Abstract
In vertebrates, molecular mechanisms dictate angioblasts' migration and subsequent differentiation into arteries and veins. In this study, we used a microarray screen to identify a novel member of the sucrose nonfermenting related kinase (snrk-1) family of serine/threonine kinases expressed specifically in the embryonic zebrafish vasculature and investigated its function in vivo. Using gain- and loss-of-function studies in vivo, we show that Snrk-1 plays an essential role in the migration, maintenance, and differentiation of angioblasts. The kinase function of Snrk-1 is critical for migration and maintenance, but not for the differentiation of angioblasts. In vitro, snrk-1 knockdown endothelial cells show only defects in migration. The snrk-1 gene acts downstream or parallel to notch and upstream of gridlock during artery-vein specification, and the human gene compensates for zebrafish snrk-1 knockdown, suggesting evolutionary conservation of function.
Introduction
In vertebrates, the circulatory system is one of the first networks developed by precise orchestration of signaling pathways emanating from genes and their products.1 Angioblasts arise from lateral plate mesoderm (LPM) cells and contribute to the formation of the first blood vessels by vasculogenesis and angiogenesis.2 Despite an increasing amount of knowledge on how blood vessels are formed, there is a fundamental gap in understanding how the angioblast, a presumptive dual-potential progenitor cell, gives rise to differentiated endothelial cells (ECs). Recently, an ets domain-containing related protein (Etsrp) was shown to be necessary and sufficient for angioblast specification.3 However, once an angioblast is specified, it is unclear what signals dictate its migration to the midline, maintain its proliferative state, and regulate its eventual differentiation to arterial or venous endothelial lineages
In this study, we used a microarray screen for differentially regulated genes in vascular development to identify a member of the sucrose nonfermenting kinase family (snrk-1) that acts upstream of grl and parallel or downstream of notch. Whole-mount in situ hybridization shows that snrk-1 is expressed ubiquitously until 20 hours postfertilization (hpf) when expression becomes exclusive to the dorsal aorta (DA) and posterior cardinal vein (PCV). Gene knockdown (KD) analyses with 2 different morpholinos show that snrk-1 affects etsrp+ angioblast migration and is required for maintenance of these cells in the LPM. In gain-of-function (GOF) experiments, snrk-1 mRNA ectopically induces etsrp+ angioblasts in the LPM at 6 somite (som), which complements loss-of-function (LOF) analyses in which knockdown of snrk-1 results in fewer etsrp+ cells. A point mutation in Snrk-1 (P142A) in the Ser/Thr kinase domain (Snrk-1 KM) results in a hypoactive kinase, and snrk-1 KM–injected GOF embryos show fewer angioblasts, which migrate prematurely from the LPM to the midline. Migration defects in vivo are recapitulated in snrk-1 siRNA KD ECs in vitro. In addition to angioblast migration and maintenance, Snrk-1 also plays an important role in A/V specification. Wild-type snrk-1 and the point mutant both induce arterial and repress venous marker expression. Snrk-1 GOF embryos show high grl expression, suggesting that snrk-1 works upstream of grl in A/V specification. In addition, notch induction of arterial markers and repression of venous markers are abolished in snrk-1 KD embryos, suggesting that snrk-1 acts downstream or parallel of notch in A/V specification. Finally, snrk-1 MO–induced vascular defects are rescued by introducing the human snrk-1 gene, demonstrating orthology and evolutionarily conserved function. Together, these results suggest that snrk-1 plays a critical role in the early development of angioblasts and their differentiation into arteries and veins.
Methods
Zebrafish stocks
Zebrafish were grown and maintained at 28.5°C.4 All procedures were performed according to Medical College of Wisconsin animal protocol guidelines (ASP no. 312-06-2). Mating was routinely carried out at 28.5°C and embryos were staged according to established protocols.5 WT fish used in this study include TL, AB, and TuAB strains.
Molecular biology
Details are provided in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Morpholino reagents and microinjections
Based on the ATG and intron-exon boundary sequences, snrk-1 morpholino phosphorodiamidate oligonucleotides were designed by Gene Tools (Philomath, OR). The morpholino sequences are as follows: MO1 (splice MO): CAGATGGTATACATACTCTTTGCAC; MO2 (ATG-MO): GTAACCGCTTTTGGTGGCAGCCATT. Microinjections of 1-cell stage zebrafish embryos with RNA or morpholinos (MOs) were carried out as described before.4 MOs were reconstituted in nuclease-free water to 2 mM stock concentration (16 ng/nL). Appropriate dilutions were made in 5× injection dye (100 mM HEPES, 1 M KCl, 1% phenol red) and approximately 2 to 3 nL MOs (8-12 ng) was injected at the 1-cell stage. RNA overexpression was performed with 150 pg capped sense RNA generated from a ScaI linearized template, pBluescriptII-SNRK-1, using T3 RNA polymerase. Snrk-1KM mRNA was generated from a backbone vector similar to WT except that mutant cDNA replaced the WT. For rescue experiments, 100 pg hsnrk-1 capped RNA transcribed by T7 RNA polymerase from a EcoRI linearized vector containing full-length hsnrk-1 cDNA was injected along with the MO.
Image acquisition
Except where noted, photomicrographs were taken using a steromicroscope, Leica MZ 16FA (Leica Microsystems) with 1× planapochromatic objective and 10× eyepiece in 100× zoom. The image acquisition software used was Image-Pro AMS 6.0 (Media Cybernetics, Bethesda, MD). All images were corrected for brightness and contrast in Photoshop 7.0 (Adobe Systems, San Jose, CA).
Migration assay
The in vitro migration assay was performed using modified Boyden chamber filter inserts (8-μm pore size; BD Biosciences, San Jose, CA) as described previously,6 with modifications described in Document S1.
Whole-mount in situ hybridization and section
Whole-mount in situ hybridization was carried out as described.7 Details of probes used are available in Document S1.
Results
Identifying and characterizing the zebrafish ortholog of human snrk-1
In a microarray transcript analysis of genes expressed in the developing zebrafish vasculature, we identified several expressed sequence tags (ESTs) or their orthologs that showed and/or were reported to have vascular expression (data not shown). One EST showed both robust vascular expression and was identified in a previous screen for transcripts in lung ECs.8 Due to its sequence similarity to other members of the sucrose nonfermenting kinase family, we followed the previous nomenclature and called it sucrose nonfermenting related kinase-1 (snrk-1). The zebrafish protein is 45% and 46% identical to human and mouse proteins, respectively (Figure S1A). The zebrafish protein contains an ATP-binding domain (23-45 AAs), serine/threonine kinase active site (135-147 AAs), ubiquitin associated domain (291-335 AAs), and a bipartite nuclear localization signal (458-480 AAs), all of which are conserved in human and mouse proteins. Based on the current zebrafish genome assembly, snrk-1 is located on linkage group 25 and spans 5 exons and 4 introns of the genome. Microarray data from this study have been deposited in Gene Expression Omnibus (GEO) under accession number GSE12516.9
Snrk-1 morpholino targets and knocks down endogenous snrk-1 transcript
To investigate snrk-1 function during embryonic zebrafish development, we designed 2 morpholinos (MOs). MO1 targets the exon 2–intron 2 splice junction, whereas MO2 targets the ATG in the 5′UTR of the snrk-1 transcript (Figure S1B). Reverse-transcription–polymerase chain reaction (RT-PCR) across exons 1 and 3 shows that the snrk-1 transcript is reduced in MO1-injected 24 hpf embryos (Figure S1C lanes MO1) compared with uninjected embryos (Figure S1C lane WT). Actin transcripts were not affected in either sample (Figure S1C actin gel). A dose-dependent decrease in snrk-1 transcript expression is observed as the concentration of MO2 increased (Figure S1C lanes MO2). Together, these results suggest that the MOs selectively target endogenous snrk-1 mRNA in vivo.
Expression of snrk-1 in embryonic zebrafish development
To determine the expression profile of snrk-1, we performed RT-PCR across several stages of embryonic zebrafish development (Figure S1D). The snrk-1 transcript is maternally expressed since it is present in 3 hpf embryos and continues until 5 hpf (Figure S1D lanes 3 and 5). The transcript is absent at 9 hpf and reappears after gastrulation at 10 hpf (Figure S2A 10 hpf), continuing into 30 hpf (Figure S1D lane 30). We also checked human placenta and hemangioma samples that are thought to be of placental origin10 for snrk-1 transcripts. RT-PCR shows strong snrk-1 expression in both samples (Figure S1E). These results suggest that the snrk-1 transcript is expressed in maternal tissues of both zebrafish and humans.
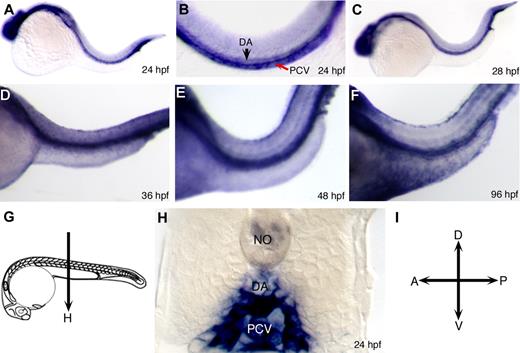
Whole-mount in situ hybridization using snrk-1 antisense digoxigenin-labeled RNA probes showed that snrk-1 is expressed ubiquitously from 10 to 20 hpf (Figure S2A-D). At 24 hpf (Figure 1A), snrk-1 expression is restricted to the brain and trunk vasculature. In high-power trunk images (Figure 1B), snrk-1 appears in the posterior cardinal vein (PCV, red arrow) and dorsal aorta (DA, black arrow). Sections of 26 hpf snrk-1 in situ hybridization embryos (Figure 1C) show that in addition to strong expression in the vessels, snrk-1 is also observed in tissues surrounding the DA and PCV (Figure 1H). As the embryo continues to develop, snrk-1 becomes highly restricted to vascular structures (Figure 1D-F). In addition to the vasculature, snrk-1 is also expressed in the neural tube and the pronephric tubules (data not shown). Interestingly, both human11 and mouse12 snrk-1 orthologs are expressed in mesodermal-derived ECs and hematopoietic progenitors.
Expression of snrk-1 during embryonic zebrafish development. (A-F) Whole-mount in situ hybridizations using digoxigenin-labeled snrk-1 antisense probes. At 24 hpf (A,B) and beyond (C-E) until 96 hpf (F), snrk-1 expression is restricted to head and trunk regions. In the trunk, section of 24 hpf snrk-1 in situ embryo (H) show expression in the dorsal aorta (DA), posterior cardinal vein (PCV), and the regions surrounding these 2 structures. No indicates notochord. Red arrow points to PCV and black arrow points to DA. The plane of the section in panel H is shown in panel G. (I) The orientation of the embryos.
Expression of snrk-1 during embryonic zebrafish development. (A-F) Whole-mount in situ hybridizations using digoxigenin-labeled snrk-1 antisense probes. At 24 hpf (A,B) and beyond (C-E) until 96 hpf (F), snrk-1 expression is restricted to head and trunk regions. In the trunk, section of 24 hpf snrk-1 in situ embryo (H) show expression in the dorsal aorta (DA), posterior cardinal vein (PCV), and the regions surrounding these 2 structures. No indicates notochord. Red arrow points to PCV and black arrow points to DA. The plane of the section in panel H is shown in panel G. (I) The orientation of the embryos.
Loss of snrk-1 results in fewer endothelial cells and angioblasts
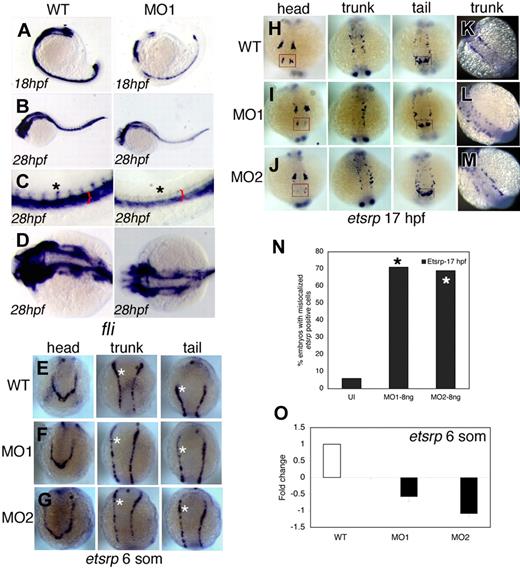
To analyze changes in vascular structures following snrk-1 knockdown, we stained MO1-injected embryos for the vascular marker friend leukemia integration factor-1 (fli). Fli is expressed in developing vascular tissues such as the axial and intersomitic vessels (ISVs; Figure 2C black asterisk in WT). At 18 hpf, MO1-injected embryos (Figure 2A MO1) show fewer fli-positive (fli+) cells compared with uninjected embryos (Figure 2A WT). By 28 hpf, there is a reduction in the fli+ cells (Figure 2C,D MO1, red bracket) in the region of DA and PCV, and fli+ ISVs (Figure 2C black asterisk, MO1) in the trunk region of MO-injected embryos. We also noticed reduction of fli+ vessels in the head of MO-injected embryos (Figure 2D, compare WT with MO1). To determine whether vascular defects occur earlier, we performed in situ hybridization for the vascular marker ets1-related protein (etsrp), an early marker of angioblast differentiation from the LPM.3 As early as 6 som (12 hpf), 3 sets of etsrp-positive cells (etsrp+) appear in the head, trunk, and future tail regions of the embryo (Figure 2E). In MO1-injected (Figure 2F) and MO2-injected (Figure 2G) embryos, subtle reductions (compare white asterisks in Figure 2E-G) in all 3 etsrp+ angioblast populations were noted at 6 som compared with uninjected WT embryos (Figure 2E), suggesting a role for snrk-1 in angioblast proliferation. At 17 hpf, a decrease in etsrp+ cells in the head, trunk, and tail is evident (Figure 2H-J, compare red with black box regions in head and tail, respectively). In overstained snrk-1 MO–injected embryos (Figure 2L,M), we observe that etsrp+ cells are mislocalized compared with WT (Figure 2K). In fact, greater than 70% of MO-injected embryos (Figure 2N) show mislocalized etsrp+ cells. Quantitative real-time PCR (QPCR) demonstrated a 0.5- to 1-fold reduction of etsrp transcript levels in MO-injected embryos at 6 som (Figure 2O). MO2 showed a greater decrease in etsrp transcript levels compared with MO1 (Figure 2O). We also checked for hematopoietic markers gata1 (Figure S2E-G) and scl (Figure S2H-J) and found minimal change. Together, these results suggest that snrk-1 plays a specific role in the localization and maintenance of etsrp+ angioblasts during embryonic development.
Snrk-1 gene knockdown analysis in vivo. (A-D) Endothelial cells at 18 hpf and 28 hpf in wild-type (WT) and MO1-injected embryos are visualized by in situ hybridization using a fli antisense RNA probe. (C,D) High-magnification images of the trunk and head, respectively, of WT and MO1-injected embryos from panel B. Reduction of ECs is observed at (A) 18 hpf and (B) 28 hpf in MO1-injected embryos. Red bracket in panel C shows reduction of fli+ region composed of DA and PCV, and black asterisk shows that intersomitic vessels (ISVs) are missing at 28 hpf in MO1-injected embryos. (A-C) Lateral views. (D) Dorsal view. etsrp+ angioblasts in the head, trunk, and tail at (E-G) 6 som and (H-M) 17 hpf in WT, MO1-, and MO2-injected embryos. White asterisks in panels E through G are included for etsrp+ cell comparison across comparable regions in the 3 samples. At 17 hpf, in the head, compare 4 populations of etsrp+ angioblasts—of which the most anterior are enclosed in red box. In the trunk, 2 bilateral stripes of etsrp+ angioblasts are noticed, of which one stripe is missing in MO1-injected embryo (I, trunk). In the tail, 2 distinct etsrp+ angioblast populations merge at the tail tip indicated by the black box, and differences are noticed in their intensities and patterning in MO-injected embryos compared with WT. (K-M) Embryos overstained with etsrp probe. (L,M) Overstained embryos in which etsrp+ angioblasts are mispatterned. (N) The percentage of MO1- and MO2-injected embryos with mislocalized angioblasts at 17 hpf; *P < .001 between MO1- or MO2-injected groups compared with UI. (O) The fold change of etsrp transcript levels at 6 som in MO1- and MO2-injected embryos as analyzed by QPCR. Error bars represent SEM.
Snrk-1 gene knockdown analysis in vivo. (A-D) Endothelial cells at 18 hpf and 28 hpf in wild-type (WT) and MO1-injected embryos are visualized by in situ hybridization using a fli antisense RNA probe. (C,D) High-magnification images of the trunk and head, respectively, of WT and MO1-injected embryos from panel B. Reduction of ECs is observed at (A) 18 hpf and (B) 28 hpf in MO1-injected embryos. Red bracket in panel C shows reduction of fli+ region composed of DA and PCV, and black asterisk shows that intersomitic vessels (ISVs) are missing at 28 hpf in MO1-injected embryos. (A-C) Lateral views. (D) Dorsal view. etsrp+ angioblasts in the head, trunk, and tail at (E-G) 6 som and (H-M) 17 hpf in WT, MO1-, and MO2-injected embryos. White asterisks in panels E through G are included for etsrp+ cell comparison across comparable regions in the 3 samples. At 17 hpf, in the head, compare 4 populations of etsrp+ angioblasts—of which the most anterior are enclosed in red box. In the trunk, 2 bilateral stripes of etsrp+ angioblasts are noticed, of which one stripe is missing in MO1-injected embryo (I, trunk). In the tail, 2 distinct etsrp+ angioblast populations merge at the tail tip indicated by the black box, and differences are noticed in their intensities and patterning in MO-injected embryos compared with WT. (K-M) Embryos overstained with etsrp probe. (L,M) Overstained embryos in which etsrp+ angioblasts are mispatterned. (N) The percentage of MO1- and MO2-injected embryos with mislocalized angioblasts at 17 hpf; *P < .001 between MO1- or MO2-injected groups compared with UI. (O) The fold change of etsrp transcript levels at 6 som in MO1- and MO2-injected embryos as analyzed by QPCR. Error bars represent SEM.
Snrk-1 gain-of-function complements loss-of-function phenotype
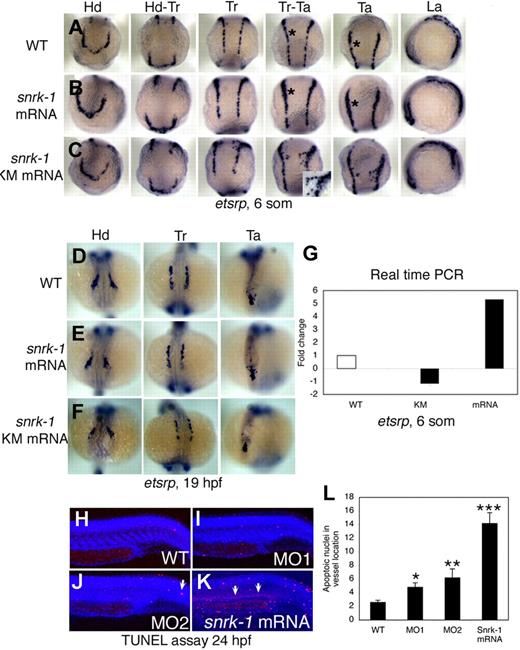
To investigate gain of snrk-1 function, we injected capped snrk-1 sense RNA into WT embryos and performed in situ hybridization for etsrp at 6 som (Figure 3A,C and Figure S4A,B) and 19 hpf (Figure 3D-F). Compared with uninjected WT embryos (Figure 3A), snrk-1 sense RNA-injected embryos (Figure 3D-F) show ectopic induction of etsrp+ angioblasts in all 3 regions of the embryo, namely the head, trunk, and tail (compare black asterisks in Figure 3A,B [Tr-Ta or Ta] or in Figure S4A,B). These effects, however, are much milder by 19 hpf (Figure 3D-F and Figure S4D-F), suggesting compensatory mechanisms or apoptosis of the ectopically induced population. High-power trunk images of snrk-1 mRNA–injected embryos show clumping of etsrp+ cells (Figure S4I) compared with a uniform distribution in uninjected WT embryos (Figure S4G), suggesting migration defects in the gain-of-function (GOF) embryos. QPCR analysis (Figure 1G) shows a 5-fold increase in etsrp transcripts in snrk-1 mRNA–injected embryos. The GOF and loss-of-function (LOF) data suggest complementary function for snrk-1 in angioblast maintenance.
Gain-of-function analysis by overexpression of snrk-1 and snrk-1 kinase mutant (snrk-1 KM). (A-C) etsrp+ cells in the head (Hd), head to trunk (Hd-Tr), trunk (Tr), trunk to tail (Tr-Ta), tail (Ta), and lateral regions (La) of WT embryos (A), snrk-1 mRNA–injected (B), and snrk-1 KM mRNA–injected (C) embryos. (B) Ectopic induction of etsrp+ cells in snrk-1 mRNA–injected embryos. (A,B) Black asterisks in Tr-Ta and Ta panels indicate thicker etsrp-stained region at LPM in panel B compared with panel A. (C) snrk-1 KM mRNA–injected embryos with early migration of angioblasts from the LPM to the midline. (C, Tr-Ta) Inset is higher magnification of prematurely migrating angioblasts at 6 som. (D-F) etsrp+ cells in Hd, Tr, and Ta regions at 19 hpf in WT, snrk-1 mRNA–, and KM mRNA–injected embryos. (G) The fold change of etsrp transcript levels in snrk-1 mRNA– and KM mRNA–injected embryos at 6 som by QPCR. (H-K) An in vivo TUNEL apoptosis assay at 24 hpf in WT, MO1-, MO2-, and snrk-1 mRNA–injected embryos. (J,K) Arrows indicate apoptotic cells. (L) The mean number of apoptotic nuclei (x-axis) in the vessel region in each sample (y-axis: WT, n = 10; MO1, n = 14; MO2, n = 18; snrk-1 mRNA, n = 11), with the error bars representing SD. Paired t test: *P < .05 between WT and MO1; **P < .05 between WT and MO2; ***P < .005 between WT and snrk-1 mRNA samples.
Gain-of-function analysis by overexpression of snrk-1 and snrk-1 kinase mutant (snrk-1 KM). (A-C) etsrp+ cells in the head (Hd), head to trunk (Hd-Tr), trunk (Tr), trunk to tail (Tr-Ta), tail (Ta), and lateral regions (La) of WT embryos (A), snrk-1 mRNA–injected (B), and snrk-1 KM mRNA–injected (C) embryos. (B) Ectopic induction of etsrp+ cells in snrk-1 mRNA–injected embryos. (A,B) Black asterisks in Tr-Ta and Ta panels indicate thicker etsrp-stained region at LPM in panel B compared with panel A. (C) snrk-1 KM mRNA–injected embryos with early migration of angioblasts from the LPM to the midline. (C, Tr-Ta) Inset is higher magnification of prematurely migrating angioblasts at 6 som. (D-F) etsrp+ cells in Hd, Tr, and Ta regions at 19 hpf in WT, snrk-1 mRNA–, and KM mRNA–injected embryos. (G) The fold change of etsrp transcript levels in snrk-1 mRNA– and KM mRNA–injected embryos at 6 som by QPCR. (H-K) An in vivo TUNEL apoptosis assay at 24 hpf in WT, MO1-, MO2-, and snrk-1 mRNA–injected embryos. (J,K) Arrows indicate apoptotic cells. (L) The mean number of apoptotic nuclei (x-axis) in the vessel region in each sample (y-axis: WT, n = 10; MO1, n = 14; MO2, n = 18; snrk-1 mRNA, n = 11), with the error bars representing SD. Paired t test: *P < .05 between WT and MO1; **P < .05 between WT and MO2; ***P < .005 between WT and snrk-1 mRNA samples.
Snrk-1's kinase function is essential for angioblast migration
To determine whether the kinase domain in Snrk-1 is important for its function, we generated a point mutation in snrk-1 that changed proline 142 within the Ser/Thr kinase domain to alanine (Figure S2K) and named the mutant snrk-1 kinase mutant (snrk-1 KM). We generated V5-tagged versions of the protein and detected proteins of the appropriate size by anti-V5 antibody (Figure S2K, V5 WB). We performed an in vitro kinase assay (Kinase Glo assay; Promega, Madison, WI) using 293 cell lysates expressing the V5 protein with recombinant histone H3 as substrate. Histone has been implicated previously as generic substrates for in vitro kinase assay of AMP activated kinase family members.13 In the Kinase Glo assay, luminescent signal correlates to the amount of ATP present and is inversely proportional to kinase activity.14 When equal amounts of Snrk-1 WT and KM protein were added to the kinase reaction, the amount of ATP in the reaction with Snrk-1 KM (Figure S2L white bar) in the presence or absence of histone substrate remained the same. In contrast, Snrk-1 WT showed a drop in ATP level in the presence of histone substrate (Figure S2L black bar), suggesting high kinase activity. These data suggest that Snrk-1 KM shows hypokinase activity compared with Snrk-1 WT, indicating that the proline to alanine substitution in Snrk-1 creates a hypoactive kinase protein.
To determine whether Snrk-1 KM retains its function in vivo, we injected snrk-1KM capped mRNA into WT embryos and performed an in situ hybridization for etsrp at 6 som (12 hpf; Figure 3C). Snrk-1 KM–injected embryos showed premature migration of etsrp+ angioblasts to the midline at 6 som (Figure 3C inset in trunk-tail panel). This effect was dose dependent (data not shown), suggesting that regulation of kinase activity is important for keeping the angioblasts from premature migration to the midline. To determine when etsrp+ cells normally migrate to the midline, we performed etsrp in situ hybridization at 2- to 5-hour intervals between 11 and 30 hpf (Figure S3A-J). At 15 hpf (Figure S3C), etsrp+ angioblasts begin to migrate to the midline and continue this process until 19 hpf (Figure S3G). Therefore, snrk-1 KM embryos are at least 3 hours ahead in their angioblast migration program. Besides premature migration of etsrp+ cells to the midline, we also noticed that the general direction of angioblast migration was altered in snrk-1 KM embryos compared with WT (compare Figure 3C inset with Figure S3D). Further, the number of etsrp+ angioblasts in snrk-1 KM–injected embryos at 6 som appears in between WT and snrk-1 mRNA–injected embryos (compare Figure S4C with Figure 4A,B) especially in the head region (Figure 3A-C). At 19 hpf, fewer etsrp+ cells are clearly noted in snrk-1 KM embryos (compare Figure 3F with 3D or 3E). We confirmed the reduction of etsrp+ cells by performing QPCR for etsrp transcripts in snrk-1 KM–injected embryos (Figure 3G), which show a 1-fold reduction in etsrp transcript levels compared with WT. The snrk-1 KM–injected embryos thus display fewer angioblasts at LPM at 6 som (Figure 3G) and angioblast migration defects (Figure 3C or Figure S4C), which are also observed in snrk-1 KD embryos (Figure 2L,O and Figure S4H), arguing for a dominant negative function for Snrk-1 KM. These results suggest that the kinase activity of Snrk-1 controls angioblast migration to LPM, and perhaps Snrk-1 KM behaves as a dominant negative that interferes with WT Snrk-1's maintenance of angioblasts at LPM.
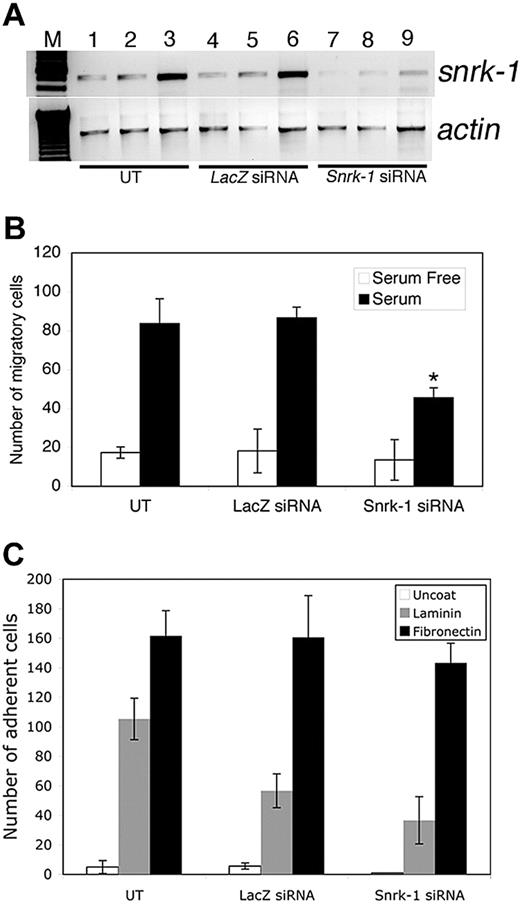
Snrk-1 gene knockdown analysis in vitro. (A) RT-PCR for snrk-1 and actin genes in snrk-1 siRNA–transfected (500 ng) and control lacZ siRNA–transfected (500 ng) HUVECs. Lanes 7 through 9 show reduced snrk-1 transcripts compared with lanes 1 through 6. (B) In vitro KD studies show that there are fewer migratory ECs in snrk-1 siRNA–transfected HUVECs. The graph shows the effect of snrk-1 gene KD on migration of transfected HUVECs to serum-free medium (□) or 10% serum (■) for 5 hours at 37°C. All migration experiments have been conducted using HUVECs from passage numbers 2 through 4. The plotted data are pooled from 3 independent experiments and the error bars represent plus or minus SD. *P < .05 between snrk-1 siRNA and lacZ siRNA cells response to serum. (C) Graphic representation of the number of adherent ECs in snrk-1 siRNA–transfected, lacZ control siRNA–transfected, or untransfected HUVECs to an uncoated surface (□), laminin ( ), or fibronectin (■). Error bars represent SD.
), or fibronectin (■). Error bars represent SD.
Snrk-1 gene knockdown analysis in vitro. (A) RT-PCR for snrk-1 and actin genes in snrk-1 siRNA–transfected (500 ng) and control lacZ siRNA–transfected (500 ng) HUVECs. Lanes 7 through 9 show reduced snrk-1 transcripts compared with lanes 1 through 6. (B) In vitro KD studies show that there are fewer migratory ECs in snrk-1 siRNA–transfected HUVECs. The graph shows the effect of snrk-1 gene KD on migration of transfected HUVECs to serum-free medium (□) or 10% serum (■) for 5 hours at 37°C. All migration experiments have been conducted using HUVECs from passage numbers 2 through 4. The plotted data are pooled from 3 independent experiments and the error bars represent plus or minus SD. *P < .05 between snrk-1 siRNA and lacZ siRNA cells response to serum. (C) Graphic representation of the number of adherent ECs in snrk-1 siRNA–transfected, lacZ control siRNA–transfected, or untransfected HUVECs to an uncoated surface (□), laminin ( ), or fibronectin (■). Error bars represent SD.
), or fibronectin (■). Error bars represent SD.
Previously, we noticed that ectopically induced etsrp+ cells in snrk-1 mRNA–injected embryos at 6 som did not result in increased etsrp+ cells at 19 hpf. We performed an in vivo TUNEL apoptosis assay in snrk-1 mRNA–injected embryos at 24 hpf and noticed a distinct increase in the number of apoptotic nuclei in regions where vessels are localized (Figure 3K, white arrow). Although an overall increase in apoptosis is observed in mRNA-injected embryos, the vascular regions clearly show a greater number of apoptotic nuclei (Figure 3K) compared with similar regions in WT (Figure 3H) or MO-injected embryos (Figure 3I or 3J). Quantitation shows a 7-fold increase in apoptotic nuclei in the trunk vessel region of snrk-1 mRNA–injected embryos compared with WT (Figure 3L). These results suggest that etsrp+ cells ectopically induced by overexpression of snrk-1 are undergoing apoptosis at 24 hpf and suggest a critical role for Snrk-1 in angioblast maintenance.
Snrk-1 knockdown endothelial cells show defective migration in vitro
To determine Snrk-1's function in vitro, we targeted endogenous snrk-1 transcripts in human umbilical vein endothelial cells (HUVECs) with siRNA technology.15 Snrk-1 siRNA–transfected cells show specific targeting of snrk-1 transcript (Figure 4A, compare lanes 7-9 with lanes 4-6) compared with cells transfected with control lacZ siRNA (Figure 4A lanes 4-6) or untransfected cells (Figure 4A lanes 1-3). Snrk-1 KD embryos show mislocalized etsrp+ cells at the midline (Figure 2L-M), and mislocalization of cells implicates a migration defect. Therefore, we investigated HUVEC migration using a modified Boyden chamber assay with serum as a stimulus in the bottom chamber. Snrk-1 siRNA–transfected ECs migrate less to serum compared with untransfected or control lacZ siRNA–transfected cells (Figure 4B, compare black bars). We also checked in vitro adhesion of snrk-1 siRNA–transfected ECs to fibronectin and laminin substrates. We did not notice a difference in adhesion between snrk-1 siRNA– and lacZ siRNA–transfected cells to either substrate (Figure 4C). In a cell cycle proliferation assay by fluorescence-activated cell sorting (FACS), snrk-1 siRNA–transfected HUVECs showed no difference in any cell cycle parameters compared with lacZ siRNA–transfected cells (data not shown). These results suggest that snrk-1 plays an exclusive role in migration of differentiated ECs in vitro.
Snrk-1 plays a specific role in Notch signaling leading to A/V specification
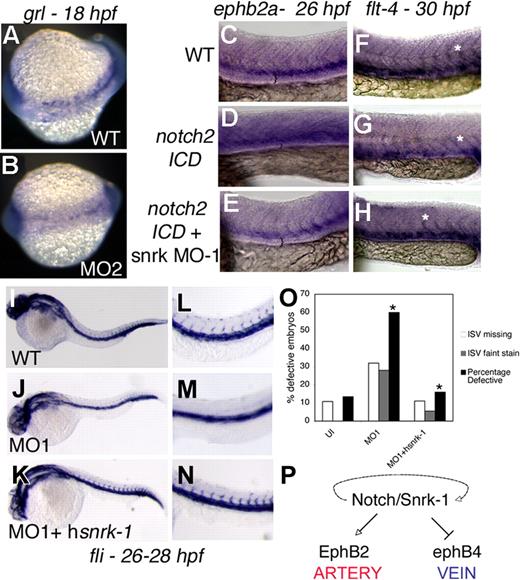
Since fli+ ECs in snrk-1 KD embryos appear to be preferentially lost in artery-vein region (Figure 2C red bracket), we investigated whether snrk-1 is involved in A/V specification by performing an in situ hybridization for the arterial marker grl in snrk-1 KD embryos. Grl guides the arterial-venous decision in angioblast precursors arising from the LPM.16 A significant percentage of snrk-1 MO–injected embryos (70%-80%, Figure S5A) show reduced grl expression (Figure 5B) compared with age-matched WT embryos (Figure 5A). Grl is a downstream target of Notch, which has also been implicated in A/V specification.16-18 To investigate whether Snrk-1 acts in Notch signaling pathway, we injected notch2 intracellular domain (notch2 ICD) mRNA into snrk-1 MO–injected embryos and performed in situ hybridization for artery marker ephrin-B2a and venous marker flt-4. Previous studies have demonstrated that notch2 ICD constitutively activates artery specification and concomitantly reduces vein specification.16,18 In our study, notch2 ICD–injected embryos also showed induced ephrin-B2a (compare Figure 5C with 5D black bracket space) and repressed flt-4 (Figure 5F,G) compared with uninjected embryos (Figure 5C,F). In snrk-1 MO–injected embryos, injection of notch2 ICD did not induce ephrin-B2a (Figure 5E) or repress flt-4 (Figure 5H). Furthermore, notch2 ICD represses flt-4 in intersomitic arteries (compare asterisks in Figure 5F,G), and flt-4 expression is restored in notch2 ICD mRNA plus snrk-1 MO–injected embryos (Figure 5H). Quantitation (Figure S5B) shows that nearly all notch2 ICD mRNA plus snrk-1 MO–injected embryos show reversed artery and venous marker induction. In addition, 45% to 65% and 50% to 65% of snrk-1 MO alone injected embryos showed reduction of ephrin-B2a (Figure S5C white bars) and an increase in flt-4+ ECs (Figure S5C black bars), respectively, compared with WT embryos. Taken together, these data suggest that the absence of snrk-1 blocks notch2 signaling, placing snrk-1 downstream or parallel to notch2 in A/V specification.
Snrk-1 functions in A/V specification. (A,B) grl in situ hybridization of 18 hpf WT and MO2-injected embryos, respectively. ephrin-B2a in situ hybridization of 26 hpf (C) WT, (D) notch2-ICD–injected, or (E) notch2-ICD + snrk-1 MO1–injected embryos. flt-4 in situ hybridization of 30 hpf (F) WT, (G) notch2-ICD–injected, and (H) notch2-ICD + snrk-1 MO1–injected embryos. (F-H) White asterisks show flt-4 expression in intersomitic arteries. (C-E) The black brackets indicate the space between the most dorsal stained regions of the embryo to the yolk extension. (D) Note the shrinkage in space in notch2-ICD–injected embryo, indicating an expansion of ephrin-B2a–stained cells. fli in situ hybridization of 26 to 28 hpf (I) WT, (J) MO1-injected, and (K) MO1 + hsnrk-1 RNA–injected embryos. (J,M) MO1-injected embryos missing ISVs compared with (I,L) WT or (K,N) MO1 + hsnrk-1–coinjected embryos. (O) Graphic representation of the number of embryos displaying faint or absent ISVs in the trunk region of samples in panels I through K. *P < .001 across MO1 and MO1 + hsnrk-1 group. (P) A model placing Snrk-1 parallel to Notch in A/V specification. The dotted line indicates that definitive evidence is needed to show that snrk-1 is downstream of notch. Images in panels C through H were taken using an Observer Z1 inverted microscope (Carl Zeiss, Thornwood, NY) with 10× objective and 10× eyepiece. The image acquisition software used was AxioVision Rel4.6 (Carl Zeiss, Oberkochen, Germany); images were corrected for brightness and contrast in Photoshop 7.0.
Snrk-1 functions in A/V specification. (A,B) grl in situ hybridization of 18 hpf WT and MO2-injected embryos, respectively. ephrin-B2a in situ hybridization of 26 hpf (C) WT, (D) notch2-ICD–injected, or (E) notch2-ICD + snrk-1 MO1–injected embryos. flt-4 in situ hybridization of 30 hpf (F) WT, (G) notch2-ICD–injected, and (H) notch2-ICD + snrk-1 MO1–injected embryos. (F-H) White asterisks show flt-4 expression in intersomitic arteries. (C-E) The black brackets indicate the space between the most dorsal stained regions of the embryo to the yolk extension. (D) Note the shrinkage in space in notch2-ICD–injected embryo, indicating an expansion of ephrin-B2a–stained cells. fli in situ hybridization of 26 to 28 hpf (I) WT, (J) MO1-injected, and (K) MO1 + hsnrk-1 RNA–injected embryos. (J,M) MO1-injected embryos missing ISVs compared with (I,L) WT or (K,N) MO1 + hsnrk-1–coinjected embryos. (O) Graphic representation of the number of embryos displaying faint or absent ISVs in the trunk region of samples in panels I through K. *P < .001 across MO1 and MO1 + hsnrk-1 group. (P) A model placing Snrk-1 parallel to Notch in A/V specification. The dotted line indicates that definitive evidence is needed to show that snrk-1 is downstream of notch. Images in panels C through H were taken using an Observer Z1 inverted microscope (Carl Zeiss, Thornwood, NY) with 10× objective and 10× eyepiece. The image acquisition software used was AxioVision Rel4.6 (Carl Zeiss, Oberkochen, Germany); images were corrected for brightness and contrast in Photoshop 7.0.
Snrk-1's gain-of-function embryos shows complementary A/V marker changes to loss-of-function embryos
To determine A/V specification in GOF embryos, we performed in situ hybridization for grl, ephrin-B2a, and flt-4 marker at 22 hpf, 26 hpf, and 30 hpf, respectively. In snrk-1 GOF embryos, 75% of snrk-1 mRNA–injected embryos showed up-regulated grl expression at 22 hpf (Figure S5C gray bars), and greater than 60% of snrk-1 mRNA–injected embryos showed up-regulation of arterial ephrin-B2a marker (Figure S5C blue bars) and reduction of venous flt-4 marker (Figure S5C green bars), which mirrors numbers observed in snrk-1 KM–injected embryos (Figure S5C arterial ephrin-B2a [blue bars] and venous flt-4 [green bars]). The ephrin-B2a, flt-4, and grl data suggest that snrk-1 specifies arterial angioblast lineages by activating grl, which in turn acts normally to repress venous fates and Snrk-1's kinase activity is dispensable for A/V specification.
Human snrk-1 rescues knockdown of zebrafish snrk-1 in vivo
To determine whether snrk-1 function is evolutionarily conserved, we coinjected human snrk-1 (hsnrk-1) sense RNA into snrk-1 KD embryos and performed an in situ hybridization for fli on uninjected, MO-injected, and MO plus hsnrk-1 mRNA–injected embryos at 26 to 28 hpf (Figure 5I-N). At 26 to 28 hpf, MO-injected embryos (Figure 5J,M) are missing fli+ ISVs as seen previously (Figure 2C). When hsnrk-1 was coinjected with the MO (Figure 5K-N), fli+ ISV cells return as seen in WT embryos (Figure 5L). It is worthwhile to note that the rescue is not complete since the ISVs and fli+ axial region appear darker (Figure 5N) compared with uninjected embryo (Figure 5L). Incomplete rescue can be explained by the fact that the amount of injected hsnrk-1 mRNA did not fully compensate for loss of snrk-1. Sixty percent of MO-injected embryos have missing or faint fli+ ISVs compared with 10% to 15% of uninjected or MO1 plus hsnrk-1 mRNA–injected embryos (Figure 5O).
Discussion
Angioblasts are multipotent progenitor cells that give rise to most vascular lineages, including arteries and veins. Angioblast specification from the LPM has recently been shown to require the expression of a single transcription factor from the ets related protein family (Etsrp).3 After specification, angioblasts migrate, proliferate, and differentiate into arteries and veins. In this study, we have identified a serine threonine kinase named after the sucrose nonfermenting kinase family called snrk-1 that plays a specific role in angioblast proliferation, migration, and specification.
Prior to this study, it was known that LPM angioblasts migrate to the midline at 12 hpf, and by 18 hpf they coalesce to form axial vessels. However, it has been difficult to parse through these 6 hours of development to understand how an undifferentiated angioblast eventually becomes a differentiated endothelial cell. Using a microarray screen for differentially expressed genes in the zebrafish vasculature, we found a novel member of the serine threonine kinase family, snrk-1, that is expressed between 12 and 18 hpf in the developing zebrafish vasculature. The mouse and human orthologs also show vascular expression.11,12
In this study, we investigated the function of snrk-1 by LOF and GOF analyses in vivo. Our experiments suggest 2 distinct roles for snrk-1 in vascular development—an early role in angioblast assembly into the DA and a later role during specification of angioblasts to arteries and veins. In early development (6-18 som), snrk-1 LOF embryos show 2 distinct features. First, angioblast migration from the LPM to the midline is impaired and second, there are fewer angioblasts in the LPM. This suggests that snrk-1 functions in angioblast migration and proliferation. In vitro, snrk-1 functions exclusively in the migration of differentiated ECs (HUVECs), suggesting that mechanisms controlling migration are shared by undifferentiated angioblasts and differentiated ECs.
A snrk-1 point mutant, snrk-1 KM shows hypophosphorylation activity in vitro, and embryos injected with this RNA show premature migration of angioblasts to the midline. Interestingly, snrk-1 KM–injected embryos have fewer angioblasts, suggesting that the kinase activity is important for angioblast proliferation and that the hypokinase activity of Snrk-1 KM and or its presumed interference (dominant negative function) with WT Snrk-1 function induces angioblasts to prematurely migrate to the midline. At present, it is less clear whether the premature migration observed in snrk-1 KM–injected embryos is exclusively due to its hypokinase activity or the dominant negative effect or both.
Angioblast numbers necessary to cover the length and breadth of a developing embryo are critical for development of DA. This task can be accomplished through proliferation and maintenance of numbers. Both of these events can occur at LPM or at the midline. Maintenance of cells would normally require a balance between proliferation and apoptosis. Our data argue that although snrk-1 overexpression increases the number of angioblasts at LPM (Figure 3B,G), these angioblasts are undergoing apoptosis by 24 hpf (Figure 3K). These data support the idea that proper maintenance of angioblasts in the LPM is critical for future angioblast migration to the midline and downstream processes of angioblast development. Snrk-1 thus likely functions as a checkpoint for maintaining angioblast numbers at the LPM.
In addition to this early role, Snrk-1 also functions later (19-22 hpf) during angioblast differentiation to arteries and veins. The Notch pathway has been implicated in A/V specification, and Notch directly activates Grl, one of the earliest markers identified in zebrafish for A/V specification.19 Grl regulates A/V specification by repressing venous fates.16 In our study, snrk-1 GOF embryos up-regulate grl expression, whereas snrk-1 KD embryos show reduced grl expression. This suggests that snrk-1 acts upstream of grl and is consistent with grl's role in A/V specification. The hypokinase mutant, however, showed no difference for well-established markers of arteries and veins, ephrin-B2a20 and flt-4, respectively, compared with snrk-1 WT mRNA–injected embryos (Figure S5C), suggesting that the hypokinase activity is less important for angioblast differentiation. In addition to grl, Notch signaling triggers A/V specification. Notch2 mRNA–injected embryos up-regulate ephrin-B2a and repress flt-4 in WT embryos. However, these functions of Notch2 are impaired in snrk-1 KD embryos, indicating that snrk-1 acts downstream or parallel to notch2 in A/V specification.
Angioblasts require specific molecular cues to target them to the midline, and during migration, they must maintain high enough numbers to develop into a mature vessel. Our study implies that Snrk-1 plays a critical role in 2 steps in this process. In the first step, wherein specified angioblasts migrate to the midline, Snrk-1 and its kinase activity are important for angioblast maintenance and migration. In the second step of angioblast specification to arteries or veins, snrk-1 works upstream of grl and downstream or parallel to notch in the signaling cascade involved in A/V specification (Figure 5P). Whether snrk-1 performs similar functions in mammalian development is not known and is currently under active investigation.
The expression pattern of snrk-1 in zebrafish is more venous than arterial (Figure 1H). In an accompanying study,21 we have identified mutations in snrk-1 in venous or lymphatic malformation patients. Therefore, it is feasible to propose that snrk-1 plays a central role in the pathogenesis of vascular anomalies. The snrk-1 gain of function in vivo leads to ectopic induction of angioblasts that undergo apoptosis by 24 hpf (Figure 3K). It is not known whether snrk-1 triggers cell survival pathways in endothelial cells, such as in the AKT pathway, but is worth investigating because akt1 overexpression in endothelial cells has resulted in the development of cutaneous vascular malformations in vivo.22 In addition, the exclusivity of the snrk-1 mutations in venous or lymphatic malformation patient samples argues that snrk-1 may be an important molecular marker like wt-1 that helps distinguish vascular malformations from proliferative endothelial lesions.23
In summary, this study reports a novel cytoplasmic signaling member of the Ser/Thr kinase family that plays a critical role in angioblast development in vertebrates in vivo.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Shobhit Singla of Cornell University for initial assistance in bioinformatic analysis of the snrk-1 gene.
This study was supported in part by research funding from National Institutes of Health (NIH, Bethesda, MD) grants HL072178 and HL70567 (D.M.) and by seed funds from the Children's Research Institute at the Medical College of Wisconsin, Milwaukee (R.R.). R.R. is a recipient of the National Cancer Institute (NCI, Bethesda, MD) Scholar Award. G.S. is a recipient of the State of Wisconsin Breast Cancer Research Tax Write-off Program Award. L.W. is a fellow of American Heart Association (Washington, DC).
National Institutes of Health
Authorship
Contribution: C.Z.C., D.M., and R.R. designed research; C.Z.C., S.K., G.V.S., L.W., K.P., M.K.G., K.L., and L.F. performed research; C.Z.C, S.K., K.P., M.K.G., L.W., G.V.S., and R.R. analyzed the data; and R.R. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ramani Ramchandran, Medical College of Wisconsin, Department of Pediatrics, CRI Developmental Vascular Biology Program, Translational and Biomedical Research Center, CRI C3420, 8701 Watertown Plank Road, PO Box 26509, Milwaukee, WI 53226; e-mail: rramchan@mcw.edu.
References
Author notes
*C.Z.C. and S.K. contributed equally to this work.