Abstract
Anaplastic large cell lymphoma (ALCL) is characterized by the presence of the t(2;5)(p23;q35) generating the nucleophosmin-anaplastic lymphoma kinase (NPM-ALK) fusion protein, a hyperactive kinase with transforming properties. Among these properties is the ability to regulate activity of the p53 tumor suppressor protein. In many human cancers, p53 is inactivated by mutation or other means, in some cases as a result of up-regulation of the negative regulator MDM2. However, the majority of ALK-expressing ALCL carry wild-type p53 and do not over express MDM2. We demonstrate a novel p53-dependent pathogenetic mechanism in ALK-expressing lymphoma. We confirm previously published reports of NPM-ALK–induced activation of the phosphoinositide (PI) 3-kinase and Jun N-terminal kinase (JNK) stress-activated protein (SAP) kinase proteins, but in this study demonstrate a role for these in the regulation of p53 activity in an intricate signaling system. Specifically, constitutive ALK signaling leads to the functional inactivation and/or degradation of p53 in JNK and MDM2 dependent manners. We also show nuclear exclusion of p53 in a PI 3-kinase–dependent manner. Furthermore, we demonstrate that reactivation of p53 in ALK-expressing cells as a result of pharmacologic inhibition of JNK, PI 3-kinase, and/or MDM2 activities results in the induction of apoptosis suggesting a novel therapeutic modality.
Introduction
Nucleophosmin-anaplastic lymphoma kinase (NPM-ALK) is generated as a result of a translocation between chromosomes 2 and 5 fusing the N-terminal domain of NPM to the entire intracellular region of ALK.1,2 NPM-ALK is a constitutively active tyrosine kinase, which generates a signalosome consisting of at least 46 proteins activating a whole plethora of downstream signal transduction events.3,4 This activity results in the generation of antiapoptotic signals as well as proliferative ones involving the phospholipase C-γ (PLC-γ), STAT3/5-JAK, phosphoinositide (PI) 3-kinase, and Ras-MAP kinase pathways among others.1,5-8
A normal cellular role for full-length ALK has yet to be fully elucidated, but current research places its expression in developing neural tissues, foregut, midgut, testes, and ovary.9,10 Ligands for ALK include pleiotrophin (PTN) and midkine, although recent data have suggested ALK is activated secondary to the receptor protein tyrosine phosphatase β/ζ (RPTPβ/ζ), which dephosphorylates and inactivates ALK in the absence of PTN.11-13 Conversely, NPM is a phosphoprotein whose functions are increasingly revealed after its discovery in a mutated form in acute myeloid leukemia (AML).14 NPM acts not only as a ribonuclear shuttle protein but also to stabilize the p53/ARF tumor suppressor pathway and as a regulator of centrosome duplication among others.15-17 The juxtaposition of the oligomerization domain of NPM to the kinase domain of ALK results in the generation of a hyperactive tyrosine kinase and is associated with the majority of cases of anaplastic large cell lymphoma (ALCL).1,2
The p53 tumor suppressor pathway is often disrupted in cancer cells, in some cases as a result of mutation of p53 itself or as a result of increased expression of the negative regulator MDM2. Neither of these events are predominant in ALK-expressing ALCL suggesting that these lymphoma cells control p53-induced apoptosis and cell cycle arrest through alternative pathways.18,19
We show that NPM-ALK regulates a dynamic equilibrium in which p53 is excluded from the nucleus and degraded. As a result of PI 3-kinase activity, p53 is associated with cellular structures in the perinucleus/cytoplasm and hence is excluded from the nucleus but primed for nuclear entry and subsequent activity. Destabilization of p53 occurs in both MDM2 and Jun N-terminal kinase (JNK)–dependent manners, effects that can be reversed on treatment with respective inhibitors. However, JNK also destabilizes MDM2, and therefore JNK inhibition results in up-regulation of p53 and MDM2 with a partially neutralizing net-effect. Inhibition of MDM2 with nutlin-3 stabilizes p53 leading to transcription of downstream targets, an effect which is antagonized on dual inhibition of MDM2 and PI 3-kinase presumably due to a defect in p53 nuclear transport. Overall, stabilization of p53 by administration of nutlin-3 acts synergistically with p53-inependent effects of JNK and PI 3-kinase inhibition to induce apoptosis of NPM-ALK–expressing cells.
Methods
Patient material
Redundant archival biopsy specimens were obtained from the Children's Cancer and Leukemia Study Group (United Kingdom) under study number 2006BS10 with approval from the appropriate research ethics committee (06/MRE04/90). All cases were reviewed by histopathology before performing fluorescence in situ hybridization (FISH) and immunohistochemistry. In total, 10 cases of ALK+ and 5 cases of ALK− ALCL were screened.
Cell lines
The human-derived NPM-ALK–expressing ALCL cell lines SUDHL-1, Karpas-299, and DEL were purchased from DSMZ (Braunschweig, Germany). These lines were cultured in RPMI 1640 supplemented with glutamine, penicillin/streptomycin, and 10% fetal calf serum (FCS; Invitrogen, Paisley, United Kingdom). The parental and NPM-ALK–expressing BaF3 cell lines were kindly provided by Prof S. Morris (St Jude Children's Research Hospital, Memphis, TN). The NPM-ALK–expressing BaF3 cells were cultured in media as described previously. The parental BaF3 cells were cultured in the presence of interleukin-3 (IL-3; 2 ng/mL; PeproTech, London, United Kingdom).
Antibodies
The phospho-MDM2 (ser166), p21, p53 (1C12), and phospho-NPM-ALK (tyr664) antibodies were purchased from Cell Signaling Technologies (Boston, MA). The MDM2 (smp14) and p53 (DO-1) antibodies were obtained from Santa Cruz Biotechnology (Heidelberg, Germany). The monoclonal ALK antibody (35-4300) was purchased from Invitrogen (Paisley, United Kingdom). α-tubulin and β-actin antibodies were purchased from Sigma-Aldrich (Poole, United Kingdom).
Inhibitors
The MDM2 antagonists nutlin-3 and NSC66811, JAK3/NPM-ALK inhibitor WHI-P154, JNK inhibitors SP600125 and JNK inhibitor I, MAP kinase inhibitor PD98059, PI 3-kinase inhibitor LY294002, and its inactive analog LY303511 were purchased from Calbiochem (San Diego, CA) and used at concentrations and for periods of time as indicated. All inhibitor compounds were resuspended in dimethyl sulfoxide (DMSO).
Knockdown of NPM-ALK by siRNA
The ON-TARGETplus SMARTpool small interfering RNA (siRNA) of NPM-ALK and negative control reagents were purchased from Dharmacon (Chicago, IL). Transient transfection of SUDHL-1 cells was performed using the Amaxa Nucleofector system and the Amaxa cell line kit T reagent (Amaxa, Cologne, Germany) according to manufacturer's instructions. Briefly, 4 × 106 cells were transfected with 1.5 μg of the siRNA in 100 μL Amaxa reagent. At 48 hours after transfection, cells were counted with trypan blue to exclude dead cells, and NPM-ALK protein and transcript levels were determined by Western blot analysis and quantitative reverse transcription polymerase chain reaction (qRT-PCR), respectively.
Western blot analysis and IP
Western blot analysis was performed as previously described.20 Briefly, cells were washed in phosphate-buffered saline (PBS), then lysed on ice for 10 minutes in Nonidet P-40 (NP-40) lysis buffer (30 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% NP-40, 5 mM ethylenediaminetetraacetic acid [EDTA], 1 mM sodium orthovanadate, 1 mM NaF, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM dithiothreitol [DTT], and protease inhibitor cocktail, EDTA-free [Roche, Basel, Switzerland]). Supernatant was harvested after centrifugation for 3 minutes at 12 000g. Nuclear extracts were prepared as indicated using a nuclear extract kit (Active Motif, Rixensart, Belgium). The supernatant was added in equal volume to 2× Laemmli sample buffer before boiling for 5 minutes. Samples were loaded onto 11% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels as indicated and proteins transferred to Immobilon polyvinylidene fluoride (PVDF) membrane (Millipore, Watford, United Kingdom) by semidry transfer (TE77, Hoefer SemiPhor; Pharmacia Biotech, San Francisco, CA). Membranes were blocked in 5% nonfat milk, followed by incubation with primary antibody (phosphospecific antibodies were applied overnight at 4°C and other antibodies were incubated at room temperature for 1 hour). After 3 washes in Tris-buffered saline with Tween 20 (TBST) of 10 minutes each, membranes were incubated with horseradish peroxidase (HRP)–conjugated secondary (Dako UK, Ely, United Kingdom) and proteins detected using Immobilon enhanced chemiluminescence (ECL) reagent (Millipore) and a FUJI LAS4000 chemiluminescent analyzer (Raytek, Sheffield, United Kingdom). Quantification of protein levels in relation to an α-tubulin or β-actin loading control was determined directly from the membranes using AIDA software (Fuji, Tokyo, Japan).
For immunoprecipitation (IP), equal quantities of cell lysates were incubated with anti-p53 (DO-1) or anti-MDM2 (smp14) antibodies, which had been cross-linked to protein G agarose beads (Santa Cruz Biotechnology), via rotation overnight at 4°C. The immunocomplexes were then washed 5 times with ice-cold NP-40 lysis buffer. Proteins were released by boiling at 95°C for 10 minutes in Laemmli sample buffer, and analyzed by Western blot analysis as described above. Quantification of protein levels was determined directly from the membrane as detailed earlier.
Quantitative RT-PCR
Total cellular RNA was extracted using the RNeasy Plus Mini kit (QIAGEN, Crawley, United Kingdom) according to the manufacturer's instructions. cDNA was synthesized from total RNA using SuperScript III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen). Gene expression levels were measured by qRT-PCR using SYBR Green PCR Master Mix (Invitrogen) and an iCycler Thermal Cycler (Bio-Rad Laboratories, Watford, United Kingdom). Primer sequences were as shown in Table 1. All PCRs were run in triplicate. After PCR amplification, melting curve analysis was used to check for contamination and primer dimerization. The mRNA levels were determined by a comparative threshold cycle (Ct) method after normalization by 18S rRNA.21
Primers used to perform qRT-PCR
| Gene . | Forward primer . | Reverse primer . |
|---|---|---|
| Noxa | 5′-TGGAAGTCGAGTGTGCTACTCAACT-3′ | 5′-AGATTCAGAAGTTTCTGCCGGAA-3′ |
| Puma | 5′-CCTGGAGGGTCCTGTACAATCT-3′ | 5′-GCACCTAATTGGGCTCCATCT-3′ |
| p21 | 5′-CCTCATCCCGTGTTCTCCTTT-3′ | 5′-GTACCACCCAGCGGACAAGT-3′ |
| MDM2 | 5′-CCCCTTCCATCACATTGCA-3′ | 5′-AGTTTGGCTTTCTCAGAGATTTCC-3′ |
| p53 | 5′-TCAACAAGATGTTTTGCCAACTG-3′ | 5′-ATGTGCTGTGACTGCTTGTAGATG-3′ |
| c-Jun | 5′-CGCCAAGAACTCGGACCTC-3′ | 5′-CCTCCTGCTCATCTGTCACG-3′ |
| S18rRNA | 5′-TGACTCAACACGGGAAACC-3′ | 5′-TCGCTCCACCAACTAAGAAC-3′ |
| CDKN2A Exon 1A | 5′-CATAGATGCCGCGGAAGGT-3′ | 5′-CCCGAGGTTTCTCAGAGCCT-3′ |
| CDKN2A Exon 2 | 5′-CCACCCTGGCTCTGACCAT-3′ | 5′-GCCACTCGGGCGCTG-3′ |
| Gene . | Forward primer . | Reverse primer . |
|---|---|---|
| Noxa | 5′-TGGAAGTCGAGTGTGCTACTCAACT-3′ | 5′-AGATTCAGAAGTTTCTGCCGGAA-3′ |
| Puma | 5′-CCTGGAGGGTCCTGTACAATCT-3′ | 5′-GCACCTAATTGGGCTCCATCT-3′ |
| p21 | 5′-CCTCATCCCGTGTTCTCCTTT-3′ | 5′-GTACCACCCAGCGGACAAGT-3′ |
| MDM2 | 5′-CCCCTTCCATCACATTGCA-3′ | 5′-AGTTTGGCTTTCTCAGAGATTTCC-3′ |
| p53 | 5′-TCAACAAGATGTTTTGCCAACTG-3′ | 5′-ATGTGCTGTGACTGCTTGTAGATG-3′ |
| c-Jun | 5′-CGCCAAGAACTCGGACCTC-3′ | 5′-CCTCCTGCTCATCTGTCACG-3′ |
| S18rRNA | 5′-TGACTCAACACGGGAAACC-3′ | 5′-TCGCTCCACCAACTAAGAAC-3′ |
| CDKN2A Exon 1A | 5′-CATAGATGCCGCGGAAGGT-3′ | 5′-CCCGAGGTTTCTCAGAGCCT-3′ |
| CDKN2A Exon 2 | 5′-CCACCCTGGCTCTGACCAT-3′ | 5′-GCCACTCGGGCGCTG-3′ |
Proliferation and apoptosis assays
Cellular metabolism was evaluated as a surrogate of proliferation by the AlamarBlue assay (AbD Serotec, Oxford, United Kingdom) according to the manufacturer's instructions. The caspase-3/7 activity of cells was measured as a marker of apoptosis using the Caspase-Glo 3/7 Assay kit (Promega, Southampton, United Kingdom) according to the manufacturer's instructions.
Flow cytometric analysis of cell cycle
Cell-cycle analysis was performed after intracellular propidium iodide staining and flow cytometry. Briefly, for each analysis, 0.5 × 106 cells were harvested after treatment and fixed using 100% ice-cold ethanol while vortex mixing. Analysis was performed on a FACSsort cytometer with an argon laser at 488 nm (Becton Dickinson, San Jose, CA). Data were analyzed using FlowJo cell-cycle analysis software (TreeStar, Ashland, OR).
Immunohistochemistry
Immunohistochemistry was performed on formalin-fixed and paraffin-embedded tissues. Briefly, 4-μm tissue sections were heat-treated in citrate buffer, pH 6.0 (Dako UK) in a pressure cooker for 3 minutes. Sections were incubated with the mouse monoclonal MDM2 (smp14) or the rabbit polyclonal phospho-MDM2 (ser166) primary antibodies mentioned above at a dilution of 1:50 for 1 hour. Tissues were then incubated with appropriate biotinylated secondary antibodies (Dako UK) followed by peroxidase-conjugated avidin (Sigma-Aldrich). Staining was visualized with 3,3-diaminobenzidine tetrahydrochloride (DAB) substrate followed by hematoxylin nuclear counterstaining.
Interphase FISH
Interphase FISH was performed on formalin-fixed, paraffin-embedded tissues as previously described.22 Chromosome translocations involving ALK were detected with a LSI ALK dual-color, break-apart rearrangement probe (Abbott Molecular, Maidenhead, United Kingdom), composed of 2 differently labeled probes located either 3′ telomeric (Spectrum Orange) or 5′ centromeric (Spectrum Green) to ALK.
Immunofluorescence and confocal microscopy
Cells were fixed in 3% formaldehyde/PBS for 10 minutes then permeabilized with 0.05% Triton X-100. After 2 washes with ice-cold PBS, cells were incubated for 2 hours with either phospho-MDM2 (ser166) or p53 antibodies. Cells were washed with PBS and then incubated with appropriate biotinylated secondary antibodies (Dako UK) for 1 hour followed by streptavidin-fluorescein isothiocyanate (FITC; Dako UK) for 30 minutes. The samples were mounted in VECTASHIELD mounting medium with 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Peterborough, United Kingdom) and examined on a Leica TCS SP confocal microscope (Leica Microsystems, Milton Keynes, United Kingdom).
Statistical analysis
One-way analysis of variance (ANOVA) was used to compare the effects of the selected inhibitors on caspase 3/7 activity and proliferation. Statistical significance was considered when P values were less than .01.
Results
NPM-ALK activity leads to phosphorylation of MDM2 on serine 166 via PI 3-kinase
We examined primary patient tissue for the presence of ALK-translocations using a dual-color ALK-specific FISH break-apart probe (Figure 1A left bottom). Of the 15 cases screened, 10 were positive for ALK translocations. Positive and negative cases were then screened by immunohistochemistry for the expression of MDM2 and the serine166 phosphorylated form of this protein (pMDM2). While MDM2 was not overexpressed in any of the patient samples, there was stronger staining of pMDM2 in ALK+ cellular nuclei precluding DAPI uptake (10/10 cases; Figure 1A top left) than in ALK− cases, where nuclei stained brighter with DAPI (5/5 cases; Figure 1A top right). However, MDM2 stained spontaneously in cytoplasm and nuclear compartments of both ALK-positive and -negative ALCL cases to a low level (Figure 1A right bottom).
NPM-ALK phosphorylates MDM2 on serine 166. (A) MDM2 is phosphorylated on serine 166 in archival biopsy specimens isolated from NPM-ALK–expressing ALCL (top left panel). Conversely, ALK− cases do not strongly express MDM2 in a phosphorylated form (top right panel). All cases were screened for the presence of a translocation involving ALK on chromosome 2 by FISH using an ALK break-apart probe (bottom left panel shows an ALK+ case) and for the expression of MDM2 by immunohistochemistry (bottom right panel shows a case representative of all samples regardless of ALK status). Representative samples are shown of the 10 ALK+ cases screened. Likewise MDM2 is phosphorylated on serine 166 in patient-derived ALK-expressing ALCL cell lines SUHL-1, Karpas-299, and DEL (B) and in BaF3 cells expressing NPM-ALK. Numbers represent arbitrary units after densitometric analysis and normalization for the tubulin loading control. (C) Treatment of SUDHL-1 cells with either (D) siRNA against ALK for 48 hours (compared with a scrambled sequence [Scr]) or (E) 10 μM of the Jak3/ALK inhibitor WHI-P154 for 2 hours, causes a reduction of pMDM2 levels in the cells. Cells were analyzed by trypan blue staining at the time of analysis to exclude significant cell death. Furthermore, 48 hours of treatment with a PI 3-kinase inhibitor (20 μM LY294002) attenuated MDM2 phosphorylation. Numbers represent arbitrary units after densitometric analysis and normalization to the loading control; vertical lines have been inserted to indicate a repositioned gel lane (F). The inhibitor compounds had no effect on MDM2 transcript levels as determined by qRT-PCR (G). Error bars represent SDs of the mean. Results are representative of at least 3 independent experiments.
NPM-ALK phosphorylates MDM2 on serine 166. (A) MDM2 is phosphorylated on serine 166 in archival biopsy specimens isolated from NPM-ALK–expressing ALCL (top left panel). Conversely, ALK− cases do not strongly express MDM2 in a phosphorylated form (top right panel). All cases were screened for the presence of a translocation involving ALK on chromosome 2 by FISH using an ALK break-apart probe (bottom left panel shows an ALK+ case) and for the expression of MDM2 by immunohistochemistry (bottom right panel shows a case representative of all samples regardless of ALK status). Representative samples are shown of the 10 ALK+ cases screened. Likewise MDM2 is phosphorylated on serine 166 in patient-derived ALK-expressing ALCL cell lines SUHL-1, Karpas-299, and DEL (B) and in BaF3 cells expressing NPM-ALK. Numbers represent arbitrary units after densitometric analysis and normalization for the tubulin loading control. (C) Treatment of SUDHL-1 cells with either (D) siRNA against ALK for 48 hours (compared with a scrambled sequence [Scr]) or (E) 10 μM of the Jak3/ALK inhibitor WHI-P154 for 2 hours, causes a reduction of pMDM2 levels in the cells. Cells were analyzed by trypan blue staining at the time of analysis to exclude significant cell death. Furthermore, 48 hours of treatment with a PI 3-kinase inhibitor (20 μM LY294002) attenuated MDM2 phosphorylation. Numbers represent arbitrary units after densitometric analysis and normalization to the loading control; vertical lines have been inserted to indicate a repositioned gel lane (F). The inhibitor compounds had no effect on MDM2 transcript levels as determined by qRT-PCR (G). Error bars represent SDs of the mean. Results are representative of at least 3 independent experiments.
We subsequently examined human patient-derived NPM-ALK–expressing ALCL cell lines for the presence of MDM2 and pMDM2 proteins by Western blot analysis. Of the human ALCL cell lines tested, all of them expressed pMDM2 and MDM2 to varying degrees (Figure 1B). In addition, we examined MDM2 expression levels and phosphorylation in an IL-3–dependent BaF3 cell line stably transfected to express NPM-ALK. MDM2 expression was not increased as a result of either IL-3 administration or NPM-ALK expression, but phosphorylation levels were increased on serine 166 in both cases (Figure 1C).
To determine whether this phosphorylation event was dependent on the activity of NPM-ALK, we knocked down expression using either siRNA against the ALK kinase domain or inhibited the kinase activity of the protein using a small molecule inhibitor known to have suppressive properties against NPM-ALK (WHI-P154, 10 μM, 2 hours treatment; Figure 1D,E, respectively). We validated the siRNA-induced silencing efficacy by densitometric analysis of protein expression levels (from 100% to 24% after 48 hours of treatment with siRNA; Figure 1D) and qRT-PCR (data not shown). In both cases (siRNA or inhibitor compound) the levels of MDM2 phosphorylation were significantly reduced. We next set out to determine whether this activity was a direct result of NPM-ALK activity or indirectly via a downstream kinase. We used the PI 3-kinase inhibitor LY294002 (20 μM, 48 hours) to inhibit the activity of this pathway and measured pMDM2 levels in comparison to cells treated with DMSO alone. SUDHL-1 cells treated with LY294002 displayed a decrease in both pMDM2 and MDM2 protein as shown by Western blot analysis (ratio of pMDM2:MDM2 decreased from 0.6 to 0.5 after LY294002 treatment), whereas transcript levels of MDM2 after qRT-PCR, showed no significant change (Figure 1F,G). These data suggest that PI 3-kinase acts to phosphorylate and stabilize MDM2 in SUDHL-1 cells.
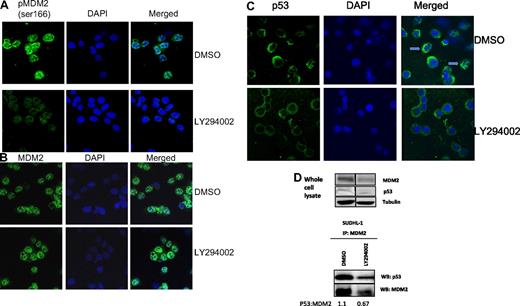
To investigate the consequences of MDM2 phosphorylation on cellular location, we performed fluorescent staining of SUDHL-1 cells after inhibitor treatment as detailed. Cells were imaged by confocal microscopy. Treatment with the PI 3-kinase inhibitor LY294002 resulted in decreased staining for pMDM2 (Figure 2A), although the cellular localization of total MDM2 protein altered only slightly, shifting to the cytoplasm (Figure 2B). However, p53 is still excluded from the nucleus in response to LY294002 suggesting a role for PI 3-kinase in the localization of p53 (Figure 2C). Specifically, p53 appeared to move to a more diffuse cytoplasmic staining pattern as opposed to attachment to cytoskeletal structures (indicated by arrows also shown in Figure S3, available on the Blood website; see the Supplemental Materials link at the top of the online article). On examining whole cell lysates prepared from LY294002-treated cells, total cellular levels of p53 protein remained unchanged, but MDM2 protein decreased as did p53:MDM2 binding, which was reduced after inhibitor treatment (p53:MDM2 decreases from 1.1 to 0.67 after inhibitor treatment; Figure 2D). NPM-ALK expressing ALCL patient samples predominantly express wild-type p53,18,19 hence it may be possible to stabilize p53 to achieve growth arrest and/or apoptosis.
Phosphorylated MDM2 resides mainly in the nucleus. SUDHL-1 cells were treated with either DMSO control or LY294002 (20 μM) for 24 hours, after which cells were prepared as cytospins and stained with fluorescently labeled antibodies against p(ser166)MDM2 and DAPI (A) or MDM2 and DAPI (B). SUDHL-1 cytospins were also analyzed for p53 expression and localization 24 hours after inhibitor treatment (C). Arrows point to cells in which p53 appears to be sequestered to cytoskeletal structures. MDM2:p53 complex formation in response to 24 hours treatment with 20 μM LY294002; vertical lines have been inserted to indicate a repositioned gel lane (D). Numbers represent the ratio of p53:MDM2 protein in arbitrary units after densitometric analysis. Equal quantities of cell lysate were entered into the IPs.
Phosphorylated MDM2 resides mainly in the nucleus. SUDHL-1 cells were treated with either DMSO control or LY294002 (20 μM) for 24 hours, after which cells were prepared as cytospins and stained with fluorescently labeled antibodies against p(ser166)MDM2 and DAPI (A) or MDM2 and DAPI (B). SUDHL-1 cytospins were also analyzed for p53 expression and localization 24 hours after inhibitor treatment (C). Arrows point to cells in which p53 appears to be sequestered to cytoskeletal structures. MDM2:p53 complex formation in response to 24 hours treatment with 20 μM LY294002; vertical lines have been inserted to indicate a repositioned gel lane (D). Numbers represent the ratio of p53:MDM2 protein in arbitrary units after densitometric analysis. Equal quantities of cell lysate were entered into the IPs.
Inhibition of MDM2 activity with nutlin-3 results in decreased cell growth concomitant with activation of the p53 tumor suppressor pathway
We treated SUDHL-1 cells with the MDM2 antagonist nutlin-3 and measured either the activity or transcription of p53 effectors by Western blot analysis and qRT-PCR, respectively. Treatment with nutlin-3 for 48 hours led to the stabilization and activation of p53 in a dose-dependent fashion (Figure 3A,B). Confocal microscopy revealed increased p53 expression in the nucleus (Figure 3B). Moreover, nutlin-3 treatment resulted in the up-regulation of several p53 transcriptional targets including NOXA (3.2 log), Puma (1.1 log), p53 itself (0.9 log), and p21 (1.4 log; Figure 3C). Interestingly, we also noted an up-regulation of CDKN2A mRNA levels (Figure 3C). Binding of MDM2 to p53 was disrupted by nutlin-3 as demonstrated by IP (Figure 3D). Consequently, SUDHL-1 cells exposed to nutlin-3 showed a decrease in proliferation in a time- and dose-dependent manner as determined by AlamarBlue assay (Figure 3E). However, the growth of SUDHL-1 cells was not completely ablated, suggesting that other cell survival mechanisms used by NPM-ALK that are not targeted by nutlin-3 treatment are still active. Alternatively, p53 activity may additionally be regulated by MDM2-independent pathways. JNK has been shown to regulate the stability of p53,23 and we therefore chose to examine the status of the JNK stress-activated protein (SAP) kinase pathway.
Treatment of ALK-expressing cells with nutlin-3 stabilizes p53 leading to growth arrest. SUDHL-1 cells were treated with increasing doses of nutlin-3 (2.5-5 μM) for 48 hours, at which point cells were lysed and assayed by Western blot analysis for the expression of p53 and its downstream target p21 (A). Confocal microscopy of SUDHL-1 cytospin preparations revealed increased nuclear localization of p53 after nutlin-3 treatment for 48 hours (B). Transcript levels of p53 downstream targets were monitored by qRT-PCR after 48 hours of 20 μM nutlin-3 treatment (C). IPs for MDM2 were performed 48 hours after treatment with 20 μM nutlin-3 and bound p53 detected by Western blot analysis. Numbers represent ratios of arbitrary units after densitometric analysis of p53 and MDM2 protein levels (D). Cells were exposed to increasing doses of nutlin-3 for 6, 24, or 48 hours and monitored for cell growth by AlamarBlue incorporation (E). Error bars represent SDs of the mean. All experiments were performed at least in triplicate.
Treatment of ALK-expressing cells with nutlin-3 stabilizes p53 leading to growth arrest. SUDHL-1 cells were treated with increasing doses of nutlin-3 (2.5-5 μM) for 48 hours, at which point cells were lysed and assayed by Western blot analysis for the expression of p53 and its downstream target p21 (A). Confocal microscopy of SUDHL-1 cytospin preparations revealed increased nuclear localization of p53 after nutlin-3 treatment for 48 hours (B). Transcript levels of p53 downstream targets were monitored by qRT-PCR after 48 hours of 20 μM nutlin-3 treatment (C). IPs for MDM2 were performed 48 hours after treatment with 20 μM nutlin-3 and bound p53 detected by Western blot analysis. Numbers represent ratios of arbitrary units after densitometric analysis of p53 and MDM2 protein levels (D). Cells were exposed to increasing doses of nutlin-3 for 6, 24, or 48 hours and monitored for cell growth by AlamarBlue incorporation (E). Error bars represent SDs of the mean. All experiments were performed at least in triplicate.
NPM-ALK inhibits p53 in a JNK-dependent manner
NPM-ALK is implicated in the activation of JNK and its downstream signal transduction pathways.24 Indeed, we show NPM-ALK dependent phosphorylation of JNK (Figure 4A,B). When NPM-ALK expression was reduced in SUDHL-1 cells, JNK phosphorylation decreased dramatically (Figure 4A). In support, when NPM-ALK was stably expressed in the BaF3 cell line, the level of JNK phosphorylation was increased compared with control cells (Figure 4A,B). Taken together, our data provide further confirmation that NPM-ALK phosphorylates JNK.24 We further investigated the role of JNK in the regulation of p53 in SUDHL-1 cells using the specific JNK inhibitors, SP600125 (Figure 4C) and JNK inhibitor I (Figure S2). Unlike LY294002, but like nutlin-3, SP600125 enabled p53 stabilization (Figure 4C and Figure S2). This activity also resulted in increased nuclear localization consistent with protein stabilization (Figure 4D). To investigate the mechanism of JNK-mediated inactivation of p53, we performed IPs, demonstrating that JNK and p53 exist in a complex (Figure 4E). Inhibition of JNK kinase activity with SP600125 resulted in a reduction of p53-bound JNK (Figure 4E) which, combined with the increase in p53 protein levels (Figure 4C), suggests that JNK acts to sequester p53 perhaps targeting it for degradation. In comparison, MDM2:p53 binding was not disrupted in the presence of SP600125 (Figure 4E), in fact more p53 was bound to MDM2 (p53:MDM2 ratio increases from 1.7 to 2.6) suggesting that MDM2 can to a certain extent compensate for JNK in the sequestration and degradation of p53. Indeed, p53 expression is stabilized to a higher extent after inhibition of MDM2 rather than after SP600125 treatment (Figure 4C). A role for JNK and MDM2 in the ubiquitination and hence degradation of p53 is confirmed by decreased levels of p53 ubiquitination in inhibitor-treated cells (68% of p53 protein present is ubiquitinated in DMSO-treated controls vs 38 and 46% in SP600125 and nutlin-3 treated cells, respectively), whereas LY294002-treated cells displayed increased levels of p53 ubiquitination (89%) without increasing the quantity of p53 protein present (densitometric analysis: p53 = 1.7 arbitrary units when cells were DMSO treated vs 1.6 units in LY294002-treated cells; Figure 4F). These data suggest that PI 3-kinase–mediated sequestration to cytoskeletal elements may protect p53 from degradation by MDM2 and JNK.
JNK destabilizes p53 in NPM-ALK–expressing cells. NPM-ALK phosphorylates JNK on Thr183/Tyr185 as demonstrated by down-regulation of NPM-ALK after transfection of SUDHL-1 cells with siRNA targeted to ALK for 48 hours (A). Likewise, NPM-ALK–expressing BaF3 cells express JNK in a phosphorylated form (B). Inhibition of JNK activity with the ATP competitive inhibitor SP600125 for 48 hours at a dose of 20 μM results in an increase in p53 expression in SUDHL-1 cells (C). Confocal microscopy examining p53 cellular location in SUDHL-1 cells treated with 20 μM SP600125 for 24 hours (D). IP of p53 or MDM2 protein after treatment of SUDHL-1 cells with SP600125 for 48 hours at 20 μM. Numbers represent ratios of arbitrary units after densitometric analysis of JNK and p53 protein levels; vertical lines have been inserted to indicate a repositioned gel lane (E). IP for p53 followed by Western blot analysis for ubiquitinated p53 using an anti-ubiquitin antibody. Numbers represent the percentage of p53 protein that is ubiquitinated as determined after densitometric analysis of protein levels (F).
JNK destabilizes p53 in NPM-ALK–expressing cells. NPM-ALK phosphorylates JNK on Thr183/Tyr185 as demonstrated by down-regulation of NPM-ALK after transfection of SUDHL-1 cells with siRNA targeted to ALK for 48 hours (A). Likewise, NPM-ALK–expressing BaF3 cells express JNK in a phosphorylated form (B). Inhibition of JNK activity with the ATP competitive inhibitor SP600125 for 48 hours at a dose of 20 μM results in an increase in p53 expression in SUDHL-1 cells (C). Confocal microscopy examining p53 cellular location in SUDHL-1 cells treated with 20 μM SP600125 for 24 hours (D). IP of p53 or MDM2 protein after treatment of SUDHL-1 cells with SP600125 for 48 hours at 20 μM. Numbers represent ratios of arbitrary units after densitometric analysis of JNK and p53 protein levels; vertical lines have been inserted to indicate a repositioned gel lane (E). IP for p53 followed by Western blot analysis for ubiquitinated p53 using an anti-ubiquitin antibody. Numbers represent the percentage of p53 protein that is ubiquitinated as determined after densitometric analysis of protein levels (F).
PI 3-kinase and JNK inhibitors sensitize NPM-ALK–expressing cells to nutlin-3–induced apoptosis
We further investigated whether apoptosis would occur in NPM-ALK–expressing cells as a result of disruption of the MDM2-p53 interaction by nutlin-3 and if so, whether this could be potentiated by inhibition of the PI 3-kinase or JNK pathways. We treated SUDHL-1 cells with a variety of inhibitors including nutlin-3, LY294002, SP600125, and a MEK inhibitor (PD98059) for 48 hours and monitored caspase 3/7 activity (Figure 5A,B) or poly(ADP-ribose) polymerase (PARP) cleavage (Figure 5C) as a measurement of apoptosis. Apoptosis in SUDHL-1 cells was induced by either nutlin-3 (1.43×) or SP600125 (1.55×). Moreover, the combinatorial activities of nutlin-3 and SP600125 had a less than additive effect (2.17× vs 2.98×), while nutlin-3 and LY294002 had an almost additive effect on apoptosis (2.42× vs 2.6×). PD 98059 had no effect on apoptosis in the absence of nutlin-3, suggesting that inhibition of the mitogen-activated protein kinase (MAPK) pathway is not crucial for the induction of p53-induced apoptosis in this cell line (Figure 5A).
Treatment of NPM-ALK expressing cells with an MDM2 inhibitor in combination with either JNK or PI 3-kinase inhibitors sensitizes cells to apoptosis and growth arrest. Induction of apoptosis as determined by the presence of caspase 3/7 in SUDHL-1 cells treated with combinations of 20 μM nutlin-3, 20 μM LY294002, and/or 20 μM SP600125 for 48 (A), 6, or 24 hours (B). Apoptosis was confirmed after Western blot analysis of cell lysates for PARP (C). Cellular proliferation is detected after incorporation of AlamarBlue 48 (D), 6, or 24 (E) hours after treatment of SUDHL-1 cells as indicated above. Percentages of cells in each stage of the cell cycle after treatment with inhibitors for 48 hours as indicated above (F). A representative dataset of triplicate experiments is shown in panels C and F. Error bars in panels A, B, D, and E represent SD of the mean of at least 3 independent experiments.
Treatment of NPM-ALK expressing cells with an MDM2 inhibitor in combination with either JNK or PI 3-kinase inhibitors sensitizes cells to apoptosis and growth arrest. Induction of apoptosis as determined by the presence of caspase 3/7 in SUDHL-1 cells treated with combinations of 20 μM nutlin-3, 20 μM LY294002, and/or 20 μM SP600125 for 48 (A), 6, or 24 hours (B). Apoptosis was confirmed after Western blot analysis of cell lysates for PARP (C). Cellular proliferation is detected after incorporation of AlamarBlue 48 (D), 6, or 24 (E) hours after treatment of SUDHL-1 cells as indicated above. Percentages of cells in each stage of the cell cycle after treatment with inhibitors for 48 hours as indicated above (F). A representative dataset of triplicate experiments is shown in panels C and F. Error bars in panels A, B, D, and E represent SD of the mean of at least 3 independent experiments.
We also demonstrate that nutlin-3 inhibits proliferation of SUDHL-1 cells (100% to 50%), and furthermore we show that in response to either a PI 3-kinase or JNK inhibitor, cellular proliferation is likewise inhibited, but to a lower level than observed with nutlin-3 (100% to 78% or 75%, respectively; Figure 5D). However, nutlin-3 in combination with LY294002 has an additive effect in the inhibition of proliferation (50% nutlin-3 alone to 34% in combination with LY294002; Figure 5D). Conversely, nutlin-3 in combination with the JNK inhibitor did not significantly increase the impact on proliferation that is observed on nutlin-3 treatment alone (Figure 5D). Likewise, SP600125 and LY294002 do not act synergistically to significantly inhibit tumor cell proliferation (Figure 5D).
We further investigated the effect of a triple-cocktail (20 μM each of nutlin-3 + LY294002 + SP600125) treatment. After 24 hours, the triple-cocktail treatment of SUDHL-1 cells induced higher caspase 3/7 activity (3.31×) compared with dual treatment with nutlin-3 plus SP600125 (2.45×) or nutlin-3 plus LY294002 (2.62×; Figure 5B). The triple inhibitor combination did not have a similar synergistic effect on proliferation (Figure 5E).
We also analyzed cell cycle after 48 hours of treatment of SUDHL-1 cells with combinations of inhibitors (Figure 5F). Unlike the JAK3/NPM-ALK inhibitor, WHI-P154, which induced arrest at G0/G1, nutlin-3 had no effect on cell cycle except for an increase in the sub-G1 peak indicating apoptosis (not shown). In contrast, treatment of SUDHL-1 cells with the JNK inhibitor SP600125 induced G2/M arrest (22% control treatment to 29%) in keeping with previously published data24 (Figure 5F). Conversely, treatment of cells with the PI 3-kinase inhibitor LY294003 did not induce p53 and G2/M arrest but presumably a p53-independent G0/G1 cell-cycle arrest (38% control treated to 49%). Therefore, NPM-ALK promotes cell-cycle progression mainly through activation of PI 3-kinase and JNK pathways and their p53-independent mechanisms of action.
The stabilization and transcriptional activation of p53 in response to inhibitor treatment in NPM-ALK–expressing cells
It is well documented that p53 induces transcription of MDM2, which in turn targets p53 for degradation in an autoregulatory feedback loop. We further investigated the extent of involvement of the PI 3-kinase/AKT or JNK pathways in the MDM2-p53 feedback loop (Figure 6A). Nutlin-3 treatment stabilized p53, enabling transcription of MDM2 and p21 as previously demonstrated (Figures 6A,B and 3A). The combinatorial treatments of nutlin-3 and LY294002, SP600125, or PD98059 did not show a further increase in MDM2 expression compared with nutlin-3 treatment alone (Figure 6A). In fact, treatment with nutlin-3 combined with LY294002 resulted in decreased levels of MDM2 and p21 compared with nutlin-3 treatment alone, providing further evidence of a role for PI 3-kinase in the stabilization of MDM2. However, LY294002 in combination with nutlin-3 significantly increased transcription of Noxa (2.4 log vs 1.03 log nutlin-3 alone; Figure 6B) perhaps accounting for the increase in apoptosis observed in Figure 5. Conversely, although treatment with the JNK inhibitor SP600125 enabled stabilization of p53 (Figure 5A), this did not result in an increase in transcription of the p53 target genes MDM2 and Noxa or c-Jun (Figure 6B). However, nutlin-3 treatment did cause an increase in transcription of c-Jun; an effect that was not enhanced on combining nutlin-3 treatment with the other inhibitors, suggesting a role for p53 but not JNK in the transcription of c-Jun (Figure 6B).
Transcriptional activation of p53 after inhibitor treatment. Treatment of cells with either inhibitor alone or in combination for 48 hours results in some cases in the stabilization of p53 protein levels and transcription of the p53 target proteins MDM2 and p21, as demonstrated by Western blot analysis (A). The transcription of MDM2, Noxa, and c-Jun were monitored by qRT-PCR after treatment of cells with a combination of 20 μM LY294002 and 20 μM nutlin-3 for 48 hours (B). Error bars represent SDs of the mean. SUDHL-1 cells were treated with combinations of inhibitors (at 20 μM each) for 12 hours in the presence of either leptomycin B or MG132 as indicated (C). Arbitrary units under the blots represent protein levels normalized for the actin control by densitometric analysis.
Transcriptional activation of p53 after inhibitor treatment. Treatment of cells with either inhibitor alone or in combination for 48 hours results in some cases in the stabilization of p53 protein levels and transcription of the p53 target proteins MDM2 and p21, as demonstrated by Western blot analysis (A). The transcription of MDM2, Noxa, and c-Jun were monitored by qRT-PCR after treatment of cells with a combination of 20 μM LY294002 and 20 μM nutlin-3 for 48 hours (B). Error bars represent SDs of the mean. SUDHL-1 cells were treated with combinations of inhibitors (at 20 μM each) for 12 hours in the presence of either leptomycin B or MG132 as indicated (C). Arbitrary units under the blots represent protein levels normalized for the actin control by densitometric analysis.
Furthermore, treatment of cells for 12 hours with the proteosome inhibitor MG132 stabilized p53 expression consistent with ubiquitination and degradation through the proteosome as the major route of p53 inactivation in SUDHL-1 cells (Figure 6C). Combinatorial treatment with MG132 and either LY294002 or SP600125 treatment did not further stabilize p53. However, treatment with SP600125 in combination with MG132 did result in an increase in MDM2 protein levels suggesting that MDM2 is degraded through the proteosome in a JNK-dependent manner. A combination of nutlin-3 and MG132 resulted in rapid cell death (data not shown), presumably due to a massive increase in p53 activity.
Inhibition of nuclear export with leptomycin B likewise resulted in the stabilization of p53, suggesting the cytoplasm as the major site of proteosomal degradation of p53 in these cells (Figure 6C). Treatment of cells with LY294002 in combination with leptomycin B demonstrated decreased levels of p53 in comparison to DMSO and leptomycin B-treated cells (0.6 vs 1.1), whereas nutlin-3 treatment in combination with leptomycin B resulted in stabilization of p53 and increased transcriptional activity as demonstrated by enhanced MDM2 expression levels (1.7 vs 0.7), an effect that was again decreased after combined treatment with LY294002 (0.8). Again, these data support a role for PI 3-kinase in the cellular localization of p53 and in the stabilization of MDM2.
Discussion
The majority of NPM-ALK+ ALCL cases express wild-type p53,18,19 suggesting that either NPM-ALK down-regulates the activity of the p53 tumor suppressor or that p53 is not the major pathway regulating cell cycle in the lymphoid cells from which the cancer arose. Equally, there is no evidence for significant overexpression of the p53 antagonist MDM2,18 an E3 ubiquitin ligase that is known for its essential function in the negative regulation of p53.25,26 We therefore examined the mechanisms by which NPM-ALK regulates the p53 tumor suppressor pathway and its negative regulator, MDM2.
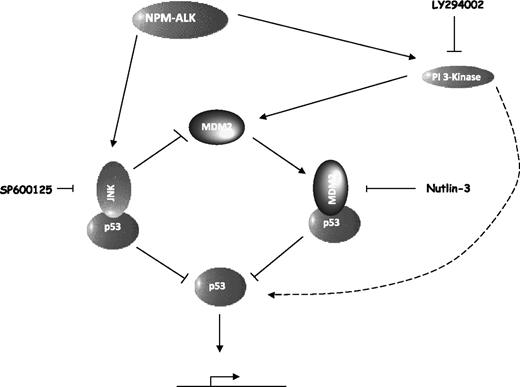
NPM-ALK controls p53 stability and activity on multiple levels as a result of highly regulated and intricate signaling pathways (Figure 7). The first step toward p53 inactivation is via enhanced degradation as a consequence of JNK and MDM2 activities (Figures 3,Figure 4,Figure 5-6). Both JNK and MDM2 destabilize p53 in an ubiquitin-mediated manner, targeting p53 to the proteosome (Figures 4,6). This is counterintuitive to the apportioned proapoptotic role of JNK as a result of p53 activation in stressed cells.27,28 However, it has also been shown in nonstressed cells that JNK binds p53 targeting it for ubiquitination and degradation23 in support of our data. It follows that antagonism of MDM2 activity or inhibition of JNK kinase activity with specific inhibitors results in the stabilization of p53. This activity is more pronounced in nutlin-3–treated cells, which display higher levels of p53 protein than SP600125-treated cells, although this could be a result of differing inhibitor efficiencies in target silencing and/or the effects of JNK inhibition on MDM2 stability (Figure 6). In substantiation of the latter case, JNK targets MDM2 to the proteosome for degradation, an effect that is only noticeable after combined inhibition of the proteosome and JNK or IP of MDM2 (Figures 4E,6C). Stabilization of p53 results in the transcription of target proteins, in the case of MDM2 antagonism, MDM2, p21, Noxa, and c-jun, and in the case of JNK inhibition, increased transcription of p21 alone perhaps reflective of a p53 dosage effect in the selection of transcriptional targets. Leventaki et al have previously hypothesized that c-Jun transcribed as a result of JNK activity plays a role in cellular survival by inhibiting p21.24 However, our data show that p21 transcription is inhibited as a result of p53 destabilization in NPM-ALK–expressing cells as opposed to c-jun activity, which in our hands is not a transcriptional target of JNK but of p53 (Figure 6B).
NPM-ALK regulates a dynamic equilibrium to inactivate the p53 tumor suppressor pathway. NPM-ALK activates the JNK SAP kinase and PI 3-kinase/Akt pathways as previously reported.6,24 These pathways act to sequester and inactivate p53 via direct binding to JNK or MDM2 leading to degradation via the proteosome. JNK also targets MDM2 for degradation. PI 3-kinase activity leads to tethering of p53 protein to cytoskeletal structures in the cytoplasm, where it is primed for nuclear entry (represented by a curved dashed arrow).
NPM-ALK regulates a dynamic equilibrium to inactivate the p53 tumor suppressor pathway. NPM-ALK activates the JNK SAP kinase and PI 3-kinase/Akt pathways as previously reported.6,24 These pathways act to sequester and inactivate p53 via direct binding to JNK or MDM2 leading to degradation via the proteosome. JNK also targets MDM2 for degradation. PI 3-kinase activity leads to tethering of p53 protein to cytoskeletal structures in the cytoplasm, where it is primed for nuclear entry (represented by a curved dashed arrow).
The second step toward p53 inactivation is via cytoplasmic sequestration in a PI 3-kinase–dependent manner. Inhibition of PI 3-kinase signaling shifts p53 from attachment to cytoskeletal structures to a more diffuse pattern within the cytoplasm (Figure 2C), but at all times, for the most part, excluded from the nucleus. Exclusion of p53 from the nucleus is one strategy adopted by cancer cells and is a result of association with microtubules or vimentin in the cytoplasm.29-32 It has been reported that association of p53 with microtubules creates a reservoir of p53 in the cytoplasm that can move to the nucleus on DNA damage in a dynein-dependent manner.29,33 Indeed, treatment of SUDHL-1 cells with nutlin-3 in association with LY294002 decreases p53 activity resulting in lower levels of transcription of target proteins (Figure 6A,C) suggesting that, although p53 is now free from MDM2 inhibition, it cannot relocate to the nucleus as effectively as when PI 3-kinase is active. These data support a role for PI 3-kinase in the sequestration of p53 to cytoskeletal elements, a position primed for nuclear entry on cellular stress. In confirmation, treatment of SUDHL-1 cells with DNA damaging agents such as etoposide leads to p53 stabilization and activity (data not shown). To add further complexity, treatment of cells with LY294002 enhances ubiquitination of p53, suggesting that association of p53 with microtubules inhibits p53 degradation (Figure 4F).
These data suggest a potential therapeutic strategy in the reactivation of p53 for the treatment of ALK-expressing ALCL. We show that nutlin-3 reduces proliferation and induces apoptosis of NPM-ALK+ cells (Figures 3E, 5), demonstrating that treatment of NPM-ALK+ ALCL expressing wild-type p53 with MDM2 antagonists may be a viable therapeutic approach. Likewise, previous studies have shown that nutlin-3 is a potent inducer of apoptosis in pediatric acute lymphoblastic leukemia expressing wild-type p53.34,35 However, induction of apoptosis in response to nutlin-3 treatment in the case of ALCL is not maximal, suggesting that other p53-independent cell survival mechanisms must likewise be inactivated or that p53 is not maximally induced. In evidence of the latter case, we show that NPM-ALK activates JNK, an event associated with cellular proliferation and inhibition of apoptosis and confirming previously reported data.24,36 It therefore follows that exposure of NPM-ALK–expressing ALCL cells to the JNK inhibitor SP600125 in combination with nutlin-3 results in an increase in apoptosis, compared with nutlin-3 treatment alone, although this is likely due to p53-independent functions of JNK inhibition or off-target effects of the inhibitor compounds used in this study. However, we have used alternative inhibitors to confirm the p53 destabilizing effects of JNK and MDM2 (Figure S2).
Likewise, inhibition of PI 3-kinase results in a reduction of cellular proliferation, cell cycle arrest at G0/G1, and an increase in apoptosis (Figure 5), most likely again a combined effect with p53-independent pathways, including inhibition of Bad phosphorylation or regulation of FOXO3 transcriptional activity.6,37 In addition, we cannot exclude off-target effects of LY294002, which is also known to inhibit other cellular kinases including DNA-PK and ATM, although we have not been able to detect expression of these proteins in our cells (data not shown). We further investigated whether the simultaneous inhibition of JNK and PI 3-kinase together with antagonism of MDM2 would have combinatorial effects in the induction of apoptosis, cell-cycle arrest, and reduction of proliferation. The combinational apoptotic effect of nutlin-3, SP600125, and LY294002 was stronger than nutlin-3 with LY294002 alone, and this effect could be seen after just 6 hours of treatment, further suggesting that NPM-ALK regulates p53 function at least through 2 distinct pathways: PI 3-kinase and JNK, and that these activities act synergistically with p53-independent effects of hyperactive JNK and PI 3-kinase in cellular survival. Whether such an approach would be viable therapeutically without significant toxicity requires testing in more relevant model systems.
In summary, our data reveal a dynamic equilibrium in which NPM-ALK balances p53 and MDM2 protein expression levels and subsequent activity of p53 (Figure 7). NPM-ALK enables cytoplasmic sequestration of p53 to cytoskeletal structures, possibly microtubules via PI 3-kinase activity and enables degradation of p53 via both MDM2 and JNK activities. Both JNK and PI 3-kinase additionally induce p53-independent mechanisms for cellular survival, and hence inhibition of both of these pathways together with antagonism of MDM2:p53 binding with the specific inhibitor nutlin-3 have synergistic effects on the inhibition of cellular growth and more so the induction of apoptosis. These data present a potential therapeutic strategy for the treatment of NPM-ALK+ ALCL-expressing wild-type p53.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
S.D.T. is a Leukemia Research Fund Bennett Fellow UK. Y.-X.C. is supported with a project grant from the Kay Kendall Leukaemia Fund (London, United Kingdom). F.K.E.M. is in receipt of a Wellcome Trust studentship. A.K. and H.Y. are supported with funding from the Leukemia Research Fund UK (London, United Kingdom).
Authorship
Contribution: S.D.T. and Y.-X.C. designed the experiments and wrote the paper; and Y.-X.C., F.K.E.M., H.Y., and A.K. performed the experiments and contributed useful discussion of the data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Suzanne Dawn Turner, Division of Molecular Histopathology, Department of Pathology, Lab Block Level 3, Box 231, Addenbrooke's Hospital, Hills Rd, Cambridge CB2 0QQ, United Kingdom; e-mail: sdt36@cam.ac.uk.

![Figure 1. NPM-ALK phosphorylates MDM2 on serine 166. (A) MDM2 is phosphorylated on serine 166 in archival biopsy specimens isolated from NPM-ALK–expressing ALCL (top left panel). Conversely, ALK− cases do not strongly express MDM2 in a phosphorylated form (top right panel). All cases were screened for the presence of a translocation involving ALK on chromosome 2 by FISH using an ALK break-apart probe (bottom left panel shows an ALK+ case) and for the expression of MDM2 by immunohistochemistry (bottom right panel shows a case representative of all samples regardless of ALK status). Representative samples are shown of the 10 ALK+ cases screened. Likewise MDM2 is phosphorylated on serine 166 in patient-derived ALK-expressing ALCL cell lines SUHL-1, Karpas-299, and DEL (B) and in BaF3 cells expressing NPM-ALK. Numbers represent arbitrary units after densitometric analysis and normalization for the tubulin loading control. (C) Treatment of SUDHL-1 cells with either (D) siRNA against ALK for 48 hours (compared with a scrambled sequence [Scr]) or (E) 10 μM of the Jak3/ALK inhibitor WHI-P154 for 2 hours, causes a reduction of pMDM2 levels in the cells. Cells were analyzed by trypan blue staining at the time of analysis to exclude significant cell death. Furthermore, 48 hours of treatment with a PI 3-kinase inhibitor (20 μM LY294002) attenuated MDM2 phosphorylation. Numbers represent arbitrary units after densitometric analysis and normalization to the loading control; vertical lines have been inserted to indicate a repositioned gel lane (F). The inhibitor compounds had no effect on MDM2 transcript levels as determined by qRT-PCR (G). Error bars represent SDs of the mean. Results are representative of at least 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/21/10.1182_blood-2008-06-160168/6/m_zh89990935830001.jpeg?Expires=1769126634&Signature=SdxytiQtH2IFXA2WYkm~s-rn61rkWk~nU4D5-ssNInS3jIt3d-nS0Fce4p8hfSTUNLeJRRAJchPznTCqjy280rmwTL16Jr7fmnRFIJmBUDRmRp2CPGfmhvpPQwVCO4aCH31D-jo043cxCv2tWGPOAbSJHNqio0A-hqDPkoiVVG~ycYnWaN4DzcOSJteF1RvOXihaEGfYOobCQ1sKGOGd~ePl6eGseFSVbqH5NllMUvpevNiGukLt9MP7RCpkwusUbGIQbrPllIEm6qi0UIFmYsy8AsqEtZpECTd0-p1ZXr9ZPL9FLE2GD-io1Eksg7IBXfiQMPopMWC4MvWiIVXyrg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal