Abstract
Hepcidin plays a major role in the regulation of iron homeostasis. Several bone morphogenetic proteins (BMPs) are strong inducers of hepcidin (Hamp1, HAMP) expression. Hemojuvelin, a protein critical for maintaining appropriate levels of hepcidin, acts as a coreceptor for BMP2 and BMP4, thereby providing a link between iron homeostasis and the BMP-signaling pathway. We show that a robust BMP, hemojuvelin, and SMAD1 response by murine Hamp1 is dependent on a distal BMP responsive element (BMP-RE2), the adjacent bZIP, HNF4α/COUP binding sites, and plus or minus 50 bp of the flanking area within −1.6 to −1.7 kb of the Hamp1 promoter. Furthermore, the STAT site and the BMP responsive element (BMP-RE1) located in the proximal 260-bp region of the Hamp1 promoter are also indispensable for maximal activation of hepcidin transcription. The homologous motifs in the distal and proximal regions of the human HAMP promoter act in a manner similar to the murine Hamp1 promoter. Therefore, we propose that the regulation of hepcidin by the BMP pathway involves the formation of a complex of liver-specific and response-specific transcription factors bound to the distal BMP-RE2 /bZIP/HNF4α/COUP region and to the proximal BMP-RE1/STAT region possibly by physical association of the 2 regions.
Introduction
Hepcidin, a 25–amino acid peptide, plays a major role in regulation of iron uptake and metabolism. Hepcidin expression is up-regulated by excess iron, inflammation, and bone morphogenic proteins (BMPs), while anemia and hypoxia suppress its expression.1,2 Diminished expression of hepcidin leads to excessive iron uptake and iron overload disease, hemochromatosis, while excessive expression of hepcidin probably contributes to the anemia of chronic disease.3-7
Hepcidin expression is affected by hemojuvelin (HJV; HFE2), hemochromatosis protein (HFE), and transferrin receptor 2 (TFR2), since mutations causing their loss of function or localization result in inappropriately low levels of hepcidin and iron overload.8-10 Patients with mutations of the genes encoding hepcidin or hemojuvelin exhibit the lowest levels of hepcidin and the most severe form of iron overload, juvenile hemochromatosis, associated with an early age of onset, severe iron deposits in the liver and heart, and hypogonadism.11 It has been demonstrated that hemojuvelin is able to bind BMP2 and BMP4, and mediates BMP signaling through BMP type I receptors, ALK3 and ALK4.12-14
We recently identified a proximal BMP responsive region between −140 bp to −200 bp from the start of translation of the murine Hamp1 promoter, just upstream of the inflammatory interleukin (IL)–6 responsive STAT3 site (−130 to −143 bp from start of translation) that slightly up-regulates hepcidin reporter expression in response to BMPs.15 Verga Falzacappa et al16 identified the 6-bp nucleotide sequence, GGCGCC, located at −152 to −157 bp from start of translation in the human HAMP promoter to be the BMP responsive element. The homologous BMP responsive element in the murine Hamp1 promoter is located between −140 and −200 bp, the so-called proximal BMP responsive region.15 In addition, we identified a distal BMP responsive region located between −1.6 and −1.8 kb of the Hamp1 promoter that enhances the BMP response approximately 5-fold.15
In this report, we have performed fine-mapping of the distal BMP-responsive region and identified a BMP-responsive element (BMP-RE2), bZIP, HNF4α/COUP motifs that are required for maximal response by murine Hamp1 to BMPs as well as SMAD1 and hemojuvelin. In addition, the plus or minus 50 bp flanking the BMP-RE2/bZIP/HNF4α/COUP site are important in the regulation of baseline expression of hepcidin, as contrasted to responsiveness to specific stimuli, and are critical for the maximal expression driven by the Hamp1 promoter. We propose a model in which formation of a transcriptional complex involving the distal region −1.6 to −1.8 kb region encompassing the BMP-RE2, bZIP, HNF4α, and COUP motifs and a proximal region at −140 to −260 bp containing both the BMP-RE1 and the STAT site accounts for the interaction of these disparate regions of the hepcidin promoter.
Methods
Materials
Human recombinant IL-6, BMP2, BMP4, and BMP9 were obtained from R&D Systems (Minneapolis, MN). Minimal essential medium, Williams medium E, l-glutamine, penicillin/streptomycin solution, fetal bovine serum (FBS), and polymyxin B sulfate were from Invitrogen (Carlsbad, CA).
Cell lines
The human hepatoma cell line HepG2 was obtained from ATCC (Manassas, VA) and cultured in minimal essential medium supplemented with 5% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, 1 mM sodium pyruvate, and 2 mM l-glutamine in an incubator with humidified atmosphere at 37°C with 5% CO2 in air.
The human embryonic kidney cell line HEK293T was obtained from the ATCC and cultured in Dulbecco modified essential medium supplemented with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM l-glutamine.
Cloning of Hamp1 promoter fragments into pGL3 basic
The 2.5-, 2.0-, 1.9-, 1.8-, 1.7-, 1.6-, 1.5-, 1.0-kb, 260-bp, 200-bp, and 140-bp fragments of the murine Hamp1 promoter were cloned into the Promega (Madison, WI) pGL3 basic vector containing the firefly luciferase reporter (luc) gene as described previously.15 Numbering of nucleotides is with respect to the start of translation.
Cloning of HAMP promoter fragments into pGL3 basic
The 3.0-, 2.7-, 2.5-, 2.4-, 2.3-, and 1.3-kb fragments of the human HAMP promoter from the start of translation were cloned into the Promega (Madison, WI) pGL3 basic vector containing the firefly luciferase reporter (luc) gene (details provided on request).
Expression vectors coding for hemojuvelin, SMAD1, SMAD4, and HNF4α
Human full-length hemojuvelin cDNA was cloned into pcDNA3.1 (Invitrogen). Murine SMAD1, SMAD4, and HNF4α were cloned using a T/A cloning mammalian expression vector pTARGET (Promega) using gene specific primers mSMAD1F CCACCATGAATGTGACCAGCTTGTTTTC, mSMAD1R TTAAGACACGGATGAAATAGGATTG, mSMAD4F, CCACCATGGACAATATGTCTATAACAAATAC, mSMAD4R CTCAGTCTAAAGGCTGTGGGT, mHNF4F CCACCATGCGACTCTCTAAAACCCTT, and mHNF4R CTAGATGGCTTCTTGCTTGG. The template used for amplification was murine C57BL/6J liver cDNA. All constructs were confirmed by direct sequencing.
Mutagenesis in the critical enhancing region of the murine Hamp1 and human HAMP promoter
The murine 2.5-kb wild-type (WT) Hamp1 promoter cloned into pGL3 was used as a template, and all changes were introduced using the Quikchange Mutagenesis Kit and the QuikChange Primer Design Program (Agilent-Stratagene, La Jolla, CA; http://www.stratagene.com). Likewise, all deletions and mutations in the human HAMP promoter constructs were made using the 3.0-kb WT HAMP promoter construct as the template. The sequences of all mutant constructs were confirmed by direct sequencing.
Transfection and BMP treatment
HepG2 cells were used because they are an established hepatoma cell line responsive to both BMPs and IL-6. Cells (5 × 104 per well) were plated onto 24-well plates (Corning, Corning, NY). The next day, 100 ng selected plasmid constructs containing the firefly luciferase gene under the control of the Hamp1 or HAMP promoter and 2.5 ng per well of the normalization plasmid pGL4.73 (Promega, Madison, WI) containing the Renilla luciferase gene were transfected into the cells using HyFect transfection reagent (Denville Scientific, Metuchen, NJ) according to manufacturer's instructions. The final ratio of DNA (μg):Hyfect reagent (μL) was 1:1.6. For hemojuvelin and SMAD1/4 transfections, 100 ng expression vector was used per well. Four to 6 hours after transfection, cells were treated with 10 ng/mL human BMPs (BMP2, BMP4, BMP9) for 12 to 16 hours in the presence of 3 μg/mL polymyxin to inhibit any possible contribution by cytokines generated by contaminating lipopolysaccharide.
HEK293T cells used for gel shift assays were transfected in T75 flasks using 8 μg plasmid DNA and the FUGENE reagent (Roche Applied Science, Indianapolis, IN), according to the manufacturer's instructions. Cells were harvested 48 hours after transfection using trypsin and washed with ice-cold phosphate-buffered saline (PBS), and cell pellets were frozen at −80°C until needed.
RNA isolation, cDNA synthesis, and quantitative real-time RT-PCR
Total RNA was isolated from nonstimulated, BMP-, SMAD1-, or hemojuvelin-stimulated HepG2 cells using the Versagene RNA Purification Kit (Gentra Systems, Minneapolis, MN), including the DNAse treatment step to avoid contamination with genomic DNA. cDNA first-strand synthesis was performed with 5 to 10 μg RNA, Moloney murine leukemia virus reverse transcriptase (Invitrogen), and oligo dT15 primer (IDT, Skokie, IL) in a 20-μL reaction volume. Quantitative real-time reverse-transcriptase polymerase chain reaction (Q-RT-PCR) was carried out using the Bio-Rad iCycler (Bio-Rad, Hercules, CA) using primers specific for human hepcidin (HAMP; accession no. NC_0000019) and a normalization gene human ribosomal protein large P2 (RPLP2; accession no. NM_001004) published previously.15
iCycler amplification was performed according to the manufacturer's instructions. After 40 amplification cycles, threshold cycle values were automatically calculated, attomoles of target cDNA were deduced from the standard curve, and levels of HAMP were expressed as the human HAMP/RPLP2 cDNA ratio.
Dual luciferase assay
Transfected cells were lysed using 150 μL of the passive lysis buffer (Promega). Ten microliters of the cell lysate was injected with 35 μL LARII solution (Promega), and the light output was measured using a LB 96V microplate luminometer (Berthold Technologies, Bad Wildbad, Germany). Next, 35 μL Stop&Glow solution (Promega) was injected, and the Renilla luciferase luminescence was measured to permit normalization. Results are expressed as firefly (luc)/Renilla luminescence ratio where the value for the full-length Hamp1 2.5-kb promoter or 3.0-kb full-length HAMP promoter was set as 1.
Identification of binding motifs and electrophoretic mobility shift assays
Identification of binding motifs.
Binding motifs were identified by MatInspector software (Genomatix Software; http://www.genomatix.de), and HNF4α and SMAD4 motifs were proven to be functional sites by electrophoretic mobility shift assays (EMSA).
Nuclear extracts.
This study received Institutional Review Board (IRB) approval for the use of mice from The Scripps Research Institute. Livers from 4 to 8 mice were rinsed in ice-cold PBS, patted dry, minced, and transferred to a glass homogenizer with 1.25 mL homogenization buffer/liver. The homogenization buffer consisted of 0.3 M sucrose, 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) pH 7.5, 1 mM EDTA (ethylenediaminetetraacetic acid), 1 mM EGTA (ethyleneglycoltetraacetic acid), 10 mM KCl, 1 mM PMSF (phenylmethylsulphonyl fluoride), 0.07 trypsin inhibitory units (TIU)/mL aprotinin, and a cocktail of phosphatase inhibitors (50 mM sodium fluoride, 10 mM sodium pyrophosphate, 1 mM sodium vanadate, 10 mM β-glycerolphosphate). The livers were homogenized with 10 strokes on ice and filtered through 4 layers of cotton gauze. Two volumes of cushion buffer (2.2 M sucrose, 10 mM HEPES pH 7.5, 0.1 mM EDTA, 0.1 mM EGTA, 10 mM KCl, and phosphatase inhibitor cocktail were added to the lysate and mixed; volume = 26 mL for 4 livers). The lysate from 4 livers was divided into 2 tubes, 13 mL/tube, layered on 10 mL cushion buffer in Beckman ultracentrifuge tubes. The samples were centrifuged for 1 hour at 32 000 rpm (76 220g) in a TI70 rotor using a Beckman L90 ultracentrifuge. The nuclear pellet was resuspended in 1mL/tube of nuclear extraction buffer (10 mM HEPES pH 7.5, 1 mM EDTA, 5 mM MgCl2, 0.4 M KCl, phosphatase inhibitors, 1 mM PMSF, and 0.07 TIU/mL aprotinin, and pooled. The nuclear extract was incubated on ice for 30 minutes and centrifuged at 4°C for 20 minutes at 13 000g in an Eppendorf microfuge. Nuclear extracts were stored at −80°C until used.
Lysates from transfected HEK293T cells expressing transcription factors were prepared by thawing cells with 100 μL lysis buffer containing 50 mM Tris (tris[hydroxymethyl]aminomethane) pH 7.5, 0.36 M NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP40, 0.25% Triton X-100, 1 mM PMSF, 0.07 TIU/mL aprotinin and incubating on ice for 30 minutes to extract. The lysates were then diluted 1:10 with the same buffer with no NaCl, and immediately centrifuged for 20 minutes at 13 000g at 4°C. The clarified supernatant was stored in aliquots at −80°C when not in use.
Electrophoretic mobility shift assays.
Complementary oligomers were annealed in 10 mM Tris HCl, pH 8.0, and 0.1 mM EDTA at 95°C for 2 minutes, 50°C for 5 minutes, 37°C for 10 minutes, 22°C for 20 minutes, and 4°C for at least 5 minutes, and labeled with 32P-γ-ATP and T4 polynucleotide kinase. Nuclear extracts (5-15 μg) were incubated at room temperature for 15 minutes with 50 g/mL dI-dC in an 18 μL reaction mix containing 50 mM Tris pH 7.5, 10% glycerol, 90 mM KCl, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, phosphatase inhibitor cocktail, 0.74 mM spermidine, 0.15 mM spermine, 1 mM PMSF, and 0.07 TIU/mL aprotinin. When indicated, a competitor probe corresponding to the consensus transcription factor motif was added to a final concentration of 125 nM at the start of the incubation. The samples were then electrophoresed on a 5% polyacrylamide/0.5× Tris-borate-EDTA (TBE) gel. Blocking complementary oligomers corresponding to the consensus motifs for SMAD 3/4 (CGAGAGCCAGACAAAAAGCCAGACATTTAGCCAGACAC) and HNF4α (CTCAGCTTGTACTTTGGTACAACTA) were annealed as described.
Results
Overexpression of SMAD1 and hemojuvelin but not of SMAD4 up-regulates hepcidin expression
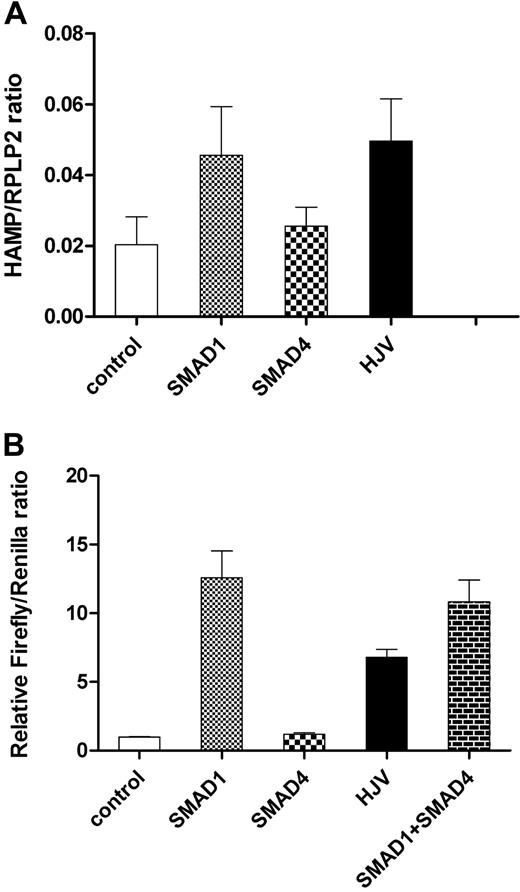
Overexpression of SMAD1 or hemojuvelin up-regulated the endogenous levels of hepcidin mRNA in HepG2 cells. In contrast, overexpression of SMAD4 did not up-regulate hepcidin expression (Figure 1A). Similarly, overexpression of SMAD1 but not SMAD4 resulted in an increased expression of the 2.5-kb Hamp1 promoter linked to the firefly luciferase reporter (Figure 1B). When hemojuvelin was transfected into HepG2 cells, a 6-fold increase in luciferase reporter expression resulted (Figure 1B), consistent with previous reports.13
The effect of SMAD1, SMAD4, and HJV on endogenous HAMP (A) or Hamp1 promoter-driven reporter expression (B) in HepG2 cells. Overexpression of SMAD1 andHJV induced expression of endogenous HAMP mRNA as assessed by Q-RT-PCR (A) as well as expression of firefly luciferase reporter driven by the 2.5-kb Hamp1 promoter (B), while the common mediator SMAD4 affected neither endogenous nor reporter expression. Values represent means and standard error of the mean (SEM) of at least 3 independent experiments.
The effect of SMAD1, SMAD4, and HJV on endogenous HAMP (A) or Hamp1 promoter-driven reporter expression (B) in HepG2 cells. Overexpression of SMAD1 andHJV induced expression of endogenous HAMP mRNA as assessed by Q-RT-PCR (A) as well as expression of firefly luciferase reporter driven by the 2.5-kb Hamp1 promoter (B), while the common mediator SMAD4 affected neither endogenous nor reporter expression. Values represent means and standard error of the mean (SEM) of at least 3 independent experiments.
The full response to SMAD1, hemojuvelin, and BMPs requires a distal part of the hepcidin promoter
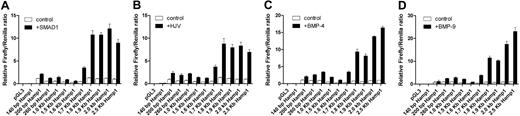
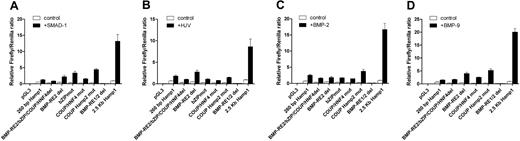
Using incrementally larger murine Hamp1 promoter constructs to drive expression of the luciferase reporter, we mapped the response induced by SMAD1 (Figure 2A) and hemojuvelin (Figure 2B) to the identical 1.6 to 1.8 kb region of Hamp1 promoter as the responsive region for BMP4 (Figure 2C) and BMP9 (Figure 2D). Presence of the 1.6 to 1.8 kb region of the Hamp1 promoter enhances the total response to all tested modulators quite significantly. The 260-bp Hamp1 promoter construct also responds to the expression of hemojuvelin and SMAD1 but to a much lower extent than with the 1.7-kb or 2.5-kb Hamp1 promoter constructs, as we have shown previously for the BMP response.15 The ability to respond to SMAD1 and HJV markedly increases with the presence of −1.6 to −1.7 kb Hamp1 region as the reporter values reach higher total values than the 260-bp Hamp1 construct. Furthermore, the 1.7-kb Hamp1 construct shows markedly lower basal expression compared with 260-bp Hamp1 construct (1.7-kb control vs 260-bp control) and thus, the ability to respond, calculated as fold induction (control versus stimuli), is significantly higher. The total SMAD1 and hemojuvelin response reaches its maximum response in constructs of 1.8 kb and larger, suggesting that the −1.6 to −1.7 kb Hamp1 region increases the actual ability to respond to the stimulus while the −1.7 to −1.8 kb Hamp1 region stabilizes or facilitates formation of a fully active complex that results in the maximal response (Figure 2A,B). The total response stimulated by the BMP4 and BMP9, although it shows a similar pattern of responsiveness, continues to gradually increase from 1.8 kb Hamp1 to 2.5 kb Hamp1 promoter constructs (1.5- to 2.0-fold higher), suggesting that there are additional sites that also contribute to the BMP responsiveness (Figure 2C,D).
Mapping of SMAD1, HJV, BMP4, and BMP9 responsive region of Hamp1. Constructs containing various fragments of the murine Hamp1 promoter linked to a firefly reporter were transfected into HepG2 cells and either cotransfected with SMAD1 (A) or hemojuvelin (B) or treated with 10 ng/mL BMP4 (C) and BMP9 (D) for 12 to 16 hours. Firefly/Renilla ratio of untreated 2.5-kb Hamp1 WT construct was set as 1.00. Mean and SEM of the relative firefly/Renilla ratios of at least 3 independent experiments are shown.
Mapping of SMAD1, HJV, BMP4, and BMP9 responsive region of Hamp1. Constructs containing various fragments of the murine Hamp1 promoter linked to a firefly reporter were transfected into HepG2 cells and either cotransfected with SMAD1 (A) or hemojuvelin (B) or treated with 10 ng/mL BMP4 (C) and BMP9 (D) for 12 to 16 hours. Firefly/Renilla ratio of untreated 2.5-kb Hamp1 WT construct was set as 1.00. Mean and SEM of the relative firefly/Renilla ratios of at least 3 independent experiments are shown.
Alignment of the −1.6 to −1.8 kb region of the murine Hamp1 promoter with the −2.3 to −2.5 kb region of the human HAMP promoter reveal significant homology (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). In the same manner used to map the murine Hamp1 promoter, incrementally larger human HAMP promoter constructs were tested to drive expression of the luciferase reporter gene to map the region of the human HAMP promoter responsive to BMPs, SMAD1, and hemojuvelin. We found that the homologous region of the human HAMP promoter (−2.3 to −2.5 kb) was required for up-regulation of reporter expression by BMPs, SMAD1, and hemojuvelin. Furthermore, the response of human HAMP to all tested stimuli was greater than the response of murine Hamp1 (Figure S2).
The BMP-RE2/bZIP/HNF4/COUP motifs are critical for the SMAD1, hemojuvelin, and BMP response of murine Hamp1
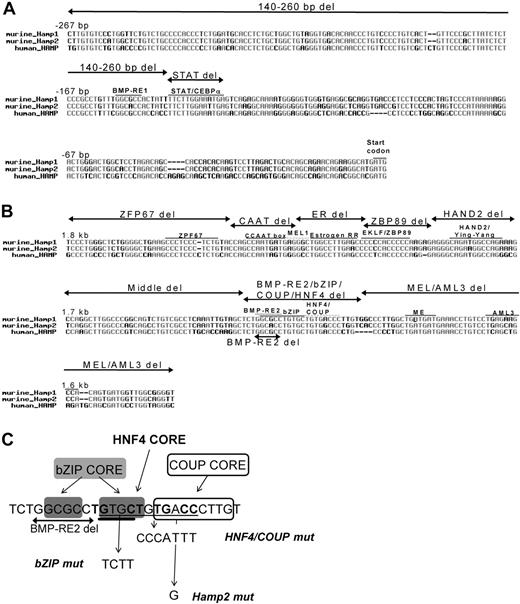
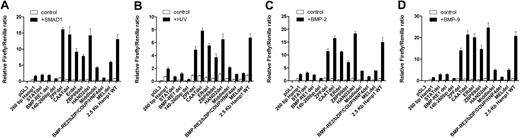
Alignment of murine and human hepcidin promoter sequences for the proximal and the distal regulatory regions revealed high homology and conserved transcription factor binding motifs (Figure 3). To determine the importance of each of the transcription factor motifs in the regulation of murine Hamp1 by the hemojuvelin/BMP/SMAD pathway, constructs were made in which specific consensus motifs or regions were either deleted or mutated. Deletion of the BMP-RE2/bZIP/HNF4/COUP motifs (−1681 to −1654 bp) resulted in profound loss of the ability to respond to hemojuvelin, BMPs, and SMAD1 (Figure 4A-D). Deletion of the regions immediately flanking the BMP-RE2/bZIP/HNF4/COUP motifs, “Middle del” that has no identifiable transcription factor consensus motifs, and “MEL” that has a MEL consensus motif, also severely affected the response to hemojuvelin, BMPs, and SMAD1. Deletions of the flanking region primarily affected basal level of expression resulting in a blunted “total” response, rather than affecting the ability to respond, defined as the fold induction over untreated controls. Deletions within the −1.7 to −1.8 kb region, notably of motifs for ZFP67, estrogen related receptor, and ZBP89, also affected the total response to the tested stimuli but did not significantly change the ability to respond. (Figure 4A-D). Further characterization of the BMP-RE2/bZIP/HNF4/COUP region was performed. Deletion of BMP-RE2, mutation of the bZIP, HNF4, or COUP motifs each resulted in the loss of responsiveness to SMAD1, HJV, BMP2, and BMP9 similar to the large BMP-RE2/bZIP/HNF4/COUP deletion (Figure 5A-D). Transition of A to G at position −1663 bp (HNF4/COUP core) to reflect the Hamp2 sequence affects the responsiveness to each stimuli but seems to be less severe compared with the other mutations.
Scheme of deletions and mutations introduced into the murine Hamp1 promoter. Murine Hamp1 and Hamp2 and human HAMP were aligned, and conserved transcriptional motifs were identified. Deletions in the proximal Hamp1 promoter region (A) and deletions in the distal −1.6 to −1.8 kb promoter region (B) are indicated. After narrowing to the BMP-RE2/bZIP/COUP/HNF4α region, further deletions and mutations were introduced to identify the critical motif (C). The 6-bp BMP-RE deletion (2-sided arrow), the 7-bp mutation of the HNF4α/COUP site (HNF4/COUP mut; underlined), the single nucleotide mutation to reflect the sequence in Hamp2 (Hamp2 mut), and the 4-bp mutation of the bZIP site (bZIP mut; double-underlined line) are shown.
Scheme of deletions and mutations introduced into the murine Hamp1 promoter. Murine Hamp1 and Hamp2 and human HAMP were aligned, and conserved transcriptional motifs were identified. Deletions in the proximal Hamp1 promoter region (A) and deletions in the distal −1.6 to −1.8 kb promoter region (B) are indicated. After narrowing to the BMP-RE2/bZIP/COUP/HNF4α region, further deletions and mutations were introduced to identify the critical motif (C). The 6-bp BMP-RE deletion (2-sided arrow), the 7-bp mutation of the HNF4α/COUP site (HNF4/COUP mut; underlined), the single nucleotide mutation to reflect the sequence in Hamp2 (Hamp2 mut), and the 4-bp mutation of the bZIP site (bZIP mut; double-underlined line) are shown.
The effect of various Hamp1 promoter deletions on the SMAD1, HJV, BMP2, and BMP9 induced response. Firefly reporter constructs driven by the murine Hamp1 promoter were used, 2.5 kb Hamp1 WT was the positive full-length promoter control, 260 bp Hamp1 represented the minimal promoter, and pGL3 represented the negative promoterless control. Constructs containing deletions in the proximal region of Hamp1 promoter (STAT del and 140-260 bp del) as well as deletions in the distal −1.6 to −1.8 kb region (all others) were transfected into HepG2 cells and either cotransfected with SMAD1 (A) and hemojuvelin (B) or treated with 10 ng/mL BMP4 (C) and BMP9 (D) for 12 to 16 hours. Firefly/Renilla ratio of nontreated 2.5-kb Hamp1 WT construct was set as 1.00. Mean and SEM of the relative firefly/Renilla ratios of at least 3 independent experiments are shown.
The effect of various Hamp1 promoter deletions on the SMAD1, HJV, BMP2, and BMP9 induced response. Firefly reporter constructs driven by the murine Hamp1 promoter were used, 2.5 kb Hamp1 WT was the positive full-length promoter control, 260 bp Hamp1 represented the minimal promoter, and pGL3 represented the negative promoterless control. Constructs containing deletions in the proximal region of Hamp1 promoter (STAT del and 140-260 bp del) as well as deletions in the distal −1.6 to −1.8 kb region (all others) were transfected into HepG2 cells and either cotransfected with SMAD1 (A) and hemojuvelin (B) or treated with 10 ng/mL BMP4 (C) and BMP9 (D) for 12 to 16 hours. Firefly/Renilla ratio of nontreated 2.5-kb Hamp1 WT construct was set as 1.00. Mean and SEM of the relative firefly/Renilla ratios of at least 3 independent experiments are shown.
Finer mapping of the SMAD1, hemojuvelin, BMP2, and BMP9 response in the critical BMP-RE2/bZIP/HNF-4/COUP region. Constructs containing mutations in critical distal BMP-RE2/bZIP/COUP/HNF4 region and also the double mutant BMP-RE1/2 were introduced into the 2.5-kb Hamp1 promoter reporter construct as described in Figure 3. Murine 2.5-kb Hamp1 WT was the positive full-length promoter control, 260 bp Hamp1 represented the minimal promoter, and pGL3 represented the negative promoterless control. Control and mutant constructs were transfected into HepG2 cells and either cotransfected with SMAD1 (A) or hemojuvelin (B) or treated with 10 ng/mL BMP4 (C) or BMP9 (D) for 12 to16 hours. Firefly/Renilla ratio of untreated 2.5-kb Hamp1 WT construct was set as 1.00. Means and SEM of the relative firefly/Renilla ratios are shown.
Finer mapping of the SMAD1, hemojuvelin, BMP2, and BMP9 response in the critical BMP-RE2/bZIP/HNF-4/COUP region. Constructs containing mutations in critical distal BMP-RE2/bZIP/COUP/HNF4 region and also the double mutant BMP-RE1/2 were introduced into the 2.5-kb Hamp1 promoter reporter construct as described in Figure 3. Murine 2.5-kb Hamp1 WT was the positive full-length promoter control, 260 bp Hamp1 represented the minimal promoter, and pGL3 represented the negative promoterless control. Control and mutant constructs were transfected into HepG2 cells and either cotransfected with SMAD1 (A) or hemojuvelin (B) or treated with 10 ng/mL BMP4 (C) or BMP9 (D) for 12 to16 hours. Firefly/Renilla ratio of untreated 2.5-kb Hamp1 WT construct was set as 1.00. Means and SEM of the relative firefly/Renilla ratios are shown.
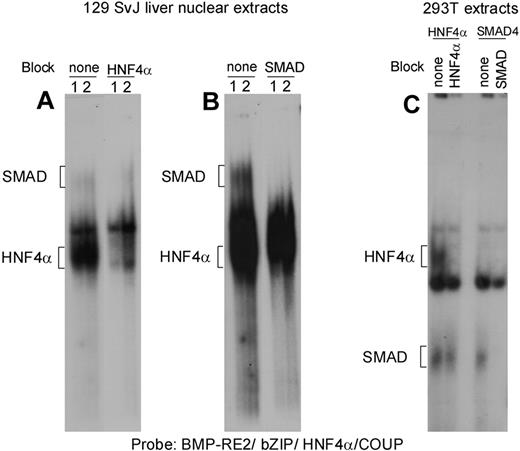
Gel shift analyses confirmed that endogenous HNF4α and SMAD4 from liver nuclear extracts as well as HEK293T-expressed recombinant HNF4α and SMAD4 were able to bind a probe corresponding to the BMP-RE2/bZIP/HNF4/COUP region (Figure 6). Furthermore, specific binding of liver nuclear extracts to the BMP-RE2/bZIP/HNF4/COUP probe was blocked by the addition of excess cold competitors corresponding to the consensus HNF4α and SMAD4 motifs (Figure 6). The specificity of the binding has been also confirmed by supershift analyses using antibodies specific for HNF4α and SMAD4 (data not shown).
Electrophoretic mobility shift assay of BMPRE2/bZIP/HNF4α/COUP. A probe corresponding to the BMP-RE2/ bZIP/HNF-4/COUP region was 32P labeled and nuclear extracts from 2 independent preparations (numbered 1 and 2) of 129 SvJ liver nuclear extracts (A,B) or lysates from HNF4α or SMAD4 overexpressing HEK293T cells (293T) were added (C). Each mouse liver sample was prepared from a group of 4 mice on an iron-deficient diet. (A) Nuclear extracts were either preincubated with buffer (none) or DNA competitor corresponding to the consensus motif of HNF4α (HNF4). (B) Nuclear extracts were either preincubated with buffer (none) or DNA competitor corresponding to the consensus motif of SMAD 3/4. Panels A and B were part of the same gel but panel B required a longer exposure to visualize the SMAD bands. This experiment is representative of more than 3 experiments.
Electrophoretic mobility shift assay of BMPRE2/bZIP/HNF4α/COUP. A probe corresponding to the BMP-RE2/ bZIP/HNF-4/COUP region was 32P labeled and nuclear extracts from 2 independent preparations (numbered 1 and 2) of 129 SvJ liver nuclear extracts (A,B) or lysates from HNF4α or SMAD4 overexpressing HEK293T cells (293T) were added (C). Each mouse liver sample was prepared from a group of 4 mice on an iron-deficient diet. (A) Nuclear extracts were either preincubated with buffer (none) or DNA competitor corresponding to the consensus motif of HNF4α (HNF4). (B) Nuclear extracts were either preincubated with buffer (none) or DNA competitor corresponding to the consensus motif of SMAD 3/4. Panels A and B were part of the same gel but panel B required a longer exposure to visualize the SMAD bands. This experiment is representative of more than 3 experiments.
The proximal BMP-RE1 and STAT motifs play an important role in murine Hamp1 responsiveness
The STAT site (−143 to −134 nt from start of translation) located in the proximal promoter region of murine Hamp1 is essential for the maximal response since deletion of the STAT site in the context of the 2.5-kb Hamp1 promoter construct resulted in significantly lower total level of reporter expression in the presence or absence of hemojuvelin, SMAD1 or BMPs (Figure 4A-D). We also examined the importance of the −140 to −260 bp region of the murine Hamp1 promoter that exhibited a 2- to 3-fold up-regulation in expression by SMAD1, hemojuvelin, and BMPs, and has been recently identified to contain a proximal BMP-responsive element (BMP-RE1). Deletion of the −140 to −260 bp region in the 2.5-kb hepcidin promoter construct resulted in a reduction but not complete loss of inducibility as well as markedly reduced total levels of reporter expression. Deletion of the BMP-RE1 alone had a similar effect, although less severe than the −140 to −260 bp deletion (Figure 4A-D). Furthermore, the deletion of both the BMP-RE1 and BMP-RE2 resulted in complete loss of basal level expression as well as ability to respond to SMAD1, hemojuvelin, BMP-2, BMP-4, or BMP-9 (Figure 5A-D, Figure S3A,B).
Unexpectedly, we found that deletion of BMP-RE1 and BMP-RE2 either separately or together compromised the total level of response to IL-6 (ie, relative ratio) as well (Figure S3A). However, the ability to respond to IL-6 (ie, fold induction) was only slightly reduced with the deletion of −140 to −260 bp and unaffected with the deletion of the BMP-RE1, BMP-RE2, individually or in combination (Figure S3B).
Deletion and mutation analyses of the distal hemojuvelin/BMP/SMAD responsive region of the human HAMP promoter show characteristics similar to those of the murine promoter
We also performed studies on the human HAMP promoter, examining the homologous hemojuvelin/BMP/SMAD responsive region located −2.3 to −2.5 kb from the start of translation (Figures S1 and S2). The deletion of 50-bp increments across this region showed a similar pattern as murine Hamp1, showing a decrease in responsiveness to hemojuvelin, SMAD1, and BMPs with the A and B deletions within −2.4 to −2.5 kb region of the human HAMP promoter and a profound effect with C and D deletions (−2.3 to −2.4 kb) corresponding to the murine Hamp1 Middle, BMP-RE2/bZIP/HNF4/COUP, and MEL regions (−1.6 to −1.7 kb; Figure S4A-C). As with the murine Hamp1 promoter, mutation of the proximal STAT-binding motif of the human 3.0-kb HAMP promoter resulted in significantly reduced hepcidin expression in response to hemojuvelin, BMPs, and SMAD1 despite the presence of the distal regulatory region (Figure S4A-C). The proximal STAT motif in the human HAMP promoter was critical for total responsiveness but did not affect the ability to respond in terms of fold induction (Figure S4A-C).
There are 2 putative BMP-RE sites (BMP-RE2 and BMP-RE3) in the distal −2.3 to −2.5 kb region of the human HAMP promoter compared with 1 present in the distal −1.6 to −1.7 kb murine Hamp1 promoter (BMP-RE2) as depicted in Figure S1. Deletion of the human BMP-RE2 (conserved between human and mouse) almost completely abrogated the response to SMAD1, HJV, and BMP-9, whereas deletion of BMP-RE3 (not conserved between human and mice) had no effect on HAMP responsiveness (Figure S5A-C). Likewise, there are 2 HNF4α sites predicted in the 2.3 to 2.5 kb region of the human HAMP promoter. To determine whether the HNF4α/COUP motifs were important for human HAMP responsiveness, we mutated the 2 HNF4α motifs (HNF4A and HNF4B) individually or in combination (Figure S1). Mutation of HNF4A (not conserved between human and mouse) did not affect the responsiveness, while mutation of the HNF4B site (conserved between the human and mouse) decreased responsiveness (Figure S5A-C). These data demonstrate that regulation of the murine and human hepcidin genes are similar although mutation of the HNF4α motif seems to have a more profound effect in murine Hamp1 than human HAMP.
Discussion
Hemojuvelin plays a crucial role in maintaining the proper level of hepcidin expression.8,17,18 Babitt et al13 and Huang et al14 demonstrated that the hemojuvelin effect is mediated by enhancement of the BMP/SMAD-signaling pathway by increasing the sensitivity of cells to low levels of BMPs. Binding to the BMP receptor results in activation of the SMAD-signaling pathway.13,14,19 Liver-specific SMAD4-deficient mice exhibit virtually no hepcidin expression, impaired response to iron and IL-6, and an iron overload phenotype.20 SMAD4 is able to increase hepcidin expression when transfected into SMAD4-deficient cells,20 and it is likely that the lack of responsiveness to SMAD4 in HepG2 cells is due to limiting levels of endogenous SMAD1 in HepG2 cells. We demonstrated that SMAD1 is able to increase expression of endogenous hepcidin when transfected into hepatoma cells. In the same manner, hemojuvelin is also able to increase hepcidin expression when transfected into hepatoma cells.13,21
We postulated that if hemojuvelin, BMP, and SMAD are indeed on a single pathway in regulating hepcidin expression, the responsive region for the 3 stimuli should be identical. Our studies demonstrated that up-regulation of hepcidin promoter reporter expression by hemojuvelin, BMPs (BMP-4 and BMP-9) and SMAD1 all map to the identical distal robust responsive region of the hepcidin promoter located at −1.6 to −1.8 kb and a weaker proximal responsive region located at −140 bp to −260 bp. Consistent with our findings, Verga-Falzacappa et al16 recently identified the proximal BMP responsive element located at −152 to −157 bp from start of translation in the human HAMP promoter as a 6-bp long BMP-RE1 GGCGCC motif. In our system, deletion of the larger proximal region containing the BMP-RE1 or specific deletion of BMP-RE1 resulted in a reduction in total responsiveness to SMAD1, hemojuvelin, BMP-2, and BMP-9 stimuli even in the presence of the distal portion of the promoter thus confirming a critical role of BMP-RE1 in hepcidin regulation.
As shown previously, the IL-6 response is dependent on a STAT motif within the proximal 140 bp of the hepcidin promoter22-24 and an additional regulatory site between 140 and 260 bp of the hepcidin promoter (numbering from the start of translation).24 The latter site was identified by Verga-Falzacappa as the BMP-RE1 located upstream of the STAT site (−72/−64 from start of transcription or −152/−157 bp from start of translation) and seems to be critical for the IL-6 response as well as the BMP response by the human −942 bp HAMP promoter reporter construct.16 Our data using the murine 2.5-kb Hamp1 promoter and human 3.0-kb promoter reporter constructs, which include the distal regulatory region, confirms their observation. Deletion analyses demonstrated that the proximal STAT site required for IL-6 responsiveness does not affect inducibility by hemojuvelin, BMPs, and SMAD1, but strongly affects the baseline expression level and total level of response that can be achieved. In contrast, the deletion of the region between −140 to −260 bp of Hamp1 (containing the homologous BMP-RE1) and the BMP-RE1 motif alone leads to a severe reduction in total IL-6 response and also a marked reduction of the total response as well as ability to respond to BMPs, SMAD1, and hemojuvelin. On the other hand, the ability to respond to IL-6 in terms of fold induction over nontreated controls remains either unchanged when one or both BMP responsive elements (BMP-RE1 del, BMP-RE2 del, or BMP-RE1/2 del) are deleted or is only slightly decreased when the −140 to −260 bp region (140-260 bp del) is deleted. Still the phenotype of −140 to −260 bp del and BMP-RE1 del constructs are similar but not identical and suggest that there may be other regulatory sites in addition to the BMP-RE1 in the −140 to −260 region. Nevertheless, these data demonstrate that both the STAT and the BMP-RE1 regions are indispensable for a normal total response to IL-6 and BMPs, and establish a link between the inflammatory pathway and BMP pathway in regulating hepcidin expression.
Deletion and mutation analyses identified a 27-bp region encompassing BMP-RE2, bZIP, HNF4α, and COUP binding motifs within the −1.6 to −1.7 kb region of the murine Hamp1 promoter to be critical for the responsiveness to SMAD1, hemojuvelin, BMP-2, and BMP-9 as well as for maintaining the proper baseline level of hepcidin expression. BMP-RE2 has 100% sequence identity (GGCGCC) to the proximal BMP responsive element (BMP-RE1) identified by Verga Falzacappa et al.16 Direct binding of HNF4α and SMAD4 complexes to a probe corresponding to the BMP-RE2/bZIP/HNF4/COUP region was demonstrated by electrophoretic mobility shift assays thus validating the observations made using the reporter approach. The mobility of the SMAD complex bound to the BMP-RE2/bZIP/HNF4/COUP region with liver nuclear extracts differed from the mobility of the SMAD complex observed with SMAD4 transfected 293T cells, suggesting the components of the complexes were different. The plus or minus 50 bp sequence flanking the BMP-RE2/bZIP/HNF4/COUP motifs was also found to significantly contribute to the hepcidin expression but seems to affect more the baseline expression than the ability to respond.
A double mutant lacking BMP-RE1 and BMP-RE2 showed a complete loss of basal level expression as well as a loss in the ability to respond to SMAD1, HJV, and BMP stimuli, while neither BMP-RE1 alone nor BMP-RE2 alone displayed such a severe phenotype. On the other hand, the total IL-6 response of BMP-RE1 del, BMP-RE2 del, and BMP-RE1/2 del constructs was severely reduced, but the ability to respond to IL-6 was not affected. These data suggest that BMP-REs regulate the basal level of expression of hepcidin and the responsiveness to BMPs but may not be critical for the actual response to IL-6. This is not completely in agreement with Verga Falzacappa et al,16 who proposed that BMP-RE1 is critical for IL-6 responsiveness. The difference between their data and ours might be due to a different cell line used for the study, our use of the longer 2.5-kb construct, and/or differences between the mouse and human hepcidin promoters.
Alignment of the distal −1.6 to −1.8 kb region of murine Hamp1, Hamp2, and corresponding portion of human HAMP promoter (−2.3 to −2.5 kb) demonstrated this region to be highly conserved. Significant homology in this region is also evident in chimpanzee, dog, and rat as well. The lack of responsiveness of murine Hamp2 to BMPs appears to be partially due to the single nucleotide difference in the BMP-REs located at both the proximal and distal regulatory sites which is supported by the BMP-RE1/2 del phenotype. Examination of the human HAMP promoter demonstrated that this region of homology (−2.3 to −2.5 kb) is important in determining hemojuvelin, BMP, and SMAD1 responsiveness. Similar to murine Hamp1, the distal BMP-RE2 that is conserved between the human and mice is critical for the SMAD1, hemojuvelin, and BMP response of the HAMP promoter while the nonconserved BMP-RE3 motif does not appear to be functional. However, the responsiveness of HAMP promoter seems to not be dependent solely on BMP-RE2 but also on an unidentified motif between 2.30 and 2.35 kb as well. Of the 2 HNF4α motifs in the −2.3 to −2.5 kb region of the human HAMP promoter, the conserved HNF4α site was more important than the nonconserved HNF4α site in regulating human HAMP expression. Nevertheless, mutation of the conserved HNF4α site was not sufficient to totally eliminate the ability of the human 3.0-kb HAMP promoter construct from responding to SMAD1, hemojuvelin, or BMPs. Therefore, the data indicate that regulation of hepcidin transcription between mice and humans are similar but not necessarily identical.
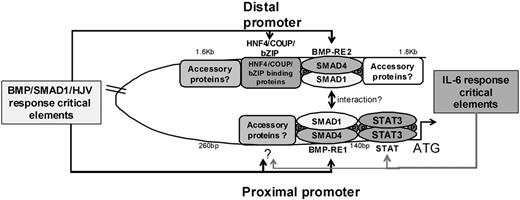
We propose that full activation of the murine Hamp1 promoter by modulators of the BMP pathway (hemojuvelin, BMPs, or SMADs) involves the formation of a transcriptional complex involving the distal region −1.6 to −1.8 kb region encompassing the BMP-RE2, bZIP, HNF4α, and COUP motifs, and the proximal region containing both the BMP-RE1 and the STAT site (Figure 7). Deletion of any of these motifs results in a markedly reduced response to hemojuvelin, BMPs, or SMAD1. The presence of a putative BMP-RE neighboring the liver-specific HNF4α consensus motif suggests that occupancy of both the liver-specific and BMP- inducible sites might be required and reflects the liver-specific expression and inducibility of hepcidin.
A hypothetical scheme of activation of the murine Hamp1 promoter. Fully active promoter complex requires the presence of both the SMAD1/4 complexes as well as STAT3. Elements depicted as critical reduce/impair the ability of the promoter to respond to such stimuli. Our data suggest that the proximal STAT site is crucial for IL-6 responsiveness, and there seems to be another weaker as yet unidentified modulatory site between proximal 140 and 260 bp (question mark). On the other hand, proximal BMP-RE1, unidentified proximal modulatory site between 140 and 260 bp (question mark) and distal BMP-RE2/bZIP/HNF4/COUP are critical for the BMP/SMAD1/HJV pathway. The absence of any critical responsive element leads to a decrease in basal expression of hepcidin promoter, thereby, affecting the total response of the promoter to all stimuli even if the actual ability to respond is not compromised. The assembly of multiple transcription factors in an activation complex would explain the crosstalk between the inflammatory pathway and BMP/SMAD1/HJV pathway even though the actual responsive elements are quite distinct. Furthermore, our data suggest that there are additional accessory proteins that facilitate formation of a fully active promoter complex by binding to the region flanking the BMPRE2/bZIP/HNF4α/COUP motifs. The scheme for the human HAMP promoter would be similar and require the distal −2.3 to −2.5 kb region.
A hypothetical scheme of activation of the murine Hamp1 promoter. Fully active promoter complex requires the presence of both the SMAD1/4 complexes as well as STAT3. Elements depicted as critical reduce/impair the ability of the promoter to respond to such stimuli. Our data suggest that the proximal STAT site is crucial for IL-6 responsiveness, and there seems to be another weaker as yet unidentified modulatory site between proximal 140 and 260 bp (question mark). On the other hand, proximal BMP-RE1, unidentified proximal modulatory site between 140 and 260 bp (question mark) and distal BMP-RE2/bZIP/HNF4/COUP are critical for the BMP/SMAD1/HJV pathway. The absence of any critical responsive element leads to a decrease in basal expression of hepcidin promoter, thereby, affecting the total response of the promoter to all stimuli even if the actual ability to respond is not compromised. The assembly of multiple transcription factors in an activation complex would explain the crosstalk between the inflammatory pathway and BMP/SMAD1/HJV pathway even though the actual responsive elements are quite distinct. Furthermore, our data suggest that there are additional accessory proteins that facilitate formation of a fully active promoter complex by binding to the region flanking the BMPRE2/bZIP/HNF4α/COUP motifs. The scheme for the human HAMP promoter would be similar and require the distal −2.3 to −2.5 kb region.
We conclude that regulation of the hepcidin promoter seems to involve complexes of tissue-specific and inducer-specific transcription factor combinations, and crosstalk between various pathways involving multiple regulatory binding motifs. The distal region of the hepcidin promoter is crucial for maximal induction by BMP, hemojuvelin, and iron, while it is not essential for the inflammatory response; however, the proximal 200 bp of the hepcidin promoter is critical for the inflammatory, BMP, and hemojuvelin response.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This is manuscript number 19557-MEM from The Scripps Research Institute.
This work was supported by grants from the National Institutes of Health (DK53505-09), the Stein Endowment Fund, and the Skaggs Scholars in Clinical Science Program from The Scripps Research Institute.
National Institutes of Health
Authorship
Contribution: J.T. helped plan and execute the experiments and write the paper; and P.L. and E.B. helped design the studies and write the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pauline Lee, PhD, Department of Molecular and Experimental Medicine, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037; e-mail: plee@scripps.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal