Abstract
Individuals with sickle cell disease (SCD) have increased inflammation, a high incidence of airway hyperreactivity (AH), and increased circulating leukotrienes (LT). We show that expression of 5-lipoxygenase and 5-lipoxygenase activating protein (FLAP), key catalytic molecules in the LT pathway, were significantly increased in peripheral blood mononuclear cells (MNCs) in patients with SCD, compared with healthy controls. Placenta growth factor (PlGF), elaborated from erythroid cells, activated MNC and THP-1 monocytic cells to induce LT production. PlGF-mediated increased FLAP mRNA expression occurred via activation of phosphoinositide-3 (PI-3) kinase, nicotinamide adenine dinucleotide phosphate (NADPH) oxidase, and hypoxia inducible factor-1α (HIF-1α). HIF-1α small interfering RNA (siRNA) reduced PlGF-induced FLAP expression. FLAP promoter-driven luciferase constructs demonstrated that PlGF-mediated luciferase induction was abrogated upon mutation of HIF-1α response element (HRE), but not the nuclear factor-κB (NF-κB) site in the FLAP promoter; a finding confirmed by chromatin immunoprecipitation (ChIP) analysis. PlGF also increased HIF-1α binding to the HRE in the FLAP promoter. Therefore, it is likely that the intrinsically elevated levels of PlGF in SCD subjects contribute to increased LT, which in turn, mediate both inflammation and AH. Herein, we identify a mechanism of increased LT in SCD and show HIF-1α as a hypoxia-independent target of PlGF. These studies provide new avenues to ameliorate these complications.
Introduction
Inflammation is increasingly recognized as central to the pathophysiology of sickle cell disease (SCD) and is manifest as leukocytosis, elevated levels of inflammatory cytokines, and activation of neutrophils, monocytes, and endothelial cells.1-4 It is present at steady state and is strongly associated with acute painful events, acute chest, and early mortality.5,6 Current evidence strongly suggests that inflammation contributes to the endothelial cell dysfunction, potentiates vasoocclusion, and may also give rise to the airway hyperreactivity (AH) that often accompanies SCD.7-10 Also intriguing is the spectrum of lung disease seen in this patient population, which spans from an increased incidence of AH and obstructive lung disease in children,11-13 to restrictive lung disease and pulmonary vascular remodeling, which is associated with pulmonary hypertension in adults.14-18
Leukotrienes (LT) mediate both inflammation and AH.19-22 5-Lipoxygenase (5-LO) and its activating partner, 5-lipoxygenase activating protein (FLAP), catalyze the production of LT from arachidonic acid (AA) by generating 5-hydroperoxyeicostatraenoic acid (5-HPETE) and leukotriene A4 (LTA4). LTA4 is the pivotal intermediate from which other LTs (ie, LTB4 and cysteinyl LT [CysLT], LTC4, LTD4, and LTE4) are formed.20 LTB4 is one of the most potent chemoattractant for neutrophils, eosinophils, and mediator of inflammation. CysLT, on the other hand, are potent bronchoconstrictors that play an important role in edema, inflammation, and mucus secretion in asthma and were previously termed “slow releasing substances.”23 LT play an important role in the pathogenesis of inflammatory disorders, specifically asthma, rheumatoid arthritis, and inflammatory bowel disease.19-21 Studies by Bigby and coworkers24,25 have shown that both tumor necrosis factor-α (TNF-α) and lipopolysaccharide (LPS) induce the expression of FLAP in THP-1 cells. These studies showed the importance of nuclear factor-κB (NF-κB) and CCAAT/enhancer binding protein (C/EBP) transcription factors in the LPS-mediated FLAP expression.24
LTB4 levels are higher in SCD patients at steady state, which are further increased in vasoocclusive pain crises (VOC) and acute chest syndrome (ACS).26 Very recently, increased LTE4 has been observed in patients with SCD, which is associated with a higher incidence of pain.27 However, less is understood about how LTs are increased in SCD at the molecular level.
Placenta growth factor (PlGF) is an angiogenic growth factor with similar effects on endothelium as vascular endothelial growth factor (VEGF) and is primarily expressed by placental trophoblasts.28-30 More recently, we and others show that erythroid cells, but not other hematopoietic cells, produce PlGF, and its expression is high in SCD and thalassemia.31,32 VEGFR1 is its cognate receptor and is expressed on endothelial cells, alveolar epithelial cells, mast cells, and monocytes. We have previously shown that plasma levels of PlGF are high in SCD patients compared with control, which correlated well with SCD severity.31 Moreover, we showed that mononuclear cells (MNCs) of SCD patients were in an activated state as demonstrated by increased levels of cytochemokines, such as interleukin-1β (IL-1β), IL-8, monocyte chemoattractant protein-1 (MCP-1), and VEGF, compared with healthy controls.31 Treatment of MNC from healthy individuals with PlGF in vitro increased expression of the same cytochemokines as was seen in SCD, strongly suggesting that PlGF may contribute to increased cytochemokine expression from monocytes. The cytochemokines induced by PlGF are potent leukocyte activators and chemoattractants.31,33 Injection of a PlGF-adenovirus vector causes leukocytosis in mice.34 These data suggest PlGF may contribute to leukocyte activation and leukocytosis in vivo. Conceivably, increased leukocytosis and leukocyte activation in SCD could result from amplified levels of LT, resulting in AH. We hypothesized that the chronic inflammation2,35 and increased incidence of AH in patients with SCD could be explained by the activation of monocytes by PlGF to induce LT production.
In the present study, we show that MNCs from SCD subjects at steady state show significantly increased expression of 5-LO and FLAP mRNA, both key catalytic components of the LT pathway, compared with healthy controls. In addition, PlGF increased FLAP mRNA expression and LT production from peripheral blood monocytes (PBMs) and THP-1 monocytic cells. We find that PlGF activated phosphoinositide-3 (PI-3) kinase, nicotinamide adenine dinucleotide phosphate (NADPH) oxidase, and hypoxia-inducible factor-1α (HIF-1α) to increase FLAP expression. Our studies show for the first time, to the best of our knowledge, a hypoxia-independent role of HIF-1α in the regulation of FLAP, define some of the mechanisms behind the nonerythroid phenotypic presentations of SCD, and open new avenues for targeted therapeutic approaches.
Methods
Cells and reagents
All blood samples were obtained from children with homozygous SCD at steady state at their elective clinic appointment with routine clinical blood draws, using Institutional Review Board (IRB)–approved protocols at Cincinnati Children's Hospital and with informed consent obtained in accordance with the Declaration of Helsinki. PBM were isolated from ethylenediaminetetraacetic acid (EDTA) blood from healthy volunteers after obtaining informed consent according to a protocol approved by the IRB at University of Southern California, Los Angeles County (USC-LAC) Hospital. To ensure values represent true steady state, samples were obtained from patients who had no acute sickle events, fever, or infections 3 weeks before or 3 weeks after the blood draw and were not transfused within the last 90 days. Complete blood counts on the SCD patients revealed a white blood cell (WBC) count of 11 600 plus or minus 1400/μL, hemoglobin of 9.5 plus or minus 0.4 g/dL, and platelets 466 000 plus or minus 63 400/μL. The proportion of eosinophils in the WBC fraction and the absolute eosinophil count (340 ± 90; n = 9) were normal; and reticulocytes were elevated as expected (8.5% ± 1.7%). Complete blood counts were not performed on normal controls. Four of 9 patients were on hydroxyurea. MNC were isolated as described,31 resuspended in RPMI-1640 medium, and treated with PlGF (250 ng/mL) for 24 hours. THP-1 cells (ATCC, Manassas, VA) were cultured in RPMI containing 10% fetal bovine serum (FBS)33 and were placed in serum-free RPMI overnight before PlGF treatment.
Reagents were obtained as follows: PlGF (R&D Systems, Minneapolis, MN); LY294002, diphenyleneiodonium chloride (DPI), PD98059, SB203580, and SP600125 (Tocris Bioscience, Ellisville, MO); R59949 and PDTC (Calbiochem, Gibbstown, NJ); MK866 and U73122 (Biomol International, Plymouth Meeting, PA); 32P-UTP (MP Biomedicals, Solon, OH); antibodies against VEGFR1, HIF-1α, HIF-1β, FLAP, prolyl hydroxylase-2 (PHD-2), β-actin, and secondary antibodies conjugated to horseradish peroxidase (HRP; Santa Cruz Biotechnology, Santa Cruz, CA). The full-length −3368FLAP-pGL3 promoter construct and deletion constructs (−965FLAP-pGL3, −371FLAP-pGL3, and −134-FLAP-pGL3) were generously provided by Dr Timothy Bigby (VA Hospital San Diego, La Jolla, CA).25 The HIF-1α response element (HRE)-Luc (Dr Michael Kahn), phosphatase and tensin homolog (PTEN) plasmid (Dr Debbie Johnson), and HIF-1α and HIF-1β expression plasmids (Dr. Michael Stallcup) were obtained from investigators at USC Keck School of Medicine. HIF-1α small interfering RNA (siRNA), HIF-1α scrambled RNA (scRNA), PHD-2 siRNA, and PHD-2 scRNA36 were synthesized at the Microchemical Core facility of USC Norris Comprehensive Cancer Center. Oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA). All other reagents, unless otherwise specified, were purchased from Sigma-Aldrich (St Louis, MO).
mRNA analysis
Total RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA). Ribonuclease protection assay (RPA) was performed using a custom RiboQuant Multi-Probe template comprising 5-LO, FLAP, HIF-1α, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; BD Biosciences, San Diego, CA) as described.33,37 The intensity of the bands was analyzed using the Spot-Denso software on the Alpha Imager 2000 gel documentation system (Alpha Innotech, San Leandro, CA). Real-time polymerase chain reaction (qRT-PCR) was carried out using the iScript One-Step RT-PCR kit with SYBR Green (Bio-Rad Laboratories, Hercules, CA) as per the manufacturer's instructions on an ABI Prism 7900 (Applied Biosystems, Foster City, CA). Briefly, 40 cycles of amplification was carried out after reverse transcription at 95°C for 10 seconds and 60°C for 30 seconds, using primers listed in Table 1. Relative quantification (RQ) values of 5-LO and FLAP mRNA expression were calculated as 2−ΔΔCt by the comparative cycle threshold (Ct) method.38 Where ΔΔCt equals (Ct target gene of SCD sample − Ct GAPDH of SCD sample) minus (Ct target gene of control sample − Ct GAPDH of control sample).
Oligonucleotide primers used in this study
| Gene/fragment location (bp) . | Method . | Forward sequence (5′ to 3′) . | Reverse sequence (5′ to 3′) . |
|---|---|---|---|
| −371-FLAP promoter HRE-M1 (−170 to −167) | SDM | gtgtgctggctttgcaggctcctctgccaag | cttggcagaggagcctgcaaagccagcacac |
| −371-FLAP promoter HRE-M2 (−251 to −248) | SDM | gcctgtgctgggctcaggctggtcatggtc | gaccatgaccagcctgagcccagcacaggc |
| −371-FLAP promoter NFκB-M1 (−43 to −34) | SDM | ggcactgtgtaattgtgccgaggctctgcagaaattgtaatg | cattacaatttctgcagagcctcggcacaattacacagtgcc |
| −371-FLAP promoter NFκB-M2 (−43 to −34) | SDM | ggcactgtgtaattgtgccgggaagcgtcagaaattgtaatg | cattacaatttctgacgcttcccggcacaattacacagtgcc |
| HRE (−170 to −167) in FLAP promoter | EMSA | ctggctttgcgtgctcctctg | cagaggagcacgcaaagccag |
| HRE mutant in FLAP promoter | EMSA | ctggctttgaaagctcctctg | cagaggagctttcaaagccag |
| FLAP promoter (−310/+9) | ChIP | cagagatgatggcagcttcca | aaggggaagtgagagcttgca |
| Gene/fragment location (bp) . | Method . | Forward sequence (5′ to 3′) . | Reverse sequence (5′ to 3′) . |
|---|---|---|---|
| −371-FLAP promoter HRE-M1 (−170 to −167) | SDM | gtgtgctggctttgcaggctcctctgccaag | cttggcagaggagcctgcaaagccagcacac |
| −371-FLAP promoter HRE-M2 (−251 to −248) | SDM | gcctgtgctgggctcaggctggtcatggtc | gaccatgaccagcctgagcccagcacaggc |
| −371-FLAP promoter NFκB-M1 (−43 to −34) | SDM | ggcactgtgtaattgtgccgaggctctgcagaaattgtaatg | cattacaatttctgcagagcctcggcacaattacacagtgcc |
| −371-FLAP promoter NFκB-M2 (−43 to −34) | SDM | ggcactgtgtaattgtgccgggaagcgtcagaaattgtaatg | cattacaatttctgacgcttcccggcacaattacacagtgcc |
| HRE (−170 to −167) in FLAP promoter | EMSA | ctggctttgcgtgctcctctg | cagaggagcacgcaaagccag |
| HRE mutant in FLAP promoter | EMSA | ctggctttgaaagctcctctg | cagaggagctttcaaagccag |
| FLAP promoter (−310/+9) | ChIP | cagagatgatggcagcttcca | aaggggaagtgagagcttgca |
ChIP indicates chromatin immunoprecipitation; EMSA, electrophoretic mobility shift assay; PCR, polymerase chain reaction; and SDM, site-directed mutagenesis. The specific mutations are highlighted in bold type.
LT assay
THP-1/PBM (1.5 × 106 cells/mL) were treated with PlGF (250 ng/mL) for 24 hours, and supernatants were assayed for LT using the enzyme-linked immunosorbent assay (ELISA) kit (Cayman Chemical, Ann Arbor, MI).
Estimation of intracellular reactive oxygen species
Briefly, THP-1 (2 × 106 cells) were washed with phosphate-buffered saline (PBS), incubated with 5 μM 2′,7′-dichlorofluorescein-diacetate dye39 in RPMI at 37°C for 30 minutes in the dark, then washed 3 times with PBS to remove excess dye, and stimulated with PlGF for 4 hours. Fluorescence intensity was analyzed for 60 seconds using RF-551 spectrofluorometric detector (Shimadzu, Kyoto, Japan) with excitation at 488 nm and emission at 525 nm. THP-1 cells loaded with the dye for 30 minutes were used as a blank control.
Western blot analysis
The cytosolic and nuclear extracts were prepared from THP-1 cells as described.40 Briefly, 5 × 106 cells were washed and resuspended in 400 μL cell lysis buffer for 20 minutes. The homogenate was centrifuged at 10 000g for 30 seconds and cytosolic supernatant was collected. The nuclear extract was obtained by resuspending nuclear pellet in 50 μL nuclear extraction buffer on ice for 60 minutes. The cytosolic extracts were used to analyze FLAP and PHD-2 protein, while nuclear extracts were subjected to HIF-1α and HIF-1β analysis. The protein bands were detected with Immunobilon western reagents (Millipore, Billerica, MA).
The mutagenesis of human FLAP promoter
Mutant constructs of the human FLAP-Luc promoter were generated using wild-type −371-FLAP-Luc construct as a template by the QuikChange site-directed mutagenesis kit (Stratagene, Cedar Creek, TX), using the primers shown in Table 1. The double mutant of HIF-1α binding sites, represented as HRE-M1+2, was generated using HRE-M1 as a template. Mutations were confirmed by sequencing.
Transient transfection
THP-1 cells (106) were transfected with various siRNA constructs (50 nM) and luciferase reporter plasmids by nucleofection with the Nucleofector kit-V (Amaxa Biosystems, Cologne, Germany). The sense and antisense siRNA oligonucleotides were annealed as described.41 The β-galactosidase plasmid (0.5 μg) was cotransfected with reporter constructs (0.5 μg) to ascertain transfection efficiency. After transfection, the cells were kept in complete medium overnight, serum-free medium for 3 hours, and treated with PlGF for the indicated times. The cells were lysed and analyzed for luciferase activity and β-galactosidase activity using kits from Promega (Madison, WI). Luciferase values were normalized to β-galactosidase values. Data are expressed relative to the activity of the promoterless pGL3 basic vector.
Electrophoretic mobility shift assay for transcription factor HIF-1α
The single-stranded oligonucleotides (Table 1) were biotin-labeled using a Lightshift Chemiluminescent electrophoretic mobility shift assay (EMSA) kit (Pierce, Rockford, IL),42 and the complementary strands were annealed in equimolar ratios for 1 hour at 37°C. The DNA binding reaction included nuclear protein extract (5 μg), 5% glycerol, 5 mM MgCl2, 50 ng/μL poly(dI·dC), 0.05% Nonidet P-40 (NP-40), and 0.5 ng biotinylated probe and incubated at room temperature for 20 minutes. The specificity of DNA-protein interaction was demonstrated using 50-fold excess of unlabeled probe. For supershift assays, nuclear extracts were preincubated for 1 hour on ice with 2 μg HIF-1α antibody. The samples were then subjected to nondenaturing 6% polyacrylamide gel electrophoresis in 0.5 × Tris-borate-EDTA (TBE), transferred to a Hybond-N+ nylon membrane (Amersham Biosciences, Piscataway, NJ), followed by detection with streptavidin-HRP/chemiluminescence.
Chromatin immunoprecipitation assay
THP-1 (10 × 106 cells) were treated with PlGF in serum-free RPMI for the indicated time periods in the presence/absence of inhibitors. Chromatin immunoprecipitation (ChIP) analysis was performed using HIF-1α antibody as previously described.36 Briefly, formaldehyde-fixed cells were lysed, and chromatin was sheared by sonication (6 × 15 seconds, 40% potency). The lysates were precleared for 2 hours at 4°C with Protein A-Sepharose beads. Immunoprecipitation was carried out at 4°C overnight with HIF-1α antibody or control immunoglobulin G (IgG) antibody. Protein-A immune complexes were collected and washed sequentially with low-salt buffer, high-salt buffer, and Tris-EDTA (TE) buffer. DNA cross-links were reversed at 65°C overnight, and DNA was extracted by phenol/chloroform/isoamyl alcohol and then ethanol-precipitated. Immunoprecipitated DNA was subjected to PCR amplification for 30 cycles under the following conditions; 95°C for 30 seconds, 58°C for 60 seconds, and 72°C for 120 seconds, using primers shown in Table 1. The PCR products were subjected to agarose gel electrophoresis followed by densitometric analysis. The values were normalized to input DNA.
Statistical analysis
Control and PlGF-treated cells were compared by Student t test. One-way analysis of variance (ANOVA) followed by Turkey-Kramer test was used for multiple comparisons using the Instat-2 software program (GraphPad, San Diego, CA). Values of P less than .05 were considered statistically significant.
Results
MNC of SCD subjects express increased levels of 5-LO and FLAP mRNA at steady state
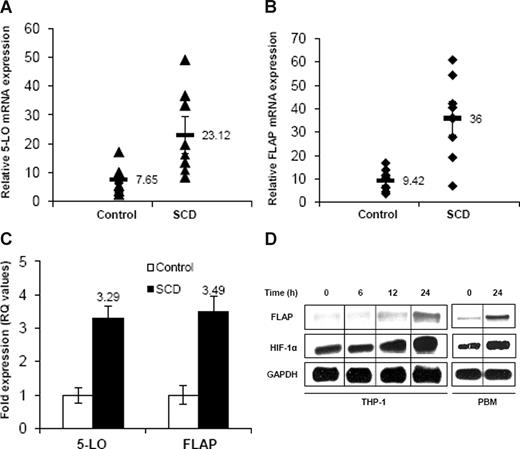
We first examined whether expression of 5-LO and FLAP, key regulatory enzymes in LT formation, were affected in MNC from SCD subjects. As shown in Figure 1A, there was approximately a 3-fold increase in 5-LO mRNA expression in SCD compared with healthy controls by qRT-PCR (ΔCt, 23.12 ± 6.49 vs 7.65 ± 1.5; n = 9; P < .05). In addition, there was a 3.8-fold increase in FLAP mRNA expression in SCD compared with healthy controls (ΔCt, 36.0 ± 7.25 vs 9.42 ± 2.12; n = 9; P < .013) as shown in Figure 1B. There was also approximately a 3-fold change in RQ values of 5-LO and FLAP mRNA expression in SCD compared with control subjects (Figure 1C). PlGF concentrations in the plasma of the same samples were significantly higher in SCD patients (24.7 ± 1.6 pg/mL; n = 9) than in control (13.7 ± 0.4; n = 9; P < .001), consistent with previously reported values.31
5-LO and FLAP mRNA expression in MNC, PBM, and THP-1. (A,B) qRT-PCR analysis of mRNA in MNC isolated from SCD subjects at steady state (n = 9) and healthy controls (n = 9). Each data point represents ΔCt values of 5-LO and FLAP expression from individual subjects. Mean values are represented as black bars. Error bars represent SEM. (C) ΔΔCt (RQ) values show fold expression of 5-LO and FLAP mRNA in SCD compared with healthy controls. (D) RPA analysis of FLAP, HIF-1α, and GAPDH in PlGF-treated THP-1 cells and PBM for the indicated time periods. Data are representative of 3 independent experiments. Where indicated, the vertical lines show repositioned gel lanes.
5-LO and FLAP mRNA expression in MNC, PBM, and THP-1. (A,B) qRT-PCR analysis of mRNA in MNC isolated from SCD subjects at steady state (n = 9) and healthy controls (n = 9). Each data point represents ΔCt values of 5-LO and FLAP expression from individual subjects. Mean values are represented as black bars. Error bars represent SEM. (C) ΔΔCt (RQ) values show fold expression of 5-LO and FLAP mRNA in SCD compared with healthy controls. (D) RPA analysis of FLAP, HIF-1α, and GAPDH in PlGF-treated THP-1 cells and PBM for the indicated time periods. Data are representative of 3 independent experiments. Where indicated, the vertical lines show repositioned gel lanes.
PlGF induced FLAP mRNA expression in THP-1 and PBM cells
THP-1 cells showed maximal increase in FLAP mRNA at 250 ng/mL, as previously observed for expression of cytochemokines,33 therefore 250 ng/mL PlGF were used in all further experiments (see Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). The FLAP mRNA expression peaked at 24 hours (Figure S2). Although this in vitro PlGF concentration used herein are similar to those previously reported,43,44 they are much higher than the estimated levels of plasma PlGF in SCD patients. However, these concentrations may not reflect the local concentrations of PlGF in the microenvironment to which the cells are exposed, as seen with other hematopoietic growth factors. Besides, PlGF has different splice variants, some with high basic residues with heparin/matrix binding activity; where it may be presented to cells directly at high local concentrations. The ELISA used to estimate plasma PlGF only detects free circulating PlGF and may underestimate the concentration presented to cells.
Next, we determined the time course of FLAP induction. As shown in Figure 1D, PlGF treatment of THP-1 cells resulted in a time-dependent (6, 12, and 24 hours) increase in mRNA expression of FLAP, as determined by RPA. There was 4.5-fold (458% ± 34%) increase in FLAP mRNA expression at 24 hours in THP-1. In addition, polymyxin B (5 μg/mL), an inhibitor of endotoxin, did not inhibit PlGF-induced FLAP mRNA expression (data not shown). Moreover, PlGF treatment of PBM from healthy controls showed approximately a 3.6-fold (365% ± 6%) increase in FLAP mRNA expression (Figure 1D). Because both THP-1 and PBM cells were responsive to PlGF in up-regulating FLAP expression, we used THP-1 monocytic cells as a model system for ease of culturing and transfection for further mechanistic studies.
PlGF causes release of LT from THP-1 cells and PBM
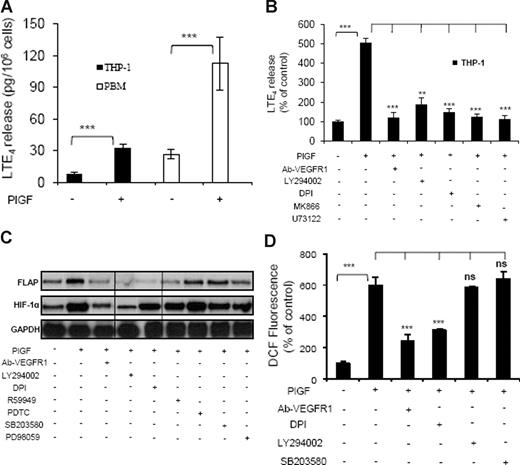
Treatment of THP-1 cells with PlGF for 24 hours resulted in a 4-fold (414% ± 13.4%) increase in the release of LTE4, as determined by ELISA (Figure 2A). However, these cells did not release LTB4, as has been previously observed.45 Moreover, pretreatment of THP-1 cells with an antibody to VEGFR1 or pharmacologic inhibitors for PI-3 kinase (LY294002), NADPH-oxidase (DPI), FLAP (MK866), and phospholipase C (PLC; U73122) reduced PlGF-mediated LTE4 release by 95% plus or minus 4%, 78% plus or minus 9%, 88% plus or minus 4%, 94% plus or minus 4%, and 97% plus or minus 5%, respectively (Figure 2B). LT agonists can release AA upon receptor activation, followed by cPLA2 activation within seconds. However, we did not observe an effect on LTE4 production at early time points (Figure S3). PlGF also caused a 4-fold (423% ± 22%) increase in the release of LTE4 from PBM (Figure 2A).
PlGF-induced LTE4 release, FLAP mRNA expression, and ROS formation. (A) PBM or THP-1 cells were treated with PlGF (250 ng/mL) for 24hours and (B) THP-1 cells were pretreated for 30 minutes with Ab-VEGFR1 (2 μg/mL), LY294002 (10 μM), DPI (10 μM), MK866 (10 μM), and U73122 (10 μM), followed by treatment with PlGF for 24 hours. The supernatants were collected and assayed for LTE4 release by ELISA. (C) RPA analysis of total RNA isolated from THP-1 cells pretreated for 30 minutes with Ab-VEGFR1, LY294002, DPI, R59949 (10 μM), PDTC (10 μM), SB203580 (10 μM), and PD98059 (10 μM) before PlGF treatment for 24 hours. (D) THP-1 cells were loaded with DCFH-DA dye for 30 minutes, incubated with indicated inhibitors for an additional 30 minutes and treated with PlGF for 4 hours. The cells were lysed, and the fluorescence was measured. Data are expressed as means plus or minus SEM of 3 independent experiments (***P < .001; **P < .01; ns, P > .05). Where indicated, the vertical lines show repositioned gel lanes.
PlGF-induced LTE4 release, FLAP mRNA expression, and ROS formation. (A) PBM or THP-1 cells were treated with PlGF (250 ng/mL) for 24hours and (B) THP-1 cells were pretreated for 30 minutes with Ab-VEGFR1 (2 μg/mL), LY294002 (10 μM), DPI (10 μM), MK866 (10 μM), and U73122 (10 μM), followed by treatment with PlGF for 24 hours. The supernatants were collected and assayed for LTE4 release by ELISA. (C) RPA analysis of total RNA isolated from THP-1 cells pretreated for 30 minutes with Ab-VEGFR1, LY294002, DPI, R59949 (10 μM), PDTC (10 μM), SB203580 (10 μM), and PD98059 (10 μM) before PlGF treatment for 24 hours. (D) THP-1 cells were loaded with DCFH-DA dye for 30 minutes, incubated with indicated inhibitors for an additional 30 minutes and treated with PlGF for 4 hours. The cells were lysed, and the fluorescence was measured. Data are expressed as means plus or minus SEM of 3 independent experiments (***P < .001; **P < .01; ns, P > .05). Where indicated, the vertical lines show repositioned gel lanes.
PlGF-induced FLAP expression involves activation of PI-3 kinase, NADPH oxidase, and HIF-1α
As shown in Figure 2C, PlGF-induced FLAP mRNA expression was inhibited by VEGFR1 antibody (93% ± 4%), LY294002 (75% ± 3%), DPI (98% ± 4%), and R59949 (84% ± 7%), the latter being a putative inhibitor of HIF-1α. However, inhibitors of NF-κB (PDTC), p38MAP kinase (SB203580), and mitogen-activated protein (MAP) kinase (PD98059) did not inhibit PlGF-induced FLAP expression. Because DPI inhibited FLAP expression, we determined whether PlGF increased the formation of reactive oxygen species (ROS) in THP-1 cells. As shown in Figure 2D, PlGF caused a 6-fold (598% ± 54%) increase in ROS formation, which was attenuated by VEGFR1 antibody (71% ± 9%) and DPI (57% ± 2%), while LY294002 and SB203580 did not affect ROS formation. These results indicate that PlGF binding to VEGFR1 causes activation of NADPH oxidase to generate ROS, which is independent of the PI-3 kinase pathway.
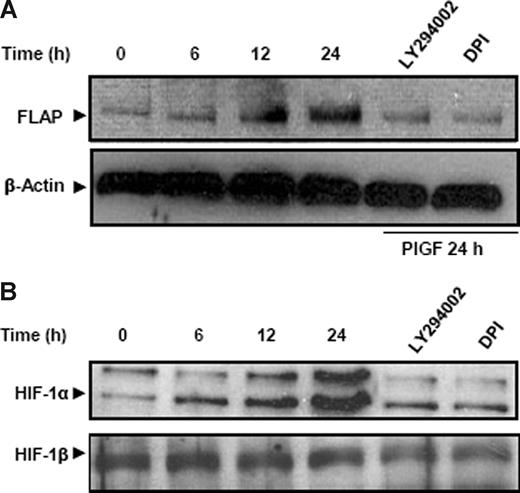
We wanted to determine whether a similar effect of PlGF can be seen on the FLAP protein levels. As shown in Figure 3A, PlGF caused a time-dependent (6-24 hours) increase in FLAP protein expression. There was a 2.6-fold (266% ± 9%) increase in FLAP protein at 24 hours, which was significantly reduced by LY294002 and DPI by approximately 80%. Taken together, PlGF-mediated FLAP expression involves activation of PI-3 kinase and NADPH oxidase.
PlGF increases FLAP and HIF-1α protein in THP-1 cells. THP-1 cells were pretreated for 30 minutes with LY294002 and DPI followed by PlGF treatment for indicated time periods. (A) Cytosolic proteins were subjected to western blot analysis using antibody to FLAP. The same membrane was reprobed with β-actin antibody to normalize protein loading. (B) Nuclear extracts were subjected to western blot analysis using antibody to HIF-1α. The same membrane was reprobed with HIF-1β antibody to normalize the protein loading. Proteins were visualized by enhanced chemiluminescence corresponding to their expected molecular weights: FLAP (18 kDa), β-actin (42 kDa), HIF-1α (120 kDa), and HIF-1β (95 kDa). Data are representative of 3 independent experiments.
PlGF increases FLAP and HIF-1α protein in THP-1 cells. THP-1 cells were pretreated for 30 minutes with LY294002 and DPI followed by PlGF treatment for indicated time periods. (A) Cytosolic proteins were subjected to western blot analysis using antibody to FLAP. The same membrane was reprobed with β-actin antibody to normalize protein loading. (B) Nuclear extracts were subjected to western blot analysis using antibody to HIF-1α. The same membrane was reprobed with HIF-1β antibody to normalize the protein loading. Proteins were visualized by enhanced chemiluminescence corresponding to their expected molecular weights: FLAP (18 kDa), β-actin (42 kDa), HIF-1α (120 kDa), and HIF-1β (95 kDa). Data are representative of 3 independent experiments.
PlGF-induced HIF-1α mRNA and protein expression
It is well established that hypoxia increases HIF-1α protein levels but not HIF-1α mRNA.46 In contrast, we observed that PlGF treatment of THP-1 cells increased HIF-1α mRNA expression in a time-dependent manner (6-24 hours; Figure 1D). There was a 3-fold (299% ± 8%) and a 2.5-fold (250% ± 5%) increase in HIF-1α mRNA levels in THP-1 and PBM, respectively, at 24 hours. As shown in Figure 2C, both VEGFR1 antibody and LY294002 reduced PlGF-induced HIF-1α mRNA expression by approximately 80%, while DPI, R59949, PDTC, SB203580, and PD98059 did not affect HIF-1α mRNA levels. Although the ROS inhibitor, DPI, attenuated FLAP mRNA expression (Figure 2C lane 5), it did not reduce HIF-1α mRNA; therefore, we determined whether ROS exerted its effect on HIF-1α protein levels. As shown in Figure 3B, PlGF led to a time-dependent (6-24 hours) increase in HIF-1α protein levels in nuclear extracts of THP-1 cells, which was attenuated by LY294002 and DPI by approximately 80% at 24 hours. It is pertinent to note that HIF-1β, a constitutive protein, remained unchanged, showing a specific induction of HIF-1α in response to PlGF (Figure 3B). These results indicate that PlGF-mediated activation of PI-3 kinase leads to increases in both HIF-1α mRNA and protein, while ROS formation through NADPH oxidase activation contributes to HIF-1α protein stabilization.
PlGF-mediated FLAP mRNA expression and LTE4 release requires HIF-1α
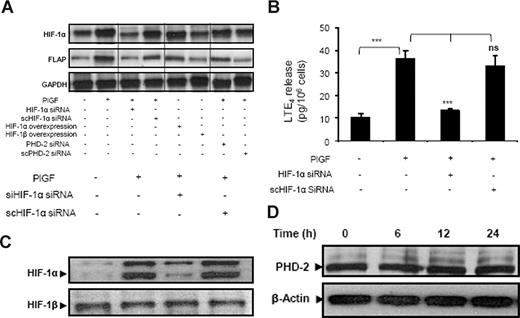
PlGF treatment of THP-1 cells transfected with HIF-1α siRNA resulted in reduction of both HIF-1α (87% ± 5%) and FLAP (78% ± 3%) mRNA expression (Figure 4A lane 3). However, scrambled (sc)HIF-1α siRNA did not affect HIF-1α and FLAP mRNA levels (Figure 4A lane 4). In addition, HIF-1α siRNA attenuated PlGF-mediated LTE4 release by 88% (± 4%), while scHIF-1α siRNA had no effect (Figure 4B). It is pertinent to note that transfection with HIF-1α siRNA, but not scHIF-1α siRNA, reduced HIF-1α protein levels (Figure 4C). Furthermore, overexpression of HIF-1α led to a 3-fold increase in FLAP mRNA in the absence of PlGF treatment (Figure 4A lane 5). However, HIF-1β overexpression did not change FLAP mRNA expression (Figure 4A lane 6). Moreover, PHD-2 siRNA increased HIF-1α protein by inhibiting its degradation, resulting in a 4-fold (395% ± 8%) increase of FLAP mRNA expression in response to PlGF (Figure 4A lane 7) compared with cells transfected with scPHD-2 siRNA (102% ± 8%; Figure 4A lane 8), supporting the role of HIF-1α in FLAP expression. Recent studies have shown that transforming growth factor-β1 (TGF-β1) induces HIF-1α stabilization through selective inhibition of PHD-2 protein levels in HepG2 cells.47 Therefore, we analyzed the effect of PlGF on PHD-2 protein levels. As shown in Figure 4D, PlGF treatment of THP-1 cells for 6, 12, and 24 hours did not change PHD-2 protein levels. Taken together, these data indicate that PlGF-mediated FLAP expression and LTE4 release involves HIF-1α.
PlGF-mediated FLAP and LTE4 expression involves HIF-1α. THP-1 cells were transfected with indicated siRNA constructs or expression plasmids, followed by PlGF treatment for 24 hours. (A) RPA, (B) LTE4 release, and (C) Western blot analysis of nuclear extract using HIF-1α antibody. (D) Western blot analysis of cytosolic extracts from THP-1 cells treated with PlGF for indicated time period (6-24 hours) using PHD-2 antibody. Data are representative of 3 independent experiments. Data are expressed as means plus or minus SEM of 3 independent experiments (***P < .001; **P < .01; ns, P > .05). Where indicated, the vertical lines show repositioned gel lanes.
PlGF-mediated FLAP and LTE4 expression involves HIF-1α. THP-1 cells were transfected with indicated siRNA constructs or expression plasmids, followed by PlGF treatment for 24 hours. (A) RPA, (B) LTE4 release, and (C) Western blot analysis of nuclear extract using HIF-1α antibody. (D) Western blot analysis of cytosolic extracts from THP-1 cells treated with PlGF for indicated time period (6-24 hours) using PHD-2 antibody. Data are representative of 3 independent experiments. Data are expressed as means plus or minus SEM of 3 independent experiments (***P < .001; **P < .01; ns, P > .05). Where indicated, the vertical lines show repositioned gel lanes.
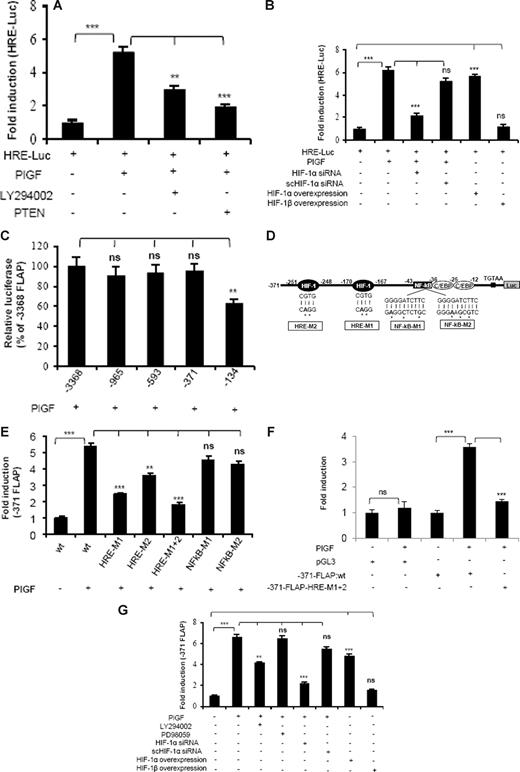
PlGF augments HRE promoter activity
Because PlGF-mediated FLAP expression was attenuated by HIF-1α siRNA, we investigated whether this occurred via HREs, which are present in promoter regions of several genes regulated by HIF-1α. As shown in Figure 5A, PlGF caused a 5-fold increase in HRE-Luc activity, which was inhibited by LY294002 (53% ± 5%) and transfection with PTEN (79% ± 4%). In addition, transfection with HIF-1α siRNA reduced HRE-Luc activity by 77% plus or minus 3%, while scHIF-1α siRNA had no effect (Figure 5B). Moreover, HIF-1α overexpression showed a 5.5-fold increase in HRE-Luc activity in the absence of PlGF. In contrast, overexpression of HIF-1β did not change HRE-Luc activity. These data show that PlGF-induced HIF-1α expression can activate HRE.
PlGF augments HRE-Luc and FLAP-Luc promoter via activation of PI-3 kinase and HIF-1α. THP-1 cells cotransfected with HRE-Luc and β-galactosidase plasmid were (A) either pretreated with LY294002 or cotransfected with PTEN and (B) cotransfected with indicated plasmids before treatment with PlGF for 24 hours. (C) Deletion analysis of FLAP promoter. THP-1 cells were cotransfected with indicated deletion construct and β-galactosidase plasmid, followed by PlGF treatment for 24 hours. (D) Schematics of FLAP promoter (−371 bp) indicating the location of HIF-1α, NF-κB, and C/EBP binding sites. (E,F) PlGF augments minimal FLAP promoter activity through HREs but not NF-κB in THP-1 (E) and PBM (F). THP-1 cells or PBM were cotransfected with indicated promoter constructs and β-galactosidase plasmid, followed by PlGF treatment for 24 hours. (G) THP-1 cells were treated with either indicated pharmacologic inhibitors or transfected with siRNA or HIF expression plasmids. These cells were then cotransfected with −371-FLAP-Luc and β-galactosidase plasmid, followed by PlGF treatment for 24 hours. Luciferase and β-galactosidase activities were measured as described in “Transient transfection.” The luciferase activity was normalized to that of the promoterless pGL3 basic vector. Data are expressed as mean plus or minus SEM of 3 independent experiments (***P < .001; **P < .01; ns, P > .05).
PlGF augments HRE-Luc and FLAP-Luc promoter via activation of PI-3 kinase and HIF-1α. THP-1 cells cotransfected with HRE-Luc and β-galactosidase plasmid were (A) either pretreated with LY294002 or cotransfected with PTEN and (B) cotransfected with indicated plasmids before treatment with PlGF for 24 hours. (C) Deletion analysis of FLAP promoter. THP-1 cells were cotransfected with indicated deletion construct and β-galactosidase plasmid, followed by PlGF treatment for 24 hours. (D) Schematics of FLAP promoter (−371 bp) indicating the location of HIF-1α, NF-κB, and C/EBP binding sites. (E,F) PlGF augments minimal FLAP promoter activity through HREs but not NF-κB in THP-1 (E) and PBM (F). THP-1 cells or PBM were cotransfected with indicated promoter constructs and β-galactosidase plasmid, followed by PlGF treatment for 24 hours. (G) THP-1 cells were treated with either indicated pharmacologic inhibitors or transfected with siRNA or HIF expression plasmids. These cells were then cotransfected with −371-FLAP-Luc and β-galactosidase plasmid, followed by PlGF treatment for 24 hours. Luciferase and β-galactosidase activities were measured as described in “Transient transfection.” The luciferase activity was normalized to that of the promoterless pGL3 basic vector. Data are expressed as mean plus or minus SEM of 3 independent experiments (***P < .001; **P < .01; ns, P > .05).
PlGF-mediated FLAP promoter activity requires HRE but not NF-κB binding site
Bigby and coworkers have shown that LPS-induced FLAP expression in THP-1 cells required the binding of both NF-κB and C/EBP to its promoter.24 As shown in Figure 5C, PlGF increased FLAP promoter activity by 5-fold (designated as 100% compared with the promoterless pGL3 vector). Analysis of serial deletion constructs of FLAP promoter showed that −371/+12-bp region of the FLAP promoter had similar activity as the full-length FLAP promoter, but −134/+12-bp promoter construct (lacking the HRE sites) showed reduced activity in response to PlGF (Figure 5C). Thus, we used −371/+12-bp construct for further studies. This region of the FLAP promoter (Figure 5D) contains 2 putative consensus HRE (RCGTG) at −170 to −167 bp and −251 to −248 bp, one NF-κB binding site located at −43 to −34 bp and 2 C/EBP consensus sites located at −36 to −28 bp and −25 to −12 bp, relative to transcriptional start site. We generated HRE mutant constructs of FLAP promoter, designated as HRE-M1 and HRE-M2 as indicated in Figure 5D. In addition, we used HRE-M1 as a template to generate a construct having mutations in both HRE sites designated as HRE-M1+2. As shown in Figure 5E, tranfections with HRE-M1 and HRE-M2 showed reduced promoter activation by 66% plus or minus 2% and 40% plus or minus 3%, respectively. However, HRE-M1+2 showed significantly higher inhibition (81% ± 3%) in response to PlGF stimulation. In contrast, 2 different mutations of NF-κB site (NFkB-M1 and NFkB-M2) did not reduce PlGF-mediated FLAP promoter activity. The role of HIF-1α in regulating FLAP promoter activity was also confirmed in PBM by using HRE-M1+2 mutant construct of FLAP promoter. As shown in Figure 5F, PBM transfected with −371-FLAP promoter showed a 3.5-fold induction, whereas HRE-M1+2 mutant FLAP promoter resulted in significant attenuation of promoter activity. It is pertinent to note that FLAP promoter construct (−134/+12 bp), lacking HRE sites, showed only a 50% reduction in PlGF-mediated luciferase activity compared with a −371/+12-bp construct (Figure 5C), indicating a possible involvement of C/EBP sites in regulating FLAP promoter activity.
Next, we examined the role of PI-3 kinase and HIF-1α in FLAP promoter activity. As shown in Figure 5G, PlGF-induced −371-FLAP promoter activity was reduced by LY294002 (44% ± 3%) but not by PD98059. Moreover, HIF-1α siRNA inhibited promoter activity by more than 80%, while scHIF-1α siRNA had no effect. In addition, overexpression of HIF-1α, but not HIF-1β, resulted in a 5-fold increase in FLAP promoter activity, independent of PlGF stimulation. Taken together, these results suggest that PlGF-mediated FLAP promoter activation requires both HRE sites, but not NF-κB site, located within the first 371 bp of the FLAP promoter.
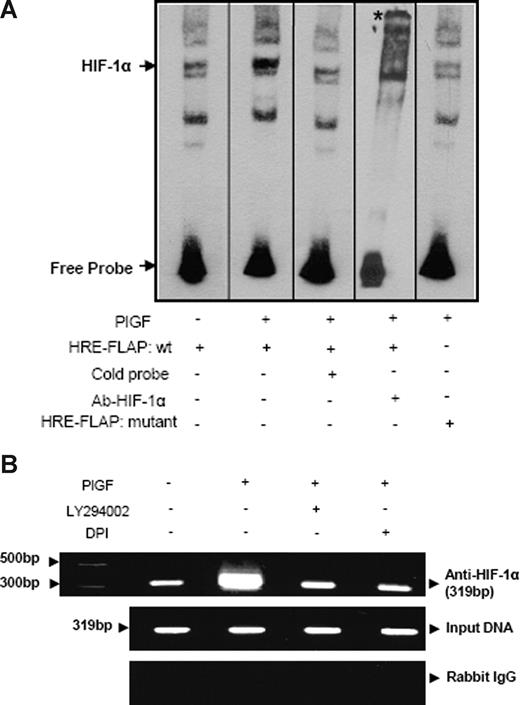
PlGF induces HIF-1α binding in vitro (EMSA) and in vivo (ChIP) to FLAP promoter
To further substantiate whether HIF-1α binds to HRE in the FLAP promoter, we used both wild-type and mutant oligonucleotides flanking the HRE site corresponding to the −170 to −167-bp region as probes for EMSA (Table 1). As shown in Figure 6A, nuclear extracts of PlGF-treated cells showed increased HIF-1α DNA binding compared with untreated cells. Moreover, 50-fold excess unlabeled probe competed out HIF-1α DNA binding (Figure 6A lane 3). In addition, HIF-1α antibody supershifted the band indicating the specificity of HIF-1α DNA binding (Figure 6A lane 4). Furthermore, mutant HRE oligonucleotide showed negligible HIF-1α DNA binding (Figure 6A lane 5) compared with wild-type probe (Figure 6A lane 2) in nuclear extracts of PlGF-treated cells. These results were also confirmed by ChIP analysis showing HIF-1α binding to the FLAP promoter in the native chromatin of THP-1 cells. PlGF-treated cells showed a 3-fold increase in expected PCR product size of 319 bp, corresponding to the FLAP promoter region (−310 to +9 bp) containing 2 HRE sites (Figure 6B), using primers listed in Table 1. Pretreatment with both LY294002 and DPI reduced the expected PCR product by approximately 80%. As shown in Figure 6B, middle panel, the amplification of input DNA before immunoprecipitation was equal in all samples. Immunoprecipitation of chromatin samples with control rabbit IgG did not show any amplification (Figure 6B bottom panel). These data indicate that PlGF increases HIF-1α binding to FLAP promoter to up-regulate the expression of FLAP in vivo.
PlGF augments HIF-1α binding to FLAP promoter in vitro (EMSA) and in vivo (ChIP). (A) Nuclear extracts from THP-1 cells (10 μg) were incubated with a biotinylated double-stranded oligonucleotide corresponding to the region (−179 to −159 bp) of the FLAP promoter containing the proximal HRE located at −170 to −167 bp. Where indicated, 50-fold excess of unlabeled wild-type probe (lane 3) or antibody to HIF-1α (lane 4) was added. EMSA analysis was also performed with a probe containing a mutation of the HRE (−170 bp to −167 bp, lane 5). * denotes supershifted band. Data are representative of 2 independent experiments. (B) THP-1 cells were pretreated with indicated pharmacologic inhibitors before PlGF stimulation for 4 hours. The soluble chromatin was isolated and immunoprecipitated with either HIF-1α antibody (top panel) or control rabbit IgG (bottom panel). The primers used to amplify the products flanking HIF-1α binding sites in the FLAP promoter are indicated in Table 1. The middle panel represents the amplification of input DNA before immunoprecipitation. Data are representative of 2 independent experiments. Where indicated, the vertical lines show repositioned gel lanes.
PlGF augments HIF-1α binding to FLAP promoter in vitro (EMSA) and in vivo (ChIP). (A) Nuclear extracts from THP-1 cells (10 μg) were incubated with a biotinylated double-stranded oligonucleotide corresponding to the region (−179 to −159 bp) of the FLAP promoter containing the proximal HRE located at −170 to −167 bp. Where indicated, 50-fold excess of unlabeled wild-type probe (lane 3) or antibody to HIF-1α (lane 4) was added. EMSA analysis was also performed with a probe containing a mutation of the HRE (−170 bp to −167 bp, lane 5). * denotes supershifted band. Data are representative of 2 independent experiments. (B) THP-1 cells were pretreated with indicated pharmacologic inhibitors before PlGF stimulation for 4 hours. The soluble chromatin was isolated and immunoprecipitated with either HIF-1α antibody (top panel) or control rabbit IgG (bottom panel). The primers used to amplify the products flanking HIF-1α binding sites in the FLAP promoter are indicated in Table 1. The middle panel represents the amplification of input DNA before immunoprecipitation. Data are representative of 2 independent experiments. Where indicated, the vertical lines show repositioned gel lanes.
Discussion
In this report, we showed that SCD patients at steady state expressed increased levels of 5-LO and FLAP mRNA in MNC compared with healthy controls. Our studies further showed that PlGF up-regulated FLAP mRNA to generate LT from PBM and THP-1 cells. A salient feature of SCD is inflammation, which manifests as leukocytosis that occurs in the absence of acute infection or inflammation.5,6 It has been a dilemma to understand how a genetic defect in sickle red blood cells (RBCs) would result in leukocytosis and leukocyte activation at baseline.2,7,31 Moreover, this leukocytosis correlates well with severity of disease and early mortality in 2 large studies,5,6 although a recent study did not find relationship.48 Also seen is increased AH in children9,11,49 and infants50 in SCD. Moreover, LTB426,51 and LTE427 levels are found to be increased in SCD at baseline, which further increased during episodes of VOC and ACS.26 Herein, we determined whether PlGF played a role in activation of monocytes to generate increased levels of LT, which would contribute to inflammation and asthma in SCD.
Styles and colleagues have shown increased levels of secretory phospholipase A2 (sPLA2) in association with development of ACS in SCD.52 sPLA2 releases AA from membrane phospholipids and has been implicated in the ACS.52,53 Increased AA can be acted upon by 5-LO and FLAP, resulting in production of LTB4 and CysLT. Because PlGF levels are highly elevated in ACS in SCD,31 sickle monocytes are activated to generate cytochemokines,31,33 and PlGF may play a role in ACS by contributing to both inflammation and AH.
Next, we examined the cellular signaling pathway for PlGF-mediated increased FLAP expression and LT release. We observed that PlGF-mediated FLAP expression and LTE4 release was attenuated by antibody to VEGFR1 and inhibition of PI-3 kinase, NADPH oxidase, and HIF-1α. PlGF-mediated LTE4 release was also inhibited by inhibitors of FLAP (MK866) and PLC (U73122). Because U73122 inhibited the LTE4 release, we examined if PlGF resulted in LTE4 release at early time points and found no effect (data not shown). Thus, it is possible that U73122 inhibits LTE4 release at 24 hours by inhibiting 5-LO, as has been previously reported.54 PlGF-mediated increase in HIF-1α mRNA occurred via activation of PI-3 kinase pathway, but not the NADPH oxidase pathway. In contrast, activation of the NADPH oxidase pathway increased HIF-1α protein levels, indicating that HIF-1α protein is stabilized by ROS pathway, as has been previously noted.55,56 However, the mechanism(s) by which PlGF stabilizes HIF-1α protein in a hypoxia-independent manner remains unknown. Recent studies have shown that TGF-β1 induces HIF-1α stabilization through selective inhibition of PHD-2,47 while nitric oxide-mediated HIF-1α degradation involves PHD-2 activation,57 indicating the role of PHD-2 in HIF-1α stabilization. However, we did not observe any change in PHD-2 protein levels in response to PlGF, suggesting that PlGF-mediated ROS generation could conceivably inhibit PHD-2 enzyme activity by modifying the levels of various intracellular cofactors such as ascorbate, Fe+2, and succinate, and thus stabilize HIF-1α.58,59
Previous studies have shown that LPS-induced FLAP expression in THP-1 cells required the binding of both NF-κB and C/EBP to its promoter,24 and deletion analysis of the full-length FLAP promoter (−3368/+12 bp; accession # 60470)60 revealed that the first 134 bp of the promoter (−134/+12 bp) were sufficient for TNF-α and LPS-induced FLAP promoter activity. Our studies of deletion constructs of the FLAP promoter showed that the −371/+12-bp region was equally effective as the full-length FLAP promoter in mediating the response to PlGF. Moreover, this effect was mediated via 2 HRE sites, unlike previously characterized NF-κB site in the FLAP promoter. Because mutations of HRE sites in the proximal promoter of FLAP (−371/+12 bp) showed residual promoter activity, we cannot rule out the activation of the C/EBP sites in the FLAP promoter by PlGF.
We confirmed our results by different approaches; we show that PlGF causes a 5-fold increase in HRE-Luc activity, which was abrogated by HIF-1α siRNA. Mutation of either HRE sites in FLAP promoter (HRE-M1 and HRE-M2) attenuated PlGF-mediated FLAP expression. In addition, mutation of both HRE (HRE-M1+2) resulted in a marked reduction in FLAP promoter activity. However, mutations of the NF-κB site in the promoter had no significant effect. We also show that silencing with HIF-1α siRNA attenuated PlGF-mediated FLAP promoter activation and downstream gene expression. Conversely, overexpression of HIF-1α, but not HIF-1β, augmented FLAP promoter activity and mRNA expression in the absence of PlGF. Overall, our results suggest that PlGF-mediated FLAP promoter activation specifically required HRE, but not the NF-κB sites. EMSA analysis showed increased HIF-1α binding to HRE in the FLAP promoter, in response to PlGF. In addition, ChIP analysis confirmed the HIF-1α binding to FLAP promoter to up-regulate FLAP expression in vivo.
Notably, PlGF may have pleiotropic effect through multiple pathways in SCD, besides increasing LT. We have shown that PlGF increases cytochemokine release from MNC,31,33 which likely contributes to inflammation. This is supported by the evidence that PlGF knockout mice display blunted inflammatory response.61,62 We recently showed that PlGF induces expression of endothelin-1 from endothelial cells and endothelin-B receptor in monocytes,42 molecules that likely contribute to the pulmonary artery hypertension seen in SCD. Notably, CysLT generated by platelet-leukocyte interactions can also stimulate endothelial cells to release von Willebrand factor, which may further potentiate vascular occlusion.63 Besides their effect on airways and smooth muscle, LT also promote adhesion of neutrophils to endothelial cells, and the neutrophil-derived LTA4 may contribute to the pool of LT.64 Thus, LT may contribute to vascular occlusion and reactive airway disease in SCD. Recently, sickle mice have been shown to have an exaggerated propensity to experimentally induced asthma.65 These results would be important to confirm in vivo. We are currently investigating the role of PlGF and the 5-LO pathway in inflammation, acute vascular occlusions, ACS, and AH in a prospective clinical study; and confirming the direct effects of PlGF in mouse models that simulate the chronic PlGF overexpression in SCD.
In conclusion, our studies show that PlGF, intrinsically released from erythroid cells caused activation of leukocytes, particularly monocytes, to generate increased levels of LT. We showed that the binding of PlGF to VEGFR1 led to activation of PI-3 kinase, NADPH oxidase, and HIF-1α. These data provide a novel mechanism of PlGF-induced FLAP expression, which involves activation of HIF-1α in a manner that is independent of hypoxia. Thus, PlGF may provide a link between increased formation of LT, inflammation, vasoocclusion, and AH seen in SCD subjects. Antagonists of the LT pathway, such as LT receptor antagonists, or zileuton, which are currently licensed for asthma,66 could be used to ameliorate inflammation and AH in SCD patients. It is pertinent to mention that zileuton is structurally similar to hydroxyurea, has been shown to increase hemoglobin F production in primary erythroid cells in vitro,67 and may have clinical utility in the treatment of SCD.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Timothy Bigby (VA Hospital San Diego, La Jolla, CA) for kindly providing FLAP promoter constructs. We thank Institutional Core of USC Research Center for Liver Disease for the use of the spectrofluorometer and sequence detection instrument (NIH-P30-DK 048522).
This work was supported by National Institutes of Health (NIH) grant HL-070595 (CSCC) and R01-HL-079916.
National Institutes of Health
Authorship
Contribution: N.P. performed most of the experiments, contributed to experimental design, analyzed the data, and contributed to the writing of the manuscript; C.G. performed experiments on LT release; M.Y. obtained patient samples, isolated MNC for quantitative RT-PCR; P.M. contributed to study design and hypothesis, obtained sickle samples, and wrote the manuscript; and V.K. contributed to the hypothesis, study design and experiments, and preparation and writing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Vijay Kalra, HMR 611, Department of Biochemistry, and Molecular Biology, USC Keck School of Medicine, Los Angeles, CA 90033; e-mail: vkalra@usc.edu; or Punam Malik, Division of Experimental Hematology, Cincinnati Children's Hospital Medical Center, MLC 7013, 3333 Burnet Avenue, Cincinnati, OH 45229-3039; e-mail: Punam.Malik@cchmc.org.
References
Author notes
*P.M. and V.K.K. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal