As key components of the erythrocyte membrane skeleton, spectrin and ankyrin specifically interact to tether the spectrin cytoskeleton to the cell membrane. The structure of the spectrin binding domain of ankyrin and the ankyrin binding domain of spectrin have been solved to elucidate the structural basis for ankyrin-spectrin recognition. The structure of repeats 14 and 15 of spectrin shows that these repeats are similar to all other spectrin repeats. One feature that could account for the preference of ankyrin for these repeats is the presence of a conserved, negatively charged patch on one side of repeat 14. The structure of the ankyrin ZU5 domain shows a novel structure containing a β core. The structure reveals that the canonical ZU5 consensus sequence is likely to be missing an important region that codes for a β strand that forms part of the core of the domain. In addition, a positively charged region is suggestive of a binding surface for the negatively charged spectrin repeat 14. Previously reported mutants of ankyrin that map to this region lie mostly on the surface of the protein, although at least one is likely to be part of the core.
Introduction
In erythrocytes, the reversible deformability facilitating passage through capillaries is largely controlled by a vast network of proteins attached to the membrane.1 Two sets of interactions can be distinguished,2 one set that is mediated by glycophorin C between the membrane and a complex formed by band 4.1 protein, spectrin, and actin3 and another set between the membrane and the spectrin-based cytoskeleton.4 In the latter case, the attachment between the membrane and the spectrin network is accomplished through an adaptor protein, ankyrin, which bridges the interaction between β-spectrin and the membrane bound anion-exchange transporter band 3.4 The greater importance of this ankyrin-mediated attachment has been demonstrated in experiments in which the glycophorin C attachment is severed without leading to loss of mechanical properties of the red blood cell.5
The human erythroid spectrin heterotetramer is formed by 2 α and 2 β subunits arranged as a head-to-head dimer of (α/β) dimers.6 The heterodimer consists of 37 triple-helical spectrin repeats, 20 complete and 1 partial repeat in the α subunit, and 16 complete and 1 partial repeat in the β subunit (Figure 1A). Although members of the spectrin superfamily, which includes not only spectrin but also α-actinin, dystrophin, and plectin, adopt a repeating triple-helical fold,7,–9 the highly specific interactions of certain spectrin repeats with other proteins suggest that the repeats have evolved to provide functional specialization while maintaining a conserved tertiary structure.7,10,11 In the case of spectrin, the ∼ 30% amino acid sequence identity among repeats7,12 illustrates the considerable variability of the different repeats within the structural constraints imposed by the conserved 3-dimensional fold. Furthermore, the structural basis for the versatility of spectrin repeats in binding different ligands with high affinity (reviewed in Djinovic-Carugo et al7 ) has not been elucidated despite the determination of structures of 2,13,–15 3,16 and 417 repeat fragments of spectrin superfamily members.
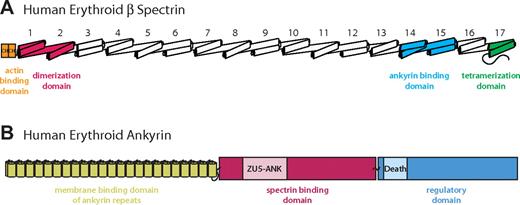
Schematic diagrams of erythrocyte β-spectrin and ankyrin. The schematic diagrams show the domain organization of the 2 molecules. (A) The intact human erythroid β-spectrin (HEβ) molecule consists mainly of tandem repeats of 3-helix bundles, each bundle known as a spectrin repeats (numbered from N to C terminus). The ankyrin binding domain comprises repeats 14 and 15 (light blue).24,25,28 Additional binding domains include 2 N-terminal calponin homology (CH) domains responsible for actin binding (orange), spectrin repeats 1 and 2, which mediate α/β dimer formation (red), and a tetramerization domain consisting of a 2-helix bundle (ie, an incomplete repeat) near the C-terminus (green). (B) Human erythroid ankyrin (Ankyrin R) consists of an N-terminal membrane protein binding domain composed of ankyrin repeats (yellow), a central spectrin binding domain that harbors a ZU5-containing subdomain identified as the minimal binding domain for β-spectrin (ZU5-ANK) (red/pink), and a C-terminal regulatory domain that modulates the affinities of the other domains and contains a Death Domain (blue).
Schematic diagrams of erythrocyte β-spectrin and ankyrin. The schematic diagrams show the domain organization of the 2 molecules. (A) The intact human erythroid β-spectrin (HEβ) molecule consists mainly of tandem repeats of 3-helix bundles, each bundle known as a spectrin repeats (numbered from N to C terminus). The ankyrin binding domain comprises repeats 14 and 15 (light blue).24,25,28 Additional binding domains include 2 N-terminal calponin homology (CH) domains responsible for actin binding (orange), spectrin repeats 1 and 2, which mediate α/β dimer formation (red), and a tetramerization domain consisting of a 2-helix bundle (ie, an incomplete repeat) near the C-terminus (green). (B) Human erythroid ankyrin (Ankyrin R) consists of an N-terminal membrane protein binding domain composed of ankyrin repeats (yellow), a central spectrin binding domain that harbors a ZU5-containing subdomain identified as the minimal binding domain for β-spectrin (ZU5-ANK) (red/pink), and a C-terminal regulatory domain that modulates the affinities of the other domains and contains a Death Domain (blue).
In humans, there are 3 major ankyrin isoforms: ankyrin 1 or R, which is found in erythrocytes,4 ankyrin 2 or B, which is localized primarily in the brain,4 and ankyrin 3 or G, which is somewhat more broadly expressed.18 In addition, several alternative splice sites are found among different tissues4,18 and result in a wide number of different ankyrin isoforms. Ankyrins have a modular structure consisting of 3 regions: an N-terminal domain composed of many ankyrin repeats that is responsible for mediating attachment to the membrane through interactions with membrane-bound proteins, a middle region whose primary role is to interact with spectrin, and a C-terminal regulatory domain that harbors a death domain and modulates the affinities of both domains N-terminal to it for other proteins.19
Specifically, human erythroid ankyrin (ankyrin R) consists of an 89-kDa N-terminal domain, a central 62-kDa spectrin-binding domain, and a 55-kDa C-terminal domain (Figure 1B).4 Many cellular roles and interactions of ankyrin R have been studied; in particular, the consequences of ankyrin mutations in cardiac and other functions20 have been widely examined. Structural work on this adaptor protein, however, has been limited mainly to the ankyrin repeats responsible for binding to band 321 and that confer spring-like flexibility to the molecule.22 Similar ankyrin repeats also have been observed in a wide variety of proteins and are generally regarded as protein–protein interaction modules.23
Given their critical role in physically linking the membrane skeleton to the cell membrane and in this way allowing the reversible deformation of red cells, it is important to understand the atomic basis for the interactions between spectrin and ankyrin. Previously, the minimal binding regions of spectrin and ankyrin were identified by a variety of biophysical and biochemical methods.24,25 The data indicate that a region of the spectrin binding domain of ankyrin consisting of a so-called ZU5 domain plus 55 residues C-terminal to it (referred to herein as “ZU5-ANK”) is the primary spectrin binding epitope of human erythroid ankyrin,24 which is consistent with previous observations of Mohler et al.26 Complementarily, β-spectrin repeats 14 and 15 were found to be the tandem repeats minimally necessary to recapitulate ankyrin binding in vitro, in good agreement with several other studies.25,27,28 The finding that these the 2 fragments comprise the ankyrin-spectrin binding site is underscored by their binding affinity of 15 nmol/L,24 similar to that in the intact complex.29
To gain an atomic-level understanding of the spectrin-ankyrin interactions, we crystallized and solved the atomic structures of the minimal binding fragments of each of these 2 proteins. The ZU5-containing ankyrin domain structure is the first structural example of this type of domain and reveals a novel fold that features a β-sheet rich core and several ordered loops. The structure of the 2 repeat spectrin fragment adopts the canonical spectrin fold with an ordered helix linking them. This fragment, however, displays a large patch of negatively charged residues not observed in other spectrin superfamily structures. The structural data provide insights regarding specialization of spectrin repeats and the general architecture of the binding domains of these proteins.
Methods
Cloning, expression, and purification of spectrin fragments
A panel of fragments coding for human erythroid β-spectrin repeats 14 and 15 (HEβ1415) was cloned into the ligation-independent vector pMCSG7 as described previously.24 The constructs had slightly different starting and terminating amino acids as it is not always possible to predict the correct phasing of spectrin repeats. Each of these constructs was overexpressed and purified as described previously,24 then concentrated to 5 to 25 mg/mL with the use of Amicon centrifugal filter devices (10-kDa molecular weight cutoff) before crystallization screening. The purified protein was aliquoted, flash frozen in liquid nitrogen, and stored at −80°C. Initial crystals and a structure solution were obtained from a fragment spanning residues 1686 to 1910. Crystals were later improved by the use of a construct encompassing only residues 1686 through 1907, but containing an additional 3 amino acids at the N-terminus from the cloning vector.
Cloning, expression, and purification of ankyrin fragments
A ZU5-containing subdomain of human erythrocyte ankyrin (residues 911-1068) was previously identified as the primary spectrin binding fragment within the spectrin binding domain of ankyrin.24,–26 This fragment, “ZU5-ANK,” was expressed and purified as previously described24 with the exception that the N-terminal His tag was removed by cleavage with TEV protease (1:20 m/m ratio construct/protease) at 4°C for ∼20 hours while the protein was simultaneously dialyzed into PED buffer (20 mmol/L sodium phosphate at pH 8.0, 1 mmol/L ethylenediamine tetraacetic acid, and 1 mmol/L dithiothreitol [DTT)] with 0.3 mol/L NaCl. Three additional nonankyrin amino acids were left on the N-terminus of the fragment after cleavage. Subsequent chromatography with the use of a sulfonyl methacrylate (Bio-Rad, Hercules, CA) column and a P-10 (Bio-Rad) size-exclusion column were performed as described previously.24 The purified ZU5-ANK protein was concentrated to ∼ 5 mg/mL for crystallization screening and subsequent crystal optimization. The purified protein was aliquoted, flash frozen in liquid nitrogen, and stored at −80°C.
Crystallization and characterization of HEβ1415
Crystals were grown by hanging drop vapor diffusion in 30% PEG 3350, 0.2 mol/L NaCl, and 100 mmol/L HEPES (pH 7.5) or 30% PEG 3350, 0.2 mol/L ammonium acetate, and 100 mmol/L Tris (pH 8.0) on siliconized glass coverslips (Hampton Research, Aliso Viejo, CA). Two microliters of purified protein solution (∼ 5 mg/mL) were mixed with an equal volume of reservoir solution and suspended over 0.75 mL of reservoir solution. Bipyramidal crystals (∼ 300 μm × 300 μm × 300 μm) grew at 22°C within 1 week. There was no apparent difference with respect to crystal size, growth rate, morphology, space group, or other crystal character between these 2 conditions. The crystals belong to space group P43212 with a = b = 65.3 Å, c = 288.9 Å, α = β = γ = 90°, and 2 molecules per asymmetric unit (∼57.0% solvent content).
Crystallization and characterization of ZU5-ANK
Crystals were grown by hanging drop vapor diffusion in 20% PEG 4000, 0.25 mol/L KBr, 0.1 mol/L MES (pH 6.5), 10 mmol/L DTT on glass coverslips (VWR) treated with Rain-X (SOPUS Products, Houston, TX). One microliter of purified protein solution (∼ 3 mg/mL) was mixed with an equal volume of reservoir solution and suspended over 0.75 mL of reservoir solution. Crystals grew as plates (∼ 300 μm × 50 μm × 10 μm) at 10°C within 24 hours. The crystals belong to the space group P21 with a = 38.0 Å, b = 204.1 Å, c = 43.0 Å, α = γ = 90° β = 113.1°, and 4 molecules per asymmetric unit (∼ 42.4% solvent content).
Data collection, structure determination, and refinement
All data were collected at 100 K with the use of synchrotron radiation at the Life Sciences Collaborative Access Team (LS-CAT) at the Advanced Photon Source (APS) at Argonne National Laboratory (Argonne, IL). Data collection statistics are shown in Table 1.30,31 All data were processed with XDS31 and scaled with SCALA.30 Further processing was done with programs from the CCP4 suite.30
Summary of crystallographic data
| . | ZU5-ANK . | ZU5-ANK SeMet . | HEβ1415 . | HEβ1415 SeMet1 . | HEβ1415 SeMet2 . |
|---|---|---|---|---|---|
| Data collection | |||||
| Detector type/source | MarCCD/APS | MarCCD/APS | MarCCD/APS | MarCCD/APS | MarCCD/APS |
| wavelength, Å | 0.918410 | 0.97620 | 0.97857 | 0.97869 | 0.97869 |
| Resolution* | 40.82-2.00 (2.09-2.00) | 40.72-2.05 (2.14-2.05) | 19.86-2.90 (3.03-2.90) | 29-00-2.95 (3.08-2.95) | 29.75-2.90 (3.03-2.90) |
| Measured reflections | 177711 (21052) | 166922 (20300) | 106581 (13259) | 59875 (5956) | 68824 (6083) |
| Unique reflections | 40191 (4810) | 37671 (4596) | 14779 (1748) | 13351 (1567) | 14184 (1538) |
| Completeness | 99.6 (97.9) | 99.9 (100.0) | 99.6 (100.0) | 97.6 (96.8) | 97.3 (90.7) |
| Anomalous completeness | 97.4 (96.0) | 98.1 (98.8) | 90.2 (89.4) | 93.3 (79.3) | |
| Multiplicity | 4.4 (4.4) | 4.4 (4.4) | 7.2 (7.6) | 4.5 (3.8) | 4.9 (4.0) |
| Anomalous multiplicity | 2.2 (2.2) | 2.2 (2.2) | 2.6 (2.1) | 2.7 (2.3) | |
| Rsym | 4.3 (29.9) | 4.8 (31.0) | 5.4 (28.6) | 6.0 (29.9) | 4.6 (30.4) |
| Rmeas | 5.7 (39.3) | 6.3 (40.5) | 5.8 (30.7) | 7.4 (38.0) | 5.6 (38.1) |
| Phasing† | |||||
| Resolution | 36.92-2.10 | 24.73-2.95 | Together with SeMet1 | ||
| Phasing powerisomorphous (acentric/centric) | 0.458/0.345 | 0.878/0.921 | 1.306/1.218 | ||
| Phasing Poweranomalous | 0.768 | 0.718 | 0.980 | ||
| Rcullis-isomorphous | 0.925/0.933 | 0.533/0.535 | 0.425/0.446 | ||
| Rcullis-anomalous | 0.887 | 0.887 | 0.824 | ||
| FOM (acentric/centric) | 0.294/0.177 | 0.316/0.184 | Together with SeMet1 | ||
| Refinement | |||||
| Resolution | 36.91-2.00 (2.05-2.00) | 19.84-2.90 (2.97-2.90) | |||
| No. of reflections working/test | 38127/2013 (2743/134) | 13968/740 (990/69) | |||
| Rfactor (%) | 21.8 (27.0) | 26.5 (36.0) | |||
| Rfree (%) | 26.4 (29.2) | 32.2 (37.0) | |||
| Protein atoms | 4982 | 3038 | |||
| Water molecules | 190 | 0 | |||
| Other atoms | Bromide - 18 | 0 | |||
| Bond lengths | 0.009 | 0.008 | |||
| Bond angles | 1.205 | 1.056 |
| . | ZU5-ANK . | ZU5-ANK SeMet . | HEβ1415 . | HEβ1415 SeMet1 . | HEβ1415 SeMet2 . |
|---|---|---|---|---|---|
| Data collection | |||||
| Detector type/source | MarCCD/APS | MarCCD/APS | MarCCD/APS | MarCCD/APS | MarCCD/APS |
| wavelength, Å | 0.918410 | 0.97620 | 0.97857 | 0.97869 | 0.97869 |
| Resolution* | 40.82-2.00 (2.09-2.00) | 40.72-2.05 (2.14-2.05) | 19.86-2.90 (3.03-2.90) | 29-00-2.95 (3.08-2.95) | 29.75-2.90 (3.03-2.90) |
| Measured reflections | 177711 (21052) | 166922 (20300) | 106581 (13259) | 59875 (5956) | 68824 (6083) |
| Unique reflections | 40191 (4810) | 37671 (4596) | 14779 (1748) | 13351 (1567) | 14184 (1538) |
| Completeness | 99.6 (97.9) | 99.9 (100.0) | 99.6 (100.0) | 97.6 (96.8) | 97.3 (90.7) |
| Anomalous completeness | 97.4 (96.0) | 98.1 (98.8) | 90.2 (89.4) | 93.3 (79.3) | |
| Multiplicity | 4.4 (4.4) | 4.4 (4.4) | 7.2 (7.6) | 4.5 (3.8) | 4.9 (4.0) |
| Anomalous multiplicity | 2.2 (2.2) | 2.2 (2.2) | 2.6 (2.1) | 2.7 (2.3) | |
| Rsym | 4.3 (29.9) | 4.8 (31.0) | 5.4 (28.6) | 6.0 (29.9) | 4.6 (30.4) |
| Rmeas | 5.7 (39.3) | 6.3 (40.5) | 5.8 (30.7) | 7.4 (38.0) | 5.6 (38.1) |
| Phasing† | |||||
| Resolution | 36.92-2.10 | 24.73-2.95 | Together with SeMet1 | ||
| Phasing powerisomorphous (acentric/centric) | 0.458/0.345 | 0.878/0.921 | 1.306/1.218 | ||
| Phasing Poweranomalous | 0.768 | 0.718 | 0.980 | ||
| Rcullis-isomorphous | 0.925/0.933 | 0.533/0.535 | 0.425/0.446 | ||
| Rcullis-anomalous | 0.887 | 0.887 | 0.824 | ||
| FOM (acentric/centric) | 0.294/0.177 | 0.316/0.184 | Together with SeMet1 | ||
| Refinement | |||||
| Resolution | 36.91-2.00 (2.05-2.00) | 19.84-2.90 (2.97-2.90) | |||
| No. of reflections working/test | 38127/2013 (2743/134) | 13968/740 (990/69) | |||
| Rfactor (%) | 21.8 (27.0) | 26.5 (36.0) | |||
| Rfree (%) | 26.4 (29.2) | 32.2 (37.0) | |||
| Protein atoms | 4982 | 3038 | |||
| Water molecules | 190 | 0 | |||
| Other atoms | Bromide - 18 | 0 | |||
| Bond lengths | 0.009 | 0.008 | |||
| Bond angles | 1.205 | 1.056 |
>Rsym indicates = Σ I−<I> /ΣI, where I is the observed intensity and <I> the average intensity obtained from multiple measurements; Rmeas, as described in Diederichs and Karplus48 ; Rfactor, Σ Fo − Fc /Σ Fo, where Fo is the observed structure factor amplitude and Fc the calculated structure factor amplitude; and Rfree, R-factor based on 5% of the data excluded from refinement.
Numbers in parentheses correspond to the highest-resolution shell throughout.
Extracted from autoSHARP log files.32
HEβ1415
Crystals were harvested directly from the drop in which they grew and immediately frozen in liquid nitrogen; the high concentration of PEG 3350 in the mother liquor served as a suitable cryoprotectant, although others were tested. An experimental map was obtained with the use of 2.9-Å data collected from seleno-methionine–derivatized crystals (protein expressed and purified as described previously) at 0.979 Å. Single-wavelength anomalous dispersion (SAD) phasing was conducted with the use of the program autoSHARP,32 which correctly located all 8 methionines in the asymmetric unit. The initial solvent flattened map was of good quality and helices could be recognized easily. An initial partial model of HEβ1415 was generated with the use of the low-resolution helix build module of ARP/wARP version 6.1.1.33 Subsequent manual model building and inspection were conducted using the program Coot.34 Model refinement was conducted with the use of REFMAC5.35
Even after refinement, not all regions of the molecule were visible, and many side chains were in poor density. In particular, the loop connecting helices A and B in repeat 15 was disordered. Despite the low resolution of the structure, proper placement of the side chains was possible because of the presence of 4 seleno-methionines, which served as guides to register the sequence. As part of the refinement, translation liberation screw (TLS) parameters were refined for each spectrin repeat. The 2 molecules were restrained by the noncrystallographic symmetry (NCS) during the refinement, although the restraints were only moderately tight because the 2 molecules are different in orientation of the 2 repeats. The final model consists of residues 1684 to 1805 and 1841 to 1896 in one chain (A), and 1684 to 1817 and 1840 to 1891 in the second chain (B). The final model has an Rfactor and Rfree of 26.5% and 32.2% respectively, with a root mean square deviation (rmsd) of 0.008 Å and 1.1° for bond lengths and bond angles, respectively, and with all residues within the favored regions of the Ramachandran plot except for one amino acid. Refinement statistics are listed in Table 1.
ZU5-ANK
Crystals were harvested and frozen after the direct addition of glycerol to a final concentration of 10% to the crystal drop. Because the crystals were best behaved in the presence of KBr, data from native crystals were collected to 2.0 Å at a wavelength of 0.918 Å (near the Br K-edge), as well as 2.1 Å data from seleno-methionine crystals at 0.976 Å wavelength (near the Se K-edge). Experimental phases were obtained by SAD with the program autoSHARP,32 which identified all 16 seleno-methionines in the asymmetric unit. The initial solvent-flattened electron density map was excellent and showed the presence of 4 monomers in the asymmetric unit. An initial model of ZU5 was built automatically with PHENIX,36 which identified 506 of 644 possible residues. As refinement progressed, additional residues were included, although some surface-exposed loops were disordered. Refinement was performed against data from the native crystals collected near the Br K-edge and, hence, it was possible to assign positions for bromide ions in the solvent model using the anomalous differences. The 4 monomers (named A, B, C, and D) form a dimer of dimers related by NCS. During the refinement, noncrystallographic symmetry restraints were applied to all the regions that were clearly identical; only some loops were allowed to deviate from the noncrystallographic symmetry. TLS parameters were refined for each monomer.
The final model consists of residues 912 to 996 and 1005 to 1068 for monomer A; 913 to 959, 961 to 996, and 1003 to 1068 for monomer B; 910 to 1000, 1004 to 1021, and 1025 to 1068 for monomer C; and 909 to 999, 1004 to 1022, and 1025 to 1068 for monomer D. In addition, 18 bromide ions and 190 water molecules are included in the model. The final model has an Rfactor and Rfree of 21.8% and 26.4%, respectively, with an rmsd of 0.009 Å and 1.2° for bonds lengths and bond angles, respectively, and with all residues within the favored regions of the Ramachandran plot. Refinement statistics are listed in Table 1. Manual model building and inspection were conducted with the use of the program Coot.34 Model refinement was conducted with the use of REFMAC5.35
Figures
Figures were made with Pymol molecular visualization system (http://www.pymol.org). Electrostatic surface calculations were performed with APBS.37 Superpositions were generated with the program lsqkab from the CCP4 software suite (Daresbury Laboratory, Warrington, United Kingdom).
Coordinates
Coordinates and structure factors discussed in this publication have been deposited in the Protein Data Bank (PDB), http://www.pdb.org/ with accession numbers 3f57 and 3f59.
Results
Structures of the minimum spectrin binding domain of ankyrin and the ankyrin binding domain of β-spectrin were solved individually by the use of X-ray crystallography. Selection and design of the ankyrin subdomain were based on previous results, which indicated ZU5-ANK as the main β-spectrin binding fragment.24,26 Similarly, the minimum 2 β-spectrin repeats required for binding ankyrin, repeats 14 and 15, were chosen as the crystallization target.24,25,27,28
Structure of ZU5-ANK
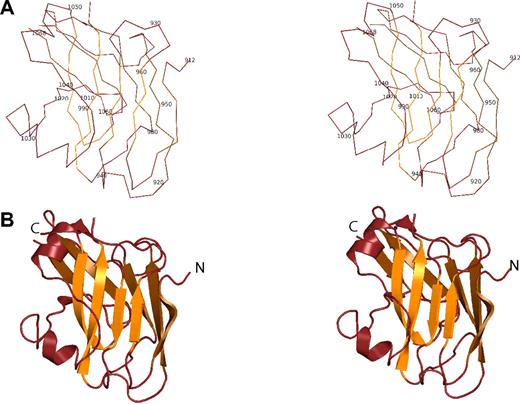
The ZU5-ANK fragment adopts a compact, well-folded structure with a β-sheet rich core and a number of loops (Figure 2). Two antiparallel β-sheets, one composed of 4 strands and the other composed of 5 strands, are juxtaposed against each other to create the core of the structure. The 2 sheets could be viewed as one large, extended antiparallel sheet folded along the middle strand. Atop the smaller sheet, 2 short helices are the only other secondary structure elements. As expected in a structure containing so many strands, there are several loops connecting the strands, all of them facing the solvent. Surprisingly, the N- and C-termini of the domain are found on opposite ends of the β-core, suggesting that the ZU5-ANK domain does not simply loop out from the spectrin binding domain of ankyrin as, for instance, the SH3 domain of spectrin does.38 Furthermore, the structure is consistent with NMR and CD measurements,24 which suggest a globular protein that is well-folded while also containing significant loop content.
Structure of the the ZU5-ANK β-spectrin binding domain. (A) Stereo diagram showing the Cα trace of ZU5-ANK. Residues are numbered corresponding to their location in the intact ankyrinR molecule. In the diagram, adjacent Cα atoms are joined by a line. (B) Ribbon stereo diagram of the ZU5-ANK subdomain in the same orientation as in panel A. The overall structure is compact and well-folded with a β-sheet–rich core and several loops. β strands are shown as arrows, helical regions as coils, and regions with no well-defined secondary structure as lines. lines. Rotating views of several representations of the molecule are shown in Video S2 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Structure of the the ZU5-ANK β-spectrin binding domain. (A) Stereo diagram showing the Cα trace of ZU5-ANK. Residues are numbered corresponding to their location in the intact ankyrinR molecule. In the diagram, adjacent Cα atoms are joined by a line. (B) Ribbon stereo diagram of the ZU5-ANK subdomain in the same orientation as in panel A. The overall structure is compact and well-folded with a β-sheet–rich core and several loops. β strands are shown as arrows, helical regions as coils, and regions with no well-defined secondary structure as lines. lines. Rotating views of several representations of the molecule are shown in Video S2 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
The presence of 4 molecules in the asymmetric unit allows ascertainment of the variability of structure. All 4 molecules superpose very well, with average rmsd values ranging between 0.24 and 0.71 Å for the main chain atoms. The tetramer can be described as a dimer of dimers, with the A/B monomers related to the C/D monomers by a 2-fold axis. The dimer of dimers superposes with an rmsd of 0.28 Å for the main chain atoms. The formation of a dimer of dimers is a crystallographic artifact and does not represent a functional form of the protein, which behaves as a monomer in solution.24 As expected, the core of the structure is hydrophobic. Comparison of the ZU5-ANK structure with other known structures in the PDB using the DALI server39 or the SSM server40 revealed no significant similarity to any other structures. Only rough similarities to other structures containing extended β-sheets were found.
There are 3 major isoforms of human ankyrin: ankyrin R, ankyrin B, and ankyrin G. Sequence alignment of the ZU5-ANK domain of these 3 isoforms (Figure 3B) indicates that most of the differences in the ZU5 region are in loops connecting secondary structure elements. The region spanning residues 956 to 972 contains one of the most variable regions in all ankyrins and forms a long loop joining 2 strands on the surface of the protein. A second variable region is disordered in the structure and spans a loop defined by residues 997 to 1004. The other highly variable region is found in the loop leading to the first α-helix and spans residues 1021 to 1027. This last loop traverses the top of one of the sheets, is completely surfaced exposed, and corresponds to the first few residues outside the canonical ZU5 region.
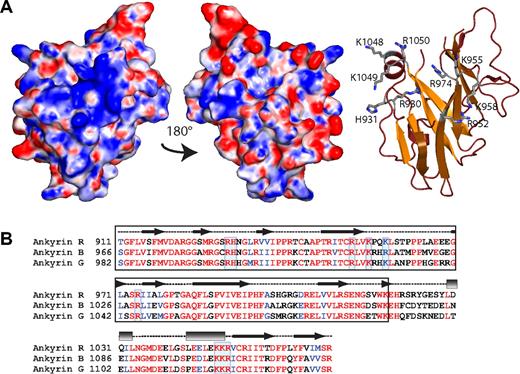
ZU5-ANK displays a conserved, positively charged surface patch. (A) The electrostatic map (left) of ZU5-ANK subdomain of ankyrin displays a positively charged patch on its surface attributable to the presence of several positively charged amino acids. The basic residues involved are shown in the ribbon diagram (right) shown in the same orientation as the electrostatic surface. Only the side chains of the positively charged residues are shown (ball-and-stick). The middle panel shows the electrostatic map rotated 180° from the left panel and serves to illustrate that only one surface shows the positively charged patch. The molecular surface of the molecule is shown with the equipotential electrostatic surface mapped onto it at ± 15 KbT/ec. (B) Sequence alignment of 3 major human ankyrin isoforms (R, B, and G) indicates that many of the charged residues, boxed in blue, are highly conserved at the sequence level and are likely to form part of similar charged surfaces. The secondary structure elements in the structure are shown above the sequence (helices are shown as rectangles, sheets as arrows, and loops as dashed lines). The black box identifies the canonical ZU5 region.46
ZU5-ANK displays a conserved, positively charged surface patch. (A) The electrostatic map (left) of ZU5-ANK subdomain of ankyrin displays a positively charged patch on its surface attributable to the presence of several positively charged amino acids. The basic residues involved are shown in the ribbon diagram (right) shown in the same orientation as the electrostatic surface. Only the side chains of the positively charged residues are shown (ball-and-stick). The middle panel shows the electrostatic map rotated 180° from the left panel and serves to illustrate that only one surface shows the positively charged patch. The molecular surface of the molecule is shown with the equipotential electrostatic surface mapped onto it at ± 15 KbT/ec. (B) Sequence alignment of 3 major human ankyrin isoforms (R, B, and G) indicates that many of the charged residues, boxed in blue, are highly conserved at the sequence level and are likely to form part of similar charged surfaces. The secondary structure elements in the structure are shown above the sequence (helices are shown as rectangles, sheets as arrows, and loops as dashed lines). The black box identifies the canonical ZU5 region.46
Overall, the structure does not show a marked charge character, although a small positively charged patch is observed on one side of the structure, comprising part of the β-core and one end of an α-helix (Figure 3). The residues in the β-core include Arg930, His931, Arg952, Lys955, Lys958, and Arg 974, whereas at the C-terminus of the second helix residues Lys1048, Lys1049, and Arg1050 are part of this positively charged area. These positively charged residues are all highly conserved in all 3 major ankyrin isoforms, suggesting that all isoforms have the same charged surface (Figure 3). Not surprisingly, these residues are not conserved in other ZU5 domains that have different functions.
Structure of HEβ1415
The structure of β-spectrin repeats 14 and 15 exhibited the canonical fold of 2 tandem spectrin repeats, which has now been observed in many members of the spectrin superfamily (Figure 4).9,13,16,17 Each repeat consists of a triple-helical bundle, with helices termed A, B, and C, with hydrophobic amino acids (positions a and d in the heptad repeat) facing the interior of the bundle and hydrophilic side chains (positions e and g) solvent-exposed. As expected based on previous multirepeat spectrin structures,9,13,16,17 the 2 repeats are connected by an α-helical linker. In addition to meeting the expectation of a helix-rich structure based on homology and structure prediction, the crystal structure also matched well with solution biophysical measurements of the fragment, which indicated a structure dominated by α-helices.24 In the crystal structure, the N-terminal half of the molecule (repeat 14) was all present and well-ordered, but the C-terminal portion (repeat 15) exhibited a disordered loop joining helices A and B and the last few amino acids of helix C. This was true, to a different extent, in both copies of the molecule in the asymmetric unit. The presence of disordered loops in spectrin structures is not unusual and has been observed before, most notably in the structure of 2 repeats of chicken brain spectrin.14 The 2 molecules in the asymmetric unit also are not identical. Superposition of the structures shows that although each repeat is basically the same (main chain atoms rmsd of 0.52 Å between repeats 14 and 0.32 Å between repeats 15), the 2 repeats have very slightly different orientations. Again, this is not unusual in spectrin structures where variability in repeat orientation is widely observed.14,–16
Structure of human β-spectrin repeats 14 and 15, the ankyrin binding domain. Stereo diagram of β-spectrin repeats 14 and 15 corresponding to the ankyrin binding domain. The structure displays a canonical spectrin fold consisting of 3-helix bundles connected by a helical linker. Repeat 14 is shown in blue and repeat 15, which contains some disordered regions distal to repeat 14, is in red. Helices are marked as A, B, or C, corresponding to the first, second, or third helix of each bundle, respectively. Rotating views of several representations of the molecule are shown in Video S1.
Structure of human β-spectrin repeats 14 and 15, the ankyrin binding domain. Stereo diagram of β-spectrin repeats 14 and 15 corresponding to the ankyrin binding domain. The structure displays a canonical spectrin fold consisting of 3-helix bundles connected by a helical linker. Repeat 14 is shown in blue and repeat 15, which contains some disordered regions distal to repeat 14, is in red. Helices are marked as A, B, or C, corresponding to the first, second, or third helix of each bundle, respectively. Rotating views of several representations of the molecule are shown in Video S1.
As mentioned previously, the overall fold of the repeats is very similar for all members of the spectrin superfamily. The arrangement and positions of the 3 helices is comparable, with the most variability in the loops joining them, especially the BC loop. Comparison of the HEβ1415 repeats with other spectrin repeats shows, for example, that repeats 14 and 15 are very similar to, yet distinguishable from, repeats 15 and 16 in chicken brain spectrin. In this case, repeat 14 of HEβ1415 superposes on repeat 15 of chicken brain spectrin with an rmsd of 3.0 Å for all equivalent main chain atoms. The other human spectrin structure, that of repeats 8 and 9, also superposes well with the HEβ1415 structure, again with a different orientation of the repeats. In this case, the repeats themselves are more variable, repeat 14 superposes on repeat 8 with an rmsd of 3.6 Å, but the overall structure is similar. This variability among the structures of tandem spectrin repeats is within that observed for previously characterized spectrin superfamily structures.
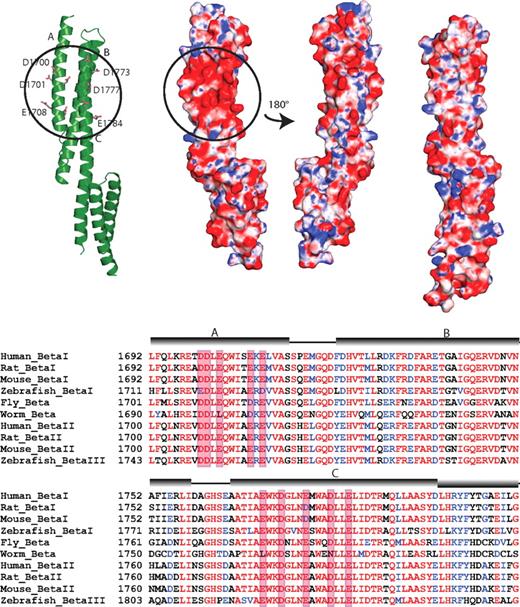
Comparison of the HEβ1415 structure with other spectrin superfamily structures also reveals that there are neither major differences in conformation among all the structures nor any additional structural features unique to HEβ1415. Analysis of the surface charge of HEβ1415 does reveal the presence of a negatively charged patch in the structure that appears to be unique to this pair (Figure 5). Although other spectrin structures do not show any large charged surface areas, HEβ1415 has a cluster of negatively charged amino acids on the face formed by helices A and C. The patch includes Asp1700, Asp1701, Glu1703, Glu1708, and Glu1710 in helix A and Glu1770, Asp1773, Glu1777, Asp1781, and Glu1784 in helix C. Many of these residues show high conservation (Figure 5B), although in general β-spectrin repeats 14 and 15 are well conserved.10 Whether some or all of these amino acids play a role in ankyrin recognition remains to be determined, but their presence does appear to be indicative of a functional role.
The ankyrin binding domain of human β-spectrin (repeats 14-15) displays a conserved, negatively charged surface patch. (A) A group of acidic residues in repeat 14 form a negatively charged patch. The ribbon diagram of repeats 14 and 15 (left) shows the acidic residues involved in forming this patch (circled). Only the side chains of the negatively charged residues are shown (ball-and-stick). The electrostatic surface of the protein shows the presence of the patch (second diagram from left with circle) in the same orientation as the ribbon diagram. The third diagram from the left shows the electrostatic surface of repeats 14 and 15 rotated approximately 180° with respect to the ribbon diagram. As can be seen, this side of the protein does not show any large charged regions. The structures of other known spectrin repeats do not display such a localized charge distribution; the electrostatic map of β-spectrin repeats 8 and 9 (right) is shown as a representative. The molecular surface of the molecule is shown with the equipotential electrostatic surface mapped onto it at ± 15 KbT/ec. (B) Sequence alignment of β-spectrin from several isoforms and species. The alignment shows that the acidic residues on helices A and C are well-conserved, although, in general, the entirety of β-spectrin repeats 14 and 15 are well-conserved. The negatively charged amino acids forming the patch are boxed in red and the position of the helices is shown by cylinders above the sequence.
The ankyrin binding domain of human β-spectrin (repeats 14-15) displays a conserved, negatively charged surface patch. (A) A group of acidic residues in repeat 14 form a negatively charged patch. The ribbon diagram of repeats 14 and 15 (left) shows the acidic residues involved in forming this patch (circled). Only the side chains of the negatively charged residues are shown (ball-and-stick). The electrostatic surface of the protein shows the presence of the patch (second diagram from left with circle) in the same orientation as the ribbon diagram. The third diagram from the left shows the electrostatic surface of repeats 14 and 15 rotated approximately 180° with respect to the ribbon diagram. As can be seen, this side of the protein does not show any large charged regions. The structures of other known spectrin repeats do not display such a localized charge distribution; the electrostatic map of β-spectrin repeats 8 and 9 (right) is shown as a representative. The molecular surface of the molecule is shown with the equipotential electrostatic surface mapped onto it at ± 15 KbT/ec. (B) Sequence alignment of β-spectrin from several isoforms and species. The alignment shows that the acidic residues on helices A and C are well-conserved, although, in general, the entirety of β-spectrin repeats 14 and 15 are well-conserved. The negatively charged amino acids forming the patch are boxed in red and the position of the helices is shown by cylinders above the sequence.
Discussion
The high affinity interaction between ankyrin and β-spectrin is of great interest not only because of the critical need of this interaction for red cell deformability and stability, but also on account of more recent findings that implicate this interaction in cellular localization,41 T-cell differentiation,42 and neuron function.43 In addition to being a high-affinity interaction, previous work from various groups who used multiple assays has demonstrated the requirement for β-spectrin repeats 14 and 15, with practically undetectable binding to other 2-repeat fragments.24 The structure of the human ankyrin R ZU5-ANK domain, which represents the first example of a structure of this motif, reveals a compact domain formed primarily by a β-strand core and several surface loops. Similar structures have not been observed before and, hence, it is not possible to identify the surface features responsible for spectrin binding by structural comparison. Although the spectrin structure, on the other hand, reveals a canonical spectrin fold, it possesses no unusual structural features, aside from a negatively charged patch.
Thus, the spectrin structure rules out the possibility that repeats 14 and 15 assume a distinctively different overall structure from other repeats that could account for its specific recognition by ankyrin. This does not exclude the possibility that repeats 14 and 15 may adopt a different conformation in the complex, but currently there are no data to support such a claim. As in many other protein complexes, recognition likely involves a combination of surface complementarity, charge complementarity, and clustering of hydrophobic residues. The calculated electrostatic map for the ZU5-ANK subdomain reveals a significant patch of positively charged residues on one face of the molecule, which, based on sequence alignment, also may be present in other ankyrin isoforms and ankyrins from other organisms (Figure 6).48,49 A similar analysis with HEβ1415 showed a considerable patch of negatively charged residues on repeat 14. Although the heptad pattern of all spectrin repeats features many charged residues on the surface of the molecule, a comparison with other spectrin structures shows that repeat 14 is unique in its inclusion of an uninterrupted negatively charged patch.
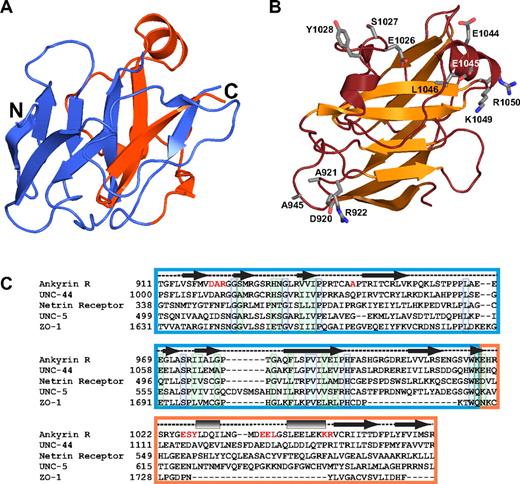
The structure of the ZU5 domain of ankyrin helps understand general features of this domain. (A) The ribbon diagram shows the ankyrin ZU5 domain (ZU5-ANK). The region corresponding to the canonical ZU5 region is shown in blue, whereas the last 55 amino acids forming this domain are shown in red. The diagram illustrates that 2 of the β strands in the β-sheet core come from residues lying outside the ZU5 consensus region, suggesting that the ZU5 consensus may extend beyond the canonical boundaries. (B) The ribbon diagram shows the functional mutants of ankyrin that map to the ZU5-ANK structure. The mutants are mostly found on the surface of a single face of the molecule. One of the mutations was mapped from an ankyrin variant implicated in hereditary spherocytosis and corresponds to L1046P,48 whereas other are functional mutants (DAR920AAA, A945P, ESY1026AAA, EE1044AA, KR1049AA) mapped from mutagenesis studies in other ankyrin isoforms that impair spectrin-binding activity in cell culture.49 Only the side chains of these mutants are shown (ball-and-stick). (C) Sequence alignment of human ankyrin R and 4 other ZU5 domain-containing proteins: Caenorhabditis elegans UNC-44, human netrin receptor, C elegans UNC-5, and human zona occludens-1. In correspondence with the color coding in panel A, the ZU5 domain46 is boxed blue whereas approximately 50 residues C-terminal to each protein's ZU5 consensus region (when available) are boxed in orange. Secondary structure assignments based on the structure of ZU5-ANK are provided above the sequence alignment as in Figure 3B. Residues printed in red indicate the locations of the functional mutants depicted in panel B. Individually boxed residues denote sites of universal (blue) or high (green) conservation among the presented sequences.
The structure of the ZU5 domain of ankyrin helps understand general features of this domain. (A) The ribbon diagram shows the ankyrin ZU5 domain (ZU5-ANK). The region corresponding to the canonical ZU5 region is shown in blue, whereas the last 55 amino acids forming this domain are shown in red. The diagram illustrates that 2 of the β strands in the β-sheet core come from residues lying outside the ZU5 consensus region, suggesting that the ZU5 consensus may extend beyond the canonical boundaries. (B) The ribbon diagram shows the functional mutants of ankyrin that map to the ZU5-ANK structure. The mutants are mostly found on the surface of a single face of the molecule. One of the mutations was mapped from an ankyrin variant implicated in hereditary spherocytosis and corresponds to L1046P,48 whereas other are functional mutants (DAR920AAA, A945P, ESY1026AAA, EE1044AA, KR1049AA) mapped from mutagenesis studies in other ankyrin isoforms that impair spectrin-binding activity in cell culture.49 Only the side chains of these mutants are shown (ball-and-stick). (C) Sequence alignment of human ankyrin R and 4 other ZU5 domain-containing proteins: Caenorhabditis elegans UNC-44, human netrin receptor, C elegans UNC-5, and human zona occludens-1. In correspondence with the color coding in panel A, the ZU5 domain46 is boxed blue whereas approximately 50 residues C-terminal to each protein's ZU5 consensus region (when available) are boxed in orange. Secondary structure assignments based on the structure of ZU5-ANK are provided above the sequence alignment as in Figure 3B. Residues printed in red indicate the locations of the functional mutants depicted in panel B. Individually boxed residues denote sites of universal (blue) or high (green) conservation among the presented sequences.
On the basis of these observations, we speculate that ankyrin and spectrin interact through charge–charge interactions mediated by their 2 oppositely charged patches. Although electrostatic interactions could certainly be required to form a stable spectrin-ankyrin complex, several biochemical and biophysical observations indicate that this may be necessary but not sufficient. For example, although β-spectrin fragments spanning repeats 13 and 14 have been shown to interact very weakly with ankyrin fragments, previous work has demonstrated that at least part of β-spectrin repeat 15 must be present to form a stable complex with ankyrin fragments.24 Presently, there is no obvious structural basis for the requirement of repeat 15, but biochemical data appear to suggest that the interaction is more extensive and complex than a simple matching of charged surfaces. Nevertheless, the presented structural data help to rule out some possibilities and to suggest possible experiments to dissect in greater detail the nature of the complex.
The ZU5 domain has a variety of functions in both vertebrate and invertebrate systems. In mammals, the ZU5 domain is present in another cytoskeletal protein, zona occludens 1 (ZO-1), which is implicated in tight junction formation.44 It is also present in netrin and netrin-like receptors involved in axon guidance and apoptosis in metazoans.47 Common features among the known proteins that contain a ZU5 domain seem to be their propensity to reside near the cell membrane and to bind other proteins with high affinity. The ankyrin ZU5 domain comprises the region defined by the canonical ZU5 domain consensus and additional C-terminal 55 amino acids (ZU5-ANK). The structure of the human ankyrin R ZU5-ANK domain represents the first example of a structure of this motif and reveals a compact domain formed primarily by a β-strand core and several surface loops.
Because similar structures have not been observed previously, it is not possible to find common features by structural comparison. Nonetheless, the structure does reveal some important characteristics that are likely to be of relevance to understanding other ZU5 domains. Sequence alignment of several proteins that contain a ZU5 domain (Figure 6C) indicate that the most conserved residues occur in the canonical ZU5 core; sequence similarity beyond this core is limited to ankyrins. Outside of the ZU5 core, the ankyrin ZU5 domain is composed of 2 short α-helices and 2 strands that form an integral part of the β-core. The last few residues in the ZU5-ANK domain form a β-strand that is sandwiched between other strands (Figure 6). Its absence would clearly affect the overall fold of the core of the molecule because the β-core would not be able to form without them, suggesting that either the structure presented here is unique to the ankyrins or that the ZU5 domain in other proteins extends beyond the consensus sequence, but with low sequence conservation. One scenario is that the last 2 strands are part of the ZU5 domain but that the loop connecting these 2 strands to the core of the ZU5 domain is of a different length, possibly even a very long loop, and with low sequence conservation.
Over the years, there have been many clinical (reviewed in Gallagher and Forget48 ) and experimental49 loss-of-function mutations reported. These include several mutations in the ankyrin ZU5 domain. Of these mutants, one, L1046P, is in one of the helices and faces the interior of the protein. This type of mutation tends to be destabilizing for the folding of the protein and, hence, it is likely that this mutant causes a partial or total loss of folding. The other mutants are more interesting because they reside in the surface of the protein and comprise, in their majority, charged hydrophilic amino acids. They cluster in 2 regions, in the helices toward the C-terminus of the protein and outside the ZU5 consensus region, and in a loop connecting strands of the core. The 2 regions are not close to each other but are on the same face of the protein. Interestingly, some of these mutations are close to the residues forming the positively charged patch in the ZU5-ANK fragment but seldom overlap with it. The role of the mutants or the charged patch is not known, but it is interesting to note the overlap. One possibility is that the charged patch is indeed involved in protein-protein interactions and the mutants directly disrupt the binding. An alternative is that the mutants do not reside near the interaction region but change the surface characteristics preventing proper binding. A greater understanding of the role of these mutants and their implications for clinical studies will hopefully be elucidated through structural studies of the complex.
In conclusion, the structures of the 2 protein domains involved in ankyrin-spectrin recognition have been solved. The structures point for the first time to some of the atomic features that may be important for this specific binding with high affinity. In addition, the structure of the ankyrin ZU5 domain provides the basis of understanding structures of other proteins containing this type of domain. Although these structures reveal some of the features important for understanding the atomic basis of ankyrin-spectrin recognition, further structural work—particularly on the complex of the 2 proteins—is needed.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank N. M. Baker and N. J. Reiter for comments and suggestions in preparation of the manuscript. We especially thank R.I. MacDonald for comments, help, and advice throughout this project. We acknowledge the use of equipment at the Center for Structural Biology at Northwestern University and LS-CAT at the APS at Argonne National Laboratory.
Research was supported by a National Institutes of Health (Bethesda, MD) grant (GM057692) to A.M. Use of the Advanced Photon Sources (APS) is supported by the Department of Energy (Washington, DC). Support from the R.H. Lurie Comprehensive Cancer Center of Northwestern University to the Structural Biology Facility is acknowledged.
National Institutes of Health
Authorship
Contribution: J.J.I. and L.H. performed research; J.J.I., L.H., and A.M. analyzed data; J.J.I. and A.M. wrote the manuscript; and J.J.I., L.H., and A.M. revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alfonso Mondragón, Department of Biochemistry, Molecular Biology, and Cell Biology, Northwestern University, 2205 Tech Dr, Evanston, IL 60208; e-mail: a-mondragon@northwestern.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal