and function of 4 novel proteins in arterial thrombus formation.5
Methods
Cell purification and purity assessment
Whole blood units (∼ 450 mL) from 7 healthy volunteer donors of the Cambridge BioResource at National Health Service (NHS) Blood and Transplant were obtained with informed consent in accordance with the Declaration of Helsinki. The study was approved by the United Kingdom National Health Service Blood and Transplant. Donors were included only if they had a hemoglobin more than 12.5 g/dL for women and13.5 g/dL for men, were negative for HepB, HepC, HIV1, and HIV2 antibodies, negative for syphilis, and negative for hepatitis C virus (HCV) by nucleic acid testing. Donor Epstein-Barr virus and cytomegalovirus status were not selection criteria. Blood was taken by venipuncture into a bag containing acid citrate dextrose anticoagulant according to the NHS Blood and Transplant procedures. CD4+ Th and CD8+ Tc lymphocytes, CD14+ monocytes, CD19+ B lymphocytes, CD56+ NK cells, and CD66b+ granulocytes were isolated using an automated magnetic labeling protocol (RoboSep; StemCell Technologies, Vancouver, BC) as described in “Supplementary Materials and Methods” in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Details of the CD markers used for cell isolation together with quality control data for the processed samples are given in Tables S1 and S2. The culture conditions of the 4 cord blood hematopoietic progenitor cell (HPC) preparations and the purification protocol of the MKs and EBs have been described previously.6
RNA purification, amplification, and hybridization
Purified cell populations were lysed in Trizol following the manufacturer's instructions (Invitrogen, Paisley, United Kingdom) using 1 mL Trizol reagent per 106 cells. Isolated total RNA was then purified further using the RNeasy MinElute Cleanup Kit (QIAGEN, Dorking, United Kingdom). Each purified RNA sample was assessed for quality and integrity using the 2100 Bioanalzyer (Agilent Technologies, Palo Alto, CA). All information on RNA processing and quality assessment is available in Table S1.
Total RNA (500 ng) was amplified using the Illumina Total Prep RNA Amplification Kit (Ambion, Austin, TX) according to the manufacturer's instructions. The biotinylated cRNA (1500 ng per sample) was applied to Illumina HumanWG-6 v2 Expression BeadChips and hybridized overnight at 58°C. Chips were washed, detected, and scanned according to the manufacturer's instructions.
Statistical analysis of gene expression data
Present genes.
The Illumina BeadStudio software calculates a Detection Score equivalent to 1 − P value for detection for each probe, which is an estimate of the confidence limit of detection relative to the local background. Probes were considered as present if they had a detection score more than 0.99 in all samples of a given cell type.
Differentially expressed genes.
We performed pairwise comparisons between one cell type and every other cell type used in the study. These comparisons are exhaustive but necessary to identify transcripts that are unique to each of the cell types or common between different cell types. In comparing the expressions between cell types, we performed a paired t test (or 2-sample t test in the case of nonpaired samples) coupled with multidimensional false-discovery control (FDR2D).7 FDR2D was used to guard against false-positive results from transcripts whose variance is underestimated by chance, whereas their fold changes are small. Analysis of the results obtained suggests that the method is effective in identifying true differentially expressed (DE) transcripts.
Cell unique and unspecific genes.
To identify transcripts that are specifically enriched or depleted in a given blood cell lineage, so-called “unique” and “unspecific” genes, respectively, we performed a comparison between the lists of DE genes using an “AND” operator. In such a way, genes that were consistently up- or down-regulated vs all other cell types were identified.
Bioinformatic analysis
Biologic processes.
The Protein Analysis Through Evolutionary Relationships (PANTHER) classification scheme (http://www.pantherdb.org/) was used to infer involvement in biologic process for the present genes as described in “Supplementary Materials and Methods” in Document S1.
IgSF proteins.
The identification of the IgSF repertoire expressed by blood cells was based on matching microarray probes to 2 existing reference sets: (1) our manually curated human IgSF reference set defined previously8 ; and (2) the subset of Homo sapiens Ensembl v46,9 gene predictions that received significant hits by either PFAM10 or SUPERFAMILY,11 hidden Markov models that represent IgSF domain sequences.
The functional presence of an IgSF gene was established using conservative signal threshold cutoff values. Furthermore, the analysis of IgSF expression was primarily focused on the identification of those cell types in which the presence of a transcript was particularly marked. This was achieved by comparing relative signal intensity values for the same probe across the cell types and, using the ratio of the mean intensity to the SD, indicative skews in the distribution were identified when such an index was less than 1.
Transcription factor networks.
We generated a dataset of transcription factors by combining (1) a manually curated list of known transcription factors and (2) sequence-specific DNA-binding transcription factors using the most recent version of our transcription factor prediction database (http://transcriptionfactor.org/).12
The combined transcription factor set contains 2528 transcripts, all of which are present on the Illumina HumanWG-6 version 2 Expression BeadChips.
Evolutionary conservation of gene expression profiles.
To identify evolutionary conservation of gene expression profiles in blood cells, we compared the human hematopoietic expression data generated in this study with that in mice obtained by Chambers et al.13 The murine study included hematopoietic stem cells, activated Tc and Th cells, in addition to the cell types analyzed here, but did not include MKs. A comparative analysis of the expression profiles for the cell types common to both studies was conducted. Notwithstanding that the tissue and membrane antigens used for cell isolation differed between the 2 studies, greater than 98% of all human-mouse orthologous transcripts were represented in both datasets.
The data described in this manuscript are available at ArrayExpress (www.ebi.ac.uk/microarray-as/ae/) under accession number E-TABM-633 or at the Bloodomics project website (www.bloodomics.org).
Results
Sample processing
In total, we purified peripheral blood cells from 43 volunteers from which 7 sets that met strict quality control criteria were selected for this study (Tables S1, S2). For each cell population, purity was more than 95% as assessed by flow cytometry together with a morphologic assessment of May-Grunwald-Giemsa (Romanovsky)–stained cytocentrifuge preparations using light microscopy (Figure 1). After cell isolation, RNA was purified and quality assessed using an Agilent BioAnalyzer before amplification. A total of 50 samples were amplified and hybridized onto the Illumina Human WG-6 version 2 Expression BeadChips as described. This represents 6 cell types isolated from peripheral blood from the 7 volunteer donors (n = 42) and MKs and EBs differentiated from CD34+ HPCs obtained from 4 umbilical cord blood samples (n = 8).6
Cells were purified to more than 95% purity as assessed by morphology and flow cytometry. After cell isolation, an aliquot of purified cells was removed and assessed for purity as described. Example of CD19+ B cells isolated from peripheral blood mononuclear cells. (A) Peripheral blood mononuclear cells assessed by Romanovsky-stained cytocentrifuge preparations and (B) phycoerythrin-labeled anti-CD19 by flow cytometry. After purification, more than 98% of cells were CD19+ as assessed by (C) a 1000 differential cell count of Romanovsky-stained cytocentrifuge preparations and (D) flow cytometry. Images and purity levels are representative of all samples processed. (A,C) Romanovsky-stained samples were visualized using an Olympus BX51 microscope (Olympus, Tokyo, Japan) with a 100×/1.30 oil objective and immersion oil (nd 1.516; Olympus). Images were captured using a Pixera Pro600ES and Penguin/Pro Application Suite version 3.0.1 (Pixera, Los Gatos, CA).
Cells were purified to more than 95% purity as assessed by morphology and flow cytometry. After cell isolation, an aliquot of purified cells was removed and assessed for purity as described. Example of CD19+ B cells isolated from peripheral blood mononuclear cells. (A) Peripheral blood mononuclear cells assessed by Romanovsky-stained cytocentrifuge preparations and (B) phycoerythrin-labeled anti-CD19 by flow cytometry. After purification, more than 98% of cells were CD19+ as assessed by (C) a 1000 differential cell count of Romanovsky-stained cytocentrifuge preparations and (D) flow cytometry. Images and purity levels are representative of all samples processed. (A,C) Romanovsky-stained samples were visualized using an Olympus BX51 microscope (Olympus, Tokyo, Japan) with a 100×/1.30 oil objective and immersion oil (nd 1.516; Olympus). Images were captured using a Pixera Pro600ES and Penguin/Pro Application Suite version 3.0.1 (Pixera, Los Gatos, CA).
Genes expressed in differentiated blood cells
For each cell type, we first determined the number of present probes by applying rigorous criteria to reduce false-positive discoveries (Figure 2; Table S3). As can be seen, the number of present probes ranged from 7302 for granulocytes to 10 314 for MKs. The lower number of present transcripts in granulocytes could not be attributed to any features of the microarrays (data not shown).
Characterization of blood cell transcriptomes and identification of differentially expressed transcripts. (A) Numbers of genes detected as present in different blood cells. (B) Clustering of samples based on genes with high precision in the dataset. (C) Overlap of present genes in human blood cells. (D) Patterns of enrichment (red) or depletion (blue) for different biologic processes of the PANTHER classification for the genes differentially expressed in different blood cell types. The color range represents Z-scores (from Z = 3 to Z = 10 for enrichment and Z = − 3 to Z = − 10 for depletion). Functional categories containing at least 20 genes were used in this analysis.
Characterization of blood cell transcriptomes and identification of differentially expressed transcripts. (A) Numbers of genes detected as present in different blood cells. (B) Clustering of samples based on genes with high precision in the dataset. (C) Overlap of present genes in human blood cells. (D) Patterns of enrichment (red) or depletion (blue) for different biologic processes of the PANTHER classification for the genes differentially expressed in different blood cell types. The color range represents Z-scores (from Z = 3 to Z = 10 for enrichment and Z = − 3 to Z = − 10 for depletion). Functional categories containing at least 20 genes were used in this analysis.
Hierarchical cluster analysis of all samples based on the probes with the highest variance across the 50 samples recapitulated the known hematopoietic differentiation pathway, with the exception that the NK-cell samples were more closely related to the Tc samples (Figure 2B) than Th. On the basis of this clustering, we defined genes that were common to (1) the 2 precursor cells, (2) granulocytes and monocytes, and (3) Tc, Th, and NK lymphoid cells and performed an overlap analysis with those present in B cells (Figure 2C). A total of 5396 genes were detected in all blood cells, with transcripts from an additional 1860 genes being present in all cells except for monocytes and granulocytes (Figure 2C). As expected, more than 1000 more transcripts were detected in the transcriptomes of the 2 precursor cells (MKs and EBs), with Gene Ontology (GO) analysis indicating enrichment for genes involved in cell cycle (GO, 0007049), mitotic cell cycle (GO, 0000278), and cell-cycle process (GO, 0022402). This observation is in agreement with the active cell proliferation and differentiation processes that are underway in these 2 elements that normally reside in the bone marrow environment.
Using PANTHER classification, we observed that genes in “nucleoside, nucleotide and nucleic acid metabolism,” “immunity and defense,” and “protein metabolism and modification” are overrepresented in hematopoietic cells but that categories such as “signal transduction” and “developmental process” are underrepresented (Figure 2D). The enrichment for genes involved in “immunity and defense” is consistent with our understanding that immune responses are one of the primary functions of the myeloid and lymphoid blood cell types. In addition, genes classified as being involved in “signal transduction” are underrepresented in all cell types studied.
Expression of CD markers
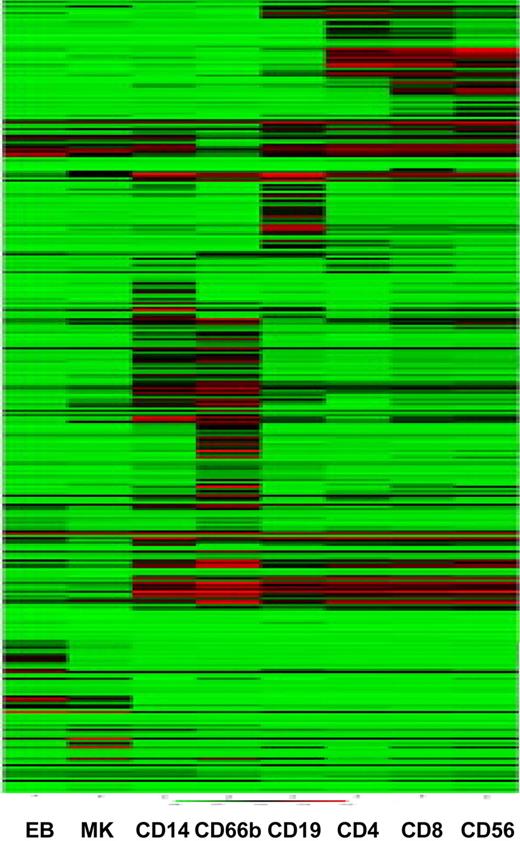
The CD markers constitute the most widely studied hematopoietic markers, with known protein expression patterns confirmed by antibody staining of normal and malignant hematopoietic cellular elements. Of the 339 currently described CD markers, 44 are not represented on the Illumina HumanWG-6 v2 Expression BeadChips (data not shown). We clustered the samples on the basis of CD marker gene expression. The samples cluster largely as expected, with a clear distinction between cells of the myeloid and lymphoid lineages (Figure 3). Interestingly, again, the CD56+ NK cells cluster most closely to the CD8+ Tc cells, confirming the results of the hierarchical clustering based on transcripts with a high variance (Figure 2), and compatible with the notion that these 2 cells are more closely related than the CD4+ and CD8+ T cells at the transcript level. Similarly, the CD66b+ granulocytes and CD14+ monocytes cluster together as do the MK and EB samples (Figure 3).
Clustering of samples on the basis of CD marker expression recapitulates cell ontogeny. Samples were clustered using the mean normalized intensity values for the 356 probes that map to CD markers.
Clustering of samples on the basis of CD marker expression recapitulates cell ontogeny. Samples were clustered using the mean normalized intensity values for the 356 probes that map to CD markers.
Combinatorial patterns of transcription factor expression
Transcriptional regulation is a key mechanism controlling fate commitment of HPCs and their progeny and ultimately in controlling intermediate phenotypes, such as volumes and numbers of the mature blood cell elements.14,15 Both gain- and loss-of-function studies have been used to demonstrate the role of transcription factors in controlling the specification and differentiation of HSCs (eg, SCL/TAL1, GATA2, c-MYB, PU.1, RUNX1, ETV6, GFI1). However, much remains to be learned about the way these key regulators interact with each other and thus form the transcriptional networks that specify the various mature lineages. We therefore explored the potential utility of our gene expression profiles to gain new insights into hematopoietic transcriptional control mechanisms.
We initially catalogued sequence-specific, DNA-binding transcription factors expressed in all cell types using the most recent version of our transcription factor prediction database DBD (http://transcriptionfactor.org/).12 The expression profiles of known key hematopoietic transcription factors and their related family members are shown in Figure S1. An immediate conclusion from this analysis was that key hematopoietic transcription factors are expressed in multiple hematopoietic lineages, consistent with the notion that hematopoietic lineages are generally not specified by single transcription factors. Instead, specific combinatorial interactions between key transcription factors are vital to control cellular identity.
To explore the combinatorial theme further, we next identified those transcription factors that share their expression profiles with 6 well-characterized transcription factors with distinct roles in the development of the 8 blood lineages profiled in this study. This approach was based on the rationale that coexpression of transcription factor pairs might reveal new regulatory links as it would be consistent with either direct regulation (transcription factors upstream or downstream of each other) or placement within the same regulatory pathways. Our analysis demonstrated that the numbers of coexpressed transcription factors (TFs) varied greatly for the 6 TFs chosen, ranging from one for GATA1 to 35 for GATA2 (Figure 4). Coexpression in several cases was consistent with known interactions (GATA2-Tal116 or GATA1-GFI1B17 ) and also suggested as yet unreported interactions between known key regulators (GATA3-GFI1; GATA2-KLF1). Moreover, many putative links with totally uncharacterized transcription factors were revealed, such as the various zinc finger families (Figure 4). Taken together, therefore, our analysis suggested that comprehensive genome-wide expression surveys provide an important resource to reveal new links in hematopoietic regulatory networks, particularly with respect to the combinatorial control of gene expression.
Transcription factor coexpression in hematopoietic lineages. Shown at the top is the hematopoietic differentiation hierarchy with key hematopoietic transcription factors GATA1, GATA2, Meis1, SPI1, GATA3, and EBF1. Only MEIS1 and EBF1 were expressed in a single lineage, whereas all other factors were expressed in 2 or more lineages. Tabulated underneath each factor are those transcription factors that share their respective expression pattern, suggesting either direct regulation or common upstream regulators. Expression of GATA1 in CD66b+ cells was an order of magnitude lower than in erythroblasts and megakaryocytes.
Transcription factor coexpression in hematopoietic lineages. Shown at the top is the hematopoietic differentiation hierarchy with key hematopoietic transcription factors GATA1, GATA2, Meis1, SPI1, GATA3, and EBF1. Only MEIS1 and EBF1 were expressed in a single lineage, whereas all other factors were expressed in 2 or more lineages. Tabulated underneath each factor are those transcription factors that share their respective expression pattern, suggesting either direct regulation or common upstream regulators. Expression of GATA1 in CD66b+ cells was an order of magnitude lower than in erythroblasts and megakaryocytes.
IgSF member expression in hematologic cells
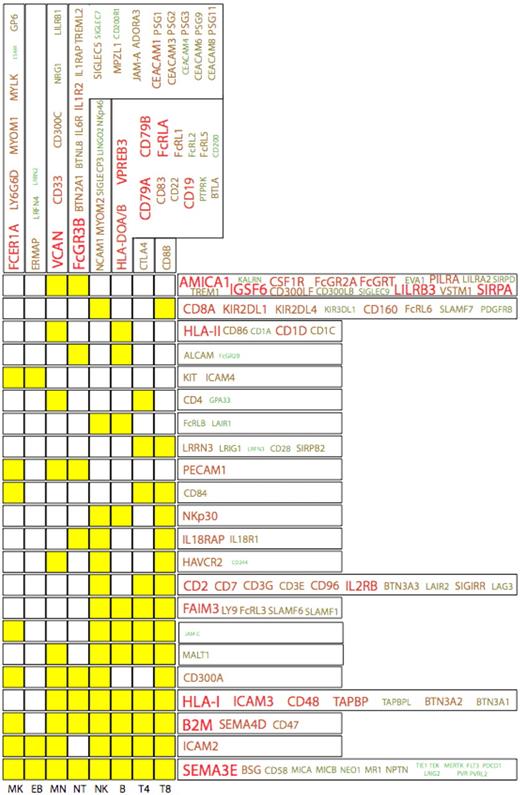
The IgSF represents a family of proteins that play key roles in hematopoiesis and blood cell function. Analysis of the HaemAtlas expression data shows that 170 (∼ 30%) of the approximately 600 known IgSF genes show significant expression across the 8 blood cell types investigated (Figure 5). The highest cumulative expression (and largest unique protein repertoire) of IgSF molecules was found in the granulocyte, whereas the erythroblast showed the lowest level of IgSF deployment. This detailed analysis of the expression of IgSF members in blood cells has identified several novel findings relating to their various functions.
The IgSF protein expression profiles in the HaemAtlas. The expression patterns of cell-specific IgSF family members (columns) together with those expressed across several cell types (rows) are depicted, with yellow boxes indicating cells in which genes are expressed. For example, CD8+ T cells are the only cell type to express CD8B, whereas FcRLB and LAIR1 are expressed in NK and B cells. The size of the font and the green-to-red color intensity are both indicative of the strength of mean expression across the cells.
The IgSF protein expression profiles in the HaemAtlas. The expression patterns of cell-specific IgSF family members (columns) together with those expressed across several cell types (rows) are depicted, with yellow boxes indicating cells in which genes are expressed. For example, CD8+ T cells are the only cell type to express CD8B, whereas FcRLB and LAIR1 are expressed in NK and B cells. The size of the font and the green-to-red color intensity are both indicative of the strength of mean expression across the cells.
IgSF members involved in boundary interactions
Several IgSF family members are involved in interactions at cell boundaries, and we investigated the expression of these in the HaemAtlas data. Blood cell-surface IgSF protein interaction with the vascular wall and epithelia is primarily mediated and modulated via molecules, such as PECAM-1, MCAM, CD47, SIRPα, and the families of the intercellular adhesion and junctional adhesion molecules, ICAM and JAM, respectively. PECAM-1,18 MCAM,19 and JAM molecules20 are crucial for interaction with the endothelium, binding both homotypically and to other non-IgSF ligands. PECAM-1 is known to interact with the non-IgSF CD177,21 which is considered to be granulocyte-specific. In our data, CD177 showed very low expression levels across the 8 cell types, whereas PECAM1 transcription was found at increasing levels in megakaryocytes, monocytes, and granulocytes. PECAM-1 is also known to play a collaborative role with JAM-A in the transmigration of granulocytes,22 and JAMA expression was predominant in the granulocyte. JAM-C, on the other hand, was well represented in all lymphoid cells as well as the megakaryocyte. JAML, which binds the Coxsackie adenovirus receptor (CxADR) during blood cell migration across the mucosal barrier,23 was among the most highly expressed cell adhesion molecules in monocytes and granulocytes. MCAM (typically up-regulated in activated T cells), VCAM1, and MADCAM1, which are known to be specifically expressed in endothelial cells,24,25 were not detected.
The ICAMs are known for their deployment on luminal endothelial and apical epithelial surfaces and for their interaction with leukocytic integrins. However, signaling from ICAMs is also known to play a role in blood cell development.26-28 In the current study, ICAM4 was exclusively expressed in megakaryocytes and erythroblasts, ICAM3 was present in all differentiated blood cells (somewhat prominently in the granulocyte), and ICAM2 had a moderate signal intensity level in all cell types except for granulocytes. Transcription levels were low for ICAMs-1 and -5 in all cells tested.
Interestingly, we observed a notable overlap between the blood cell IgSF cell surface sensors and those associated with neural development. Moderate intensity levels of NCAM-1 and low levels of LINGO2 were exclusive to the NK cell, whereas CD4+ and CD8+ T cells showed considerable specificity for LRRN3 and LRIG1, whereas granulocytes and B cells both had moderate expression of ALCAM (also known as neurolin).29 It is of interest that ALCAM has been reported to bind to EGFR,30 whereas LRIG-1 is known to inhibit the signaling of this growth factor receptor.31,32 Modest expression levels of LRFN4 and LRRN2 were observed but only in EBs. Low levels of LRIG2 transcription were observed in all 8 cell types, and the expression of schizophrenia-associated MPZL1 was restricted to granulocytes.
Finally, we observed that versican, an abundant IgSF proteoglycan in vessel walls whose expression is increased after vascular injury, is abundantly and specifically expressed in monocytes. Versican is known to accumulate in advanced atherosclerotic plaques33 and after myocardial infarction.34 These observations raise the possibility that monocyte-derived versican may play a key role in atherosclerotic plaque formation.
SLAM
The signaling lymphocytic activation molecule (SLAM) family is known to modulate the function of immune system cells through homotypic interactions and signaling through SLAM-associated protein-related adaptor molecules.35,36 In this experiment, SLAMs F1, F6, F7, and Ly9 were moderately expressed in differentiated lymphoid cells, with SLAM F7 not detectable in B cells and CD4+ T cells. SLAM CD84 was found in T cells and more strongly in megakaryocytes. CD244 was found in NK cells and was weakly expressed in monocytes and CD8+ T cells. SLAMs F8 and F9 showed a very low signal in all cell types. CD48 transcription was high in all differentiated blood cells, whereas the CD2-related CD58 was moderately expressed in all 8 cell categories. CD2 itself was restricted to NK and T cells.
Comparative analysis of gene expression in human and murine blood cells
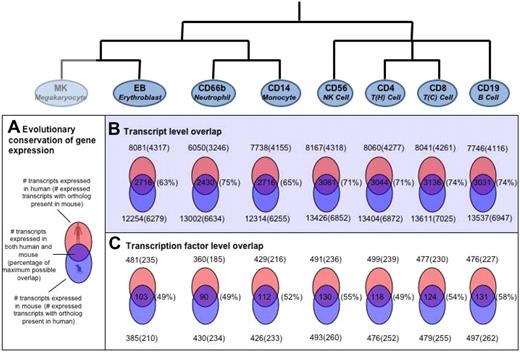
Recently, a study of blood cell gene expression in mice has been performed (http://franklin.imgen.bcm.tmc.edu/loligag/)13 and to investigate whether the gene expression patterns in hematopoietic cells remain evolutionarily conserved, we compared the expression pattern of human transcripts with that of corresponding mouse orthologs. A comparative analysis of the expression profiles for the 7 cell types common to both studies was performed (Figure 6).
Evolutionary conservation of human versus mouse gene expression in various hematopoietic cell types. (A) Schematic representation of overlap in differential gene expression between human and mouse. The percentage of maximum possible overlap, shown in parentheses, is the percentage of orthologous proteins of the lower number (human or mouse) of DE genes. For the 7 cell types with data in both human and mouse, the extent of conservation of differential gene expression is shown at the level of (B) all transcripts and (C) transcription factors only. For those genes that were detected as expressed in human blood cells, mouse orthologs were identified as described. The presence of these orthologs in the mouse data was then investigated. Venn diagrams showing the number of overlapping genes with the number of orthologs identified shown in parentheses.
Evolutionary conservation of human versus mouse gene expression in various hematopoietic cell types. (A) Schematic representation of overlap in differential gene expression between human and mouse. The percentage of maximum possible overlap, shown in parentheses, is the percentage of orthologous proteins of the lower number (human or mouse) of DE genes. For the 7 cell types with data in both human and mouse, the extent of conservation of differential gene expression is shown at the level of (B) all transcripts and (C) transcription factors only. For those genes that were detected as expressed in human blood cells, mouse orthologs were identified as described. The presence of these orthologs in the mouse data was then investigated. Venn diagrams showing the number of overlapping genes with the number of orthologs identified shown in parentheses.
The number of transcripts expressed in different hematopoietic cells in human had a range of approximately 6000 to approximately 8000, whereas the corresponding number for mouse had a range of approximately 12 000 to approximately 13 500. The apparent consistent increase in the number of transcripts in mouse over human most probably reflects the difference in platforms and expression cutoffs chosen in the 2 studies. However, despite these differences, the overall patterns of gene expression were fairly consistent between the 2 species with approximately 50% of the transcripts expressed in human having orthologs in mouse and vice versa (Figure 6). For all cell types tested, the overlaps were found to be statistically significant (P < 10−5).
The number of transcription factors found in equivalent cell types between human and mouse were also comparable. We detected between 360 and 500 transcription factors in each human cell type, of which approximately 50% (∼ 230) had orthologs in mouse, with 25% (∼ 120) being shared between species in equivalent cell types (Figure 6). This overlap of transcription factor expression was found to be significant for all cells (P < .005) except for the erythroblast samples (P = .07).
Identification of differentially expressed genes
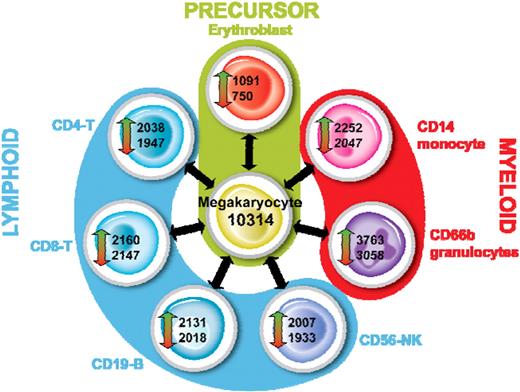
A statistical analysis was performed to identify transcripts that are differentially expressed between each cell type as described. For this analysis, we considered a transcript to be differentially expressed if it had a P value less than .05 and a fold change more than 2. The outcome of this analysis for MKs is shown in Figure 7. An average of 2206 features were up-regulated (range, 1091-3763) and 1986 down-regulated (range, 750-3058) between MKs and each other cell type. As expected when the MK was used as a reference, the smallest number of DE features was observed in the comparison with EBs. Interestingly, we observed that the CD66b+ granulocytes had the greatest number of DE features in each comparison, reflecting the significant differences between the myeloid cells and the other cells tested. The complete lists of DE features are given in Table S4.
Identification of differentially expressed genes in MKs. For each cell type, we identified transcripts that were up- or down-regulated versus all other cell types as described. The outcome for MKs is shown.
Identification of differentially expressed genes in MKs. For each cell type, we identified transcripts that were up- or down-regulated versus all other cell types as described. The outcome for MKs is shown.
Cell-specific transcripts
We also performed an overlap analysis of the lists of DE transcripts to identify those that are consistently up-regulated in one cell type compared with all others (Table S5). The lists of genes thus generated are considered “unique” for each cell type analyzed. We observed that the number of transcripts uniquely expressed in a given cell type varies by more than 2 orders of magnitude, with CD8+ cells expressing only 5 unique transcripts and CD66b+ cells expressing 878. Similarly, the CD66b+ cells have the highest number of unspecific transcripts, whereas CD8+ cells express the least (data not shown).
The CD8+ T cell–specific genes included both CD8A and CD8B, although low-level expression of CD8A was also observed in the NK-cell population, but this was in the absence of CD8B. The other CD8+ T cell–specific transcripts were CD248, DKK3 (dickkopf homolog 3), and the T-cell receptor alpha V gene segment TRAV1-2. CD248, also known as endosialin, has previously been reported as a fibroblast and pericyte marker where it plays a role in tissue remodelling and repair.37 The function of DKK3, which is divergent from the 3 other dickkopf family members (DKK1, 2, and 4), is unknown, although a role as a tumor suppressor has been suggested because it is down-regulated in several tumor cells.38 Interestingly, Dkk3 knockout mice, which do not show enhanced tumorigenesis, have several unique hematologic features compared with wild-type mice, including the frequency of NK cells and IgM levels.39 More recently, a role for DKK3 in TGF-β signaling has been identified40 ; however, its role as a secreted molecule in cytotoxic T-cell function remains to be elucidated.
We hypothesized that the identification of cell-specific transcripts would lead to the discovery of novel genes that play important roles in cellular functions. We tested this hypothesis for 2 of the cell types used in this study, CD8+ T cells and MKs. CD248 (endosialin) was identified as a CD8+ T cell–specific transcript in this study; however, it is not expressed in mouse T cells, and studies in knockout mice show no role for CD248 in T cells. Using 4 CD248-specific monoclonal antibodies, we were able to demonstrate the surface expression of CD248 on CD8+ CD45RA+ T cells (Figure 8), confirming the lineage specificity of this protein. The reason for the differential expression of CD248 between human and mouse T cells is unknown but warrants further investigation.
CD248 expression is restricted to CD8+CD45RA+ T cells. Flow cytometry with 4 different CD248 antibodies on lymphocytes from (A) peripheral blood and (B) tonsil. Lymphocytes were first gated on forward scatter and side scatter and then on the specific markers shown (CD3, CD4, CD8, CD45RO, CD45RA). All 4 CD248-specific monoclonal antibodies (B1 35.1, B1 473, 18 37.30, and B1 22.4) show that CD248 expression is restricted to CD8+CD45RA+ T cells. Nonfilled histograms represent anti-CD248; and gray-filled histograms, negative control.
CD248 expression is restricted to CD8+CD45RA+ T cells. Flow cytometry with 4 different CD248 antibodies on lymphocytes from (A) peripheral blood and (B) tonsil. Lymphocytes were first gated on forward scatter and side scatter and then on the specific markers shown (CD3, CD4, CD8, CD45RO, CD45RA). All 4 CD248-specific monoclonal antibodies (B1 35.1, B1 473, 18 37.30, and B1 22.4) show that CD248 expression is restricted to CD8+CD45RA+ T cells. Nonfilled histograms represent anti-CD248; and gray-filled histograms, negative control.
For MKs, we selected 4 MK-specific transcripts for study in a zebrafish thrombosis model. Knockdown of all 4 genes, which are uniquely expressed in MKs, significantly affected thrombus formation in the caudal artery after laser-induced vessel injury. Using this model, combined with the selection of MK-specific genes identified using the HaemAtlas, we have demonstrated a role for BAMBI and LRRC32 in promotion and DCBLD2 and ESAM in inhibition of thrombus formation.5
Discussion
In this study, we have generated a gene expression atlas for 8 cells of the hematopoietic system in what represents the most comprehensive study of gene expression in blood cells from normal healthy persons published to date. We envisage that the future use of the HaemAtlas will primarily be that of a reference resource for gene expression in blood cells.
We developed standardized protocols and stringent quality-control measures for cell isolation before microarray analysis to ensure the quality of this gene expression atlas. All cell types used in this study were more than 95% pure based on flow cytometry analysis and inspection by microscopy. The granulocyte population consists of 3 cell types (neutrophils, eosinophils, and basophils), all of which express CD66b and would therefore be copurified. Interestingly, we did observe variation in the levels of eosinophils in the granulocyte preparations from 15% to 30%. The effect of this variation on the transcriptome data is unknown, but it is probably most apparent in the determination of lineage-specific transcripts. Such an effect was observed for the single CD56+ NK sample with platelet contamination, as this NK sample showed the presence of transcripts deemed MK-specific (data not shown). This observation highlights the importance of maintaining a high level of cell purity when identifying lineage-specific genes. However, the presence of a single, platelet-contaminated sample had minimal effect because of the number of replicates used and the high purity of the other samples.
Our analysis of this comprehensive dataset was focused on transcription factors and IgSF members as these proteins play key roles in both blood cell differentiation and function. An analysis of the coexpression of all TFs with 6 well-characterized TFs that have distinct roles in blood cell development confirmed known interactions and identified as yet unreported ones between known key regulators of transcription. This analysis highlights the utility of genome-wide expression in revealing new links in hematopoietic regulatory networks. Similarly, the IgSF analysis confirmed known expression patterns and identified several previously unreported ones in hematopoietic cells. Of particular interest is the expression of many transcripts involved in neural development. Furthermore, we were able to identify genes that are unique to each cell type studied. These lists of unique genes include the classic lineage-specific CD transcripts and novel lineage-specific transcripts recently identified by both others and us (eg, G6B, G6F, LRRC32, and SUCNR1 in MKs6 ) and also identify novel lineage-specific transcripts for further study. Having an established catalog of lineage-specific transcripts is important for several reasons. First, it provides reassurance of the accuracy of the data presented in this manuscript. A close inspection of the lineage-specific transcripts encoding transmembrane proteins in EBs and MKs identified the presence of lineage-specific CD transcripts, confirming the excellent sensitivity of the array platform. Second, proteins encoded by lineage-specific transcripts are ideal drug targets allowing for pharmacologic manipulation of cell function in a cell-specific manner. Third, sequence variation of transcripts for transmembrane proteins, which alter the amino acid sequence of cell-specific membrane proteins, may be alloantigens, such as the human platelet antigens.41 It is probable that, by an approach of inverted immunology, novel clinically relevant alloantigens may be uncovered. Finally, lineage-specific transcripts may play a key role in cell function, as highlighted by the fact that all the novel MK-specific transcripts that we have tested in a zebrafish thrombosis model have a clear role in thrombus formation.5
Unlike previous studies performed with pooled cells isolated from inbred strains of mice, we performed each hybridization with RNA obtained from a single person. In addition, samples in this study were isolated from unrelated donors; hence, it is possible to ascertain the extent of biologic variation in gene expression. A parallel study, in which gene expression profiles of monocytes from 40 persons were compared, has identified those monocyte genes with the greatest variation in expression (data not shown). Such studies, combined with genome-wide genotyping, will allow the identification of cis- and trans-regulatory genetic variants that control gene expression in primary cells, as has recently been determined for immortalized lymphoblastoid B-cell lines.42,43
The analysis of the HaemAtlas data reported here is based on statistical comparisons performed on a cell-by-cell basis. It is possible to analyze the data by making use of the known hematopoietic hierarchy such that opposite “arms” in the hematopoietic lineage tree would be combined. This strategy would potentially allow the identification of DE genes in in silico–generated precursor cells that are not readily accessible for analysis.
In conclusion, the HaemAtlas that we have generated serves not only as a reference library for gene expression in human blood cells but also as a resource for identifying key genes with roles in blood cell function.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
Acknowledgments
The authors thank the staff and donors of the National Health Service Blood and Transplant, Cambridge Center, and David Bloxham, Department of Hematology, Addenbrooke's Hospital. The authors also thank the Bloodomics Consortium participants.
The Bloodomics project (www.bloodomics.org) was supported by the 6th Framework Programme of the European Union (LSHM-CT-2004-503485). N.A.W. and S.F.G. were supported by a grant from the National Institute for Health Research to National Health Service Blood and Transplant. Support for the Cambridge BioResource was obtained from the National Institute for Health Research Biomedical Research grant for Cambridge University Hospitals National Health Service Foundation Trust. C.F.L., P.D.E., and K.M.R. were supported by the Wellcome Trust.
This is an Open Access article published in accordance with the policies of the Wellcome Trust.
National Institutes of Health
Wellcome Trust
Authorship
Contribution: N.A.W. performed and designed research, analyzed data, and wrote paper; A.G. performed statistical analysis of the data; B.d.B. performed analysis of immunoglobulin superfamily expression and wrote the paper; S.D. and D.M.-S. performed analysis of transcription factor expression and wrote the paper; W.G.J.A. and A.P.A. provided critical bioinformatic support; D.L.H., C.M.I., and C.D.B. performed T-cell experiments; P.D.E. performed amplifications and microarray hybridizations; W.E. analyzed morphology of blood cells and provided critical expertise; N.S.F. isolated cells and RNA and wrote the paper; S.F.G. performed sample quality control; J.J. isolated cells and RNA; K.K. performed donor recruitment; I.C.M. performed preliminary study and analyzed data; S.L.M. provided clinical support and donor assessment; N.T. isolated cells and RNA; A.R. and L.W. performed statistical analysis; K.M.R. oversaw bioinformatics support; D.C.T.-T. and M.R.T. generated megakaryocytes and erythroblasts; C.E.v.d.S. provided critical expertise; T.W. isolated cells and RNA; F.D. provided critical statistical expertise; C.F.L. designed research and performed microarray analysis; S.T. and B.G. designed research, analyzed data, and wrote the paper; and W.H.O. designed research and wrote the paper.
A complete list of the members of the Bloodomics Consortium appears in Document S1, available on the Blood website.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nicholas A. Watkins, Department of Haematology, University of Cambridge & National Health Service Blood and Transplant Cambridge. Long Road, Cambridge, CB2 2PT United Kingdom; e-mail: naw23@cam.ac.uk.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal