Abstract
Unique and shared cytogenetic abnormalities have been documented for marginal zone lymphomas (MZLs) arising at different sites. Recently, homozygous deletions of the chromosomal band 6q23, involving the tumor necrosis factor alpha–induced protein 3 (TNFAIP3, A20) gene, a negative regulator of NF-κB, were described in ocular adnexal MZL, suggesting a role for A20 as a tumor suppressor in this disease. Here, we investigated inactivation of A20 by DNA mutations or deletions in a panel of extranodal MZL (EMZL), nodal MZL (NMZL), and splenic MZL (SMZL). Inactivating mutations encoding truncated A20 proteins were identified in 6 (19%) of 32 MZLs, including 2 (18%) of 11 EMZLs, 3 (33%) of 9 NMZLs, and 1 (8%) of 12 SMZLs. Two additional unmutated nonsplenic MZLs also showed monoallelic or biallelic A20 deletions by fluorescent in situ hybridization (FISH) and/or SNP-arrays. Thus, A20 inactivation by either somatic mutation and/or deletion represents a common genetic aberration across all MZL subtypes, which may contribute to lymphomagenesis by inducing constitutive NF-κB activation.
Introduction
Marginal zone lymphoma (MZL), a B-cell non-Hodgkin lymphoma (B-NHL) derived from marginal zone B cells, is currently classified as extranodal MZL (EMZL), nodal MZL (NMZL), and splenic MZL (SMZL).1 Four recurrent and mutually exclusive chromosomal translocations: t(11;18)(q21;q21), t(1;14)(p22;q32), t(14;18)(q32;q21), and t(3;14)(p14.1;q32), have been described in EMZLs, but not in NMZLs or SMZLs, with frequencies ranging from 0% to 40% depending on the anatomic site and geographic regions.2–4 At least 3 of these translocations, t(11;18) API2/MALT1, t(1;14) IgH/BCL10, and t(14;18) IgH/MALT, result in constitutive activation of nuclear factor-κB (NF-κB), a transcription factor complex regulating multiple cellular processes, including cell growth and survival.5 In addition, trisomy 3 or 18 has been reported in 30% to 60% of all MZLs and 7q22-32 deletions or translocations of the immunoglobulin heavy chain gene with various partners are found in 7% to 40% of SMZLs.6,7 The functional consequences of these aberrations are, however, unknown. Currently, approximately 25% of MZLs lack any recognizable recurrent genetic alteration, and evidence of lesions affecting tumor suppressor genes in MZL is limited.8,9 Recently, deletions of the 6q23.3-q24.1 region containing the tumor necrosis factor alpha–induced protein 3 (TNFAIP3, A20), a negative regulator of NF-κB, were described in ocular adnexal MZLs.10 Here, we report that A20 is targeted and inactivated by both somatic mutations and/or deletions in a significant fraction of MZL subtypes, indicating a role of NF-κB deregulation in the pathogenesis of these B-NHLs.
Methods
Case selection
Frozen samples of newly diagnosed MZL (lesional content > 70%) and matched normal control tissue were obtained from the tumor banks of the Departments of Pathology, Columbia University and the Division of Hematology, Amedeo Avogadro University of Eastern Piedmont. Hematoxylin and eosin–stained sections were used for morphologic analysis. Immunohistochemical (IHC) staining and 4-color flow cytometry were performed for phenotypic characterization using antibodies described in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). MZLs were classified according to the current WHO classification1 as EMZL (n = 11: 4 lung, 4 parotid gland, 1 skin, 1 jejunum, 1 orbit), NMZL (n = 9), and SMZL (n = 12; Table S1). The institutional review boards of Columbia University Medical Center and University of Eastern Piedmont approved this study.
Karyotype, fluorescent in situ hybridization analysis, and SNP-array
Giemsa (G)–banded karyotype analysis was performed by standard methods. Fluorescence in situ hybridization (FISH), on either methanol–acetic acid–fixed cells or formalin-fixed paraffin-embedded sections was performed as previously described.11,12 FISH probes included IgH and MALT1 dual-color break-apart probes and centromeric probes for chromosomes 3 and 18 for all cases, and IAP2/MALT1 dual-color dual-fusion probes as indicated (Vysis, Downers Grove, IL). FISH analysis of the A20 locus was performed using BAC clones RP11-703G8 and RP11-102P5 spanning the gene (BACPAC Resources, http://bacpac.chori.org). A locus-specific probe for BLIMP1 was used in all cases with A20 deletions as described.12 The sensitivity of FISH in detecting A20 deletions on paraffin sections was determined by analysis of analogously processed normal tonsils. The threshold for detecting A20 deletions was 9.7% plus or minus 4.6% and for monosomy 6 (loss of the A20 and the centromere 6 probe) was 33.5% plus or minus 12.5% of cells with signal loss. Deletion or monosomy 6 was diagnosed if the fractional signal loss exceeded the threshold mean plus 1 SD. Genome-wide DNA profiles were analyzed for 29 cases using SNP-arrays (Document S1).
DNA extraction, PCR amplification, and sequencing
Genomic DNA was extracted from frozen or paraffin-embedded tissue according to standard methods. Rearranged IgVH genes were amplified as previously described, and the mutation frequencies were determined using the IMGT (http://www.ebi.ac.uk/imgt/)13 and NCBI (http://www.ncbi.nlm.nih.gov/igblast/)14 databases.15 Primers and conditions for polymerase chain reaction (PCR) amplification of all A20 coding exons are described in Table S4. Purified amplicons were sequenced directly from both strands (Genewiz, South Plainfield, NJ) and compared with corresponding germ-line sequences (NM 006290.2). All mutations were confirmed on independent PCR products, and germ-line polymorphisms, including changes listed in the NCBI SNP database16 or present in available matched normal DNA, were excluded. Identity of matched normal DNA was verified by analyzing known polymorphisms (Table S4).
Additional methods descriptions are provided in Document S1.
Results and discussion
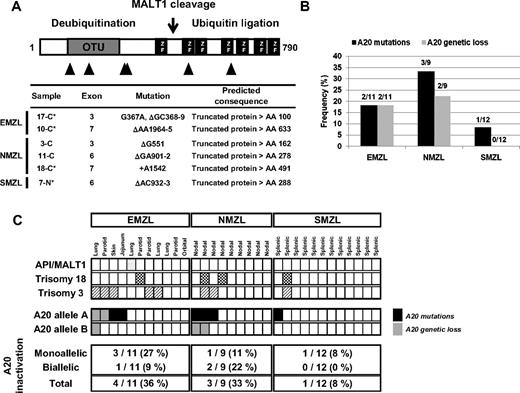
The recent description of chromosome region 6q23 deletions, targeting the A20 gene, in ocular adnexal MZL,10 prompted us to investigate the possibility that A20 might represent a candidate tumor suppressor in MZL. Thirty-two MZLs representing all WHO subtypes were analyzed by SNP-arrays; frequencies of recurrent aberrations detected are reported in Figure S1. Sequencing of the A20 coding exons showed 5 1- to 2-bp deletions and a single base pair insertion, all leading to premature stop codons, in 5 (25%) of 20 nonsplenic and 1 (8%) of 12 splenic MZLs. The somatic nature of mutations was confirmed by analyzing matched normal DNA available in 4 cases (Figure 1A; Table S2). All A20 mutations were predictive of truncated polypeptides lacking the functionally relevant domains.17 In addition, genetic loss of A20, determined by locus-specific FISH probes, was detected in 4 (20%) of 20 nonsplenic MZLs, including one with biallelic loss, but not in SMZL (Figures 1B and 2B). Two of the 4 A20 deletions seen by FISH were also detected by SNP-arrays (Table S3). No PRDM1 (Blimp1) deletions were detected by FISH in MZL with A20 deletions (data not shown). Thus, specific biallelic inactivation of A20 either via deletion of both alleles or frameshift mutations of one allele and loss of the second allele was identified in 3 (15%) of 20 nonsplenic MZLs, whereas an additional 4 (20%) of 20 nonsplenic MZLs displayed monoallelic A20 inactivation: mutations (N = 3) and deletion (N = 1; Figure 2C). No evidence for epigenetic silencing of A20 via promoter methylation of CpG islands was found in 10 of 32 cases analyzed (data not shown). In summary, our results indicate that A20 is monoallelically or biallelically inactivated in 25% of MZLs, arising in both nodal (3/9, 33%) and extranodal (4/11, 36%) sites, and in a minority of SMZLs (1/12, 8%), with evidence of a classic 2-hit mechanism observed in 3 (15%) of 20 nonsplenic MZLs.
Inactivation of A20 by mutation and deletion. (A) Representative chromatograms of A20 exon 7 genomic sequences obtained by direct sequencing of tumor and matched normal DNA from a nodal MZL (case 18-C), with a 1-bp somatic insertion leading to a frameshift. Positions according to reference sequence NM_006290.2. (B) Dual-color FISH analysis of an extranodal MZL (case 9-C), hybridized with A20 probes (red) and a chromosome 6 centromeric probe (green). Red arrows indicate cells with homozygous A20 deletions; white arrows point to cells with a normal signal pattern. (Three hundred cells were analyzed, fluorescence signals were captured after staining with 4′-6-diamidino-2-phenylindole [DAPI] using the Cytovision Imaging System [Applied Imaging, Santa Clara, CA] attached to a Nikon Eclipse 600 microscope 100×/1.40 NA oil objective [Nikon Instruments, Melville, NY.])
Inactivation of A20 by mutation and deletion. (A) Representative chromatograms of A20 exon 7 genomic sequences obtained by direct sequencing of tumor and matched normal DNA from a nodal MZL (case 18-C), with a 1-bp somatic insertion leading to a frameshift. Positions according to reference sequence NM_006290.2. (B) Dual-color FISH analysis of an extranodal MZL (case 9-C), hybridized with A20 probes (red) and a chromosome 6 centromeric probe (green). Red arrows indicate cells with homozygous A20 deletions; white arrows point to cells with a normal signal pattern. (Three hundred cells were analyzed, fluorescence signals were captured after staining with 4′-6-diamidino-2-phenylindole [DAPI] using the Cytovision Imaging System [Applied Imaging, Santa Clara, CA] attached to a Nikon Eclipse 600 microscope 100×/1.40 NA oil objective [Nikon Instruments, Melville, NY.])
Mono- and bi-allelic inactivation of A20 in MZL. (A) Distribution and features of A20 mutations in MZL. Schematic representation of the human A20 protein with its functional domains (OTU indicates ovarian tumor domain, mediating the deubiquitinating activity of A20; ZF, zinc-finger domain, exerting the ubiquitin ligase activity of A20); the cleavage site of A20 by the MALT1 protease18 is also indicated. The approximate location of A20 mutations is indicated below the map with triangles, and the types of mutations are described in detail in the table. *In these cases, the somatic origin of the mutation was confirmed by analysis of matched normal DNA. (B) Frequencies of A20 mutations and genetic loss in MZL subtypes. (C) Allelic distribution of A20 inactivation by mutations and deletions and known recurrent cytogenetic aberrations for all MZL cases analyzed. Each column represents 1 case, and the sites of the EMZL are indicated.
Mono- and bi-allelic inactivation of A20 in MZL. (A) Distribution and features of A20 mutations in MZL. Schematic representation of the human A20 protein with its functional domains (OTU indicates ovarian tumor domain, mediating the deubiquitinating activity of A20; ZF, zinc-finger domain, exerting the ubiquitin ligase activity of A20); the cleavage site of A20 by the MALT1 protease18 is also indicated. The approximate location of A20 mutations is indicated below the map with triangles, and the types of mutations are described in detail in the table. *In these cases, the somatic origin of the mutation was confirmed by analysis of matched normal DNA. (B) Frequencies of A20 mutations and genetic loss in MZL subtypes. (C) Allelic distribution of A20 inactivation by mutations and deletions and known recurrent cytogenetic aberrations for all MZL cases analyzed. Each column represents 1 case, and the sites of the EMZL are indicated.
We next determined the relationship of A20 inactivation with recurrent translocations: t(11;18), t(1;14), t(14;18), and trisomy 18. These translocations, and possibly also trisomy 18 through increased levels of MALT1, result in constitutive NF-κB activation. One case with trisomy 18 showed A20 inactivation, whereas the case with API2-MALT1 translocation did not (Figure 2C). Interestingly, a mutually exclusive pattern of A20 deletions and translocations involving MALT1 or BCL10 was observed in a recent series of EMZLs.19
A20 is known to inhibit the NF-κB pathway via its dual enzymatic activities.17 A20−/− mice develop severe multiorgan inflammation due to prolonged NF-κB activation.20 Biallelic as well as monoallelic inactivation of A20 can lead to activation of NF-κB, since NF-κB target genes are up-regulated in the liver of A20 haploinsufficient mice.21 Homozygous deletions encompassing the A20 locus were recently reported in EMZLs from various sites,10,19 but A20 inactivating mutations were either not found10 or not assessed19 in cases with heterozygous A20 deletions. Inactivation of A20 by the typical 2-hit mechanism, deletions and inactivating mutations, as observed by us in MZL has been detected in Hodgkin lymphoma, primary mediastinal B-cell lymphoma,22 and activated B-cell–like diffuse large B-cell lymphoma,23 all frequently associated with constitutive activation of the NF-κB pathway.5 Thus, A20 inactivation may represent a common mechanism for constitutive NF-κB activation, which may contribute to lymphomagenesis by stimulating cell proliferation and survival. Larger studies are warranted to determine the true frequency of A20 inactivation across MZL subtypes. The identification of genetic lesions in MZLs that deregulate NF-κB activity offers the possibility of targeted therapeutic intervention.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the Molecular Pathology Facility of the Herbert Irving Comprehensive Cancer Center for tissue procurement and preparation.
This work was supported by Oncosuisse (BIL-KLS-01522-02-2004, OCS-02034-02-2007), the Swiss National Science Foundation (1145, 205321-112430), Bernese Scholarship for Cancer Research (Bern, Switzerland), Zuppinger-Stiftung (Bern, Switzerland), Fondazione per la Ricerca e la Cura sui Linfomi (Lugano, Switzerland), Novara-AIL Onlus and Ricerca Sanitaria Finalizzata (Regione Piemonte, Italy), and grants from the National Institutes of Health (Bethesda, MD) and the Leukemia & Lymphoma Society (White Plains, NY).
National Institutes of Health
Authorship
Contribution: U.N. designed and performed most of the experiments, analyzed data, and wrote the paper; G.B. and F.B. designed research, analyzed data, and wrote the paper; A.R. performed the SNP-array analysis and assessed the methylation of the A20 promoter; S.V.N. and V.V.M. performed karyotype and FISH analyses; M. Compagno helped in the sequencing analysis; I.K., P.M.V.R., and F.B. were involved in the analysis of SNP-array data; G.B., M. Cerri, D.R., and G.G. provided and characterized MZL samples; and E.Z., L.P., and R.D.-F. provided advice and designed research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Govind Bhagat, Department of Pathology, College of Physicians and Surgeons, Columbia University, VC14-228, 630 W 168th St, New York, NY 10032; e-mail: gb96@columbia.edu; or Francesco Bertoni, Laboratory of Experimental Oncology, Oncology Institute of Southern Switzerland (IOSI), Via V Vela 6, CH-6500 Bellinzona, Switzerland; e-mail: frbertoni@mac.com.
References
Author notes
*G.B. and F.B. contributed equally to this work.

![Figure 1. Inactivation of A20 by mutation and deletion. (A) Representative chromatograms of A20 exon 7 genomic sequences obtained by direct sequencing of tumor and matched normal DNA from a nodal MZL (case 18-C), with a 1-bp somatic insertion leading to a frameshift. Positions according to reference sequence NM_006290.2. (B) Dual-color FISH analysis of an extranodal MZL (case 9-C), hybridized with A20 probes (red) and a chromosome 6 centromeric probe (green). Red arrows indicate cells with homozygous A20 deletions; white arrows point to cells with a normal signal pattern. (Three hundred cells were analyzed, fluorescence signals were captured after staining with 4′-6-diamidino-2-phenylindole [DAPI] using the Cytovision Imaging System [Applied Imaging, Santa Clara, CA] attached to a Nikon Eclipse 600 microscope 100×/1.40 NA oil objective [Nikon Instruments, Melville, NY.])](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/20/10.1182_blood-2008-08-174110/5/m_zh89990935220001.jpeg?Expires=1769274499&Signature=dURaRR36ugjD-daWIrR9gDWjrjgBzvcpol373MW1xamGQGBN~ya4R314rQjI3L~1D6EjxVssIN5NfVH2k8UAFkSB40g18jiHi9v1VKFgC-VBTAps6jD4Ebif0FY~yTHCaZ~C8WMsCVb3qm9gzqobzT~6jMClk1d~G~uQ3AzronRN6B43oFJaxnNfTJ0q4DOfl7hBko4RqoauzInsoJBxbaxGcx36BB4u~w06PaqeeWpiB6t-Qnm1CoDiM2ovrxXZ468DZ5YrgW9Ls1zqJzYsqhyuzk7I2-LARc04RtERN8Q0-cxDDdNCIbVZ9Q3prPh81XMJ9eDL9iQOAcjMupeagA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)