Abstract
One of the most fundamental questions in immunology pertains to the recognition of non-self, which for the most part means microbes. How do we initially realize that we have been inoculated with microbes, and how is the immune response ignited? Genetic studies have made important inroads into this question during the past decade, and we now know that in mammals, a relatively small number of receptors operate to detect signature molecules that herald infection. One or more of these signature molecules are displayed by almost all microbes. These receptors and the signals they initiate have been studied in depth by random germline mutagenesis and positional cloning (forward genetics). Herein is a concise description of what has been learned about the Toll-like receptors, which play an essential part in the perception of microbes and shape the complex host responses that occur during infection.
Introduction
When it was first realized that microbes are the cause of infectious diseases, a new question arose immediately. How are they detected? What molecules are seen as foreign, and why do we respond to them? This question touched on the molecular basis of self/non-self discrimination, which is often perceived as the most fundamental question of immunology. As we understand it today, self/non-self discrimination has many layers, depending upon which type of immune cell is at issue, or what type of “non-self” is to be recognized. However, activation of immune responses is hierarchical, and microbes are the “non-self” with which the immune system evolved to cope. As soon as there was consensus as to their involvement in infectious disease, there was a great void to be filled in understanding how they were initially perceived by the host
At one level the answer was obvious. In biologic systems, recognition most commonly depends upon receptors. The notion that there must be receptors for molecules of microbial origin was commonplace from an early stage, and the first step was to identify just which molecules were detected. By the 1940s, fractionation and chemical analyses of microbes were beginning to reveal the molecular components that elicited inflammatory responses in mammals. The first microbial component to be studied in detail was “endotoxin” (lipopolysaccharide [LPS]). The inflammatory character of other components (for example, dsRNA, DNA, peptidoglycan, and lipopeptides) was established in turn.
Later pronouncements that there must be receptors for “pathogen-associated microbial patterns” added no new concept, but merely restated the question.1 On the other hand, actually finding the receptors that inform us of infection and establishing their specificity was, and to some extent remains, a central challenge of immunology. Understanding the signaling pathways activated by the receptors we now know, in order that we may view immune responses in purely mechanistic terms, is an unfinished story. One day our understanding of innate immune signaling may let us modulate inflammation as suits us best, produce vaccines, and treat autoimmune diseases—all in a more rational manner than we do at present.
Finding the receptors that distinguish self from non-self was achieved through a classical genetic approach. It began with a phenotype (unresponsiveness to LPS) and ended with the identification of the signaling core of the LPS receptor. As it happened, the LPS receptor was part of a family of molecules that are dedicated to the detection of microbial infection. Not only bacteria, but viruses are recognized through activation of TLRs. Genetic analysis continues to enlighten us as to how these receptors signal. And at the same time, fundamental questions have arisen as to how these receptors and others like them maintain contact with their microbial quarry while avoiding self.
Inflammation and microbe sensing are two aspects of a single process
The earliest investigations into the taxonomically broad, intrinsic toxicity of microbes were carried out at the onset of the postmicrobial era, when it first became widely accepted that infections were the visible concomitant of host infestation by germs. Richard Pfeiffer, working with Robert Koch, coined the term “endotoxin” to refer to a heat-stable material associated with Gram-negative microbes (Vibrio cholerae) capable of producing shock and death in guinea pigs.2 With this, the basis for much future investigation was established because endotoxin, later known as LPS, could be purified and analyzed chemically.3-5 Today we know it as a structural constituent of Gram-negative bacteria, comprising most of the glycolipid of the outer leaflet of the outer membrane.
Quintessentially inflammatory, LPS was also endowed with certain protective effects. It was able to stimulate nonspecific immunity to microbial infections6 and to protect against otherwise lethal doses of radiation.7 LPS was also seen to be an adjuvant, capable of driving antibody responses to admixed protein antigens, as early as 1955.8 There followed some puzzlement over LPS. Was it intended as a weapon, or, on the other hand, did the mammalian immune system evolve so as to recognize it and mount a response to Gram-negative bacteria? And how did LPS work? The answers to these questions flowed from spontaneous mutations observed in 2 different strains of mice.
Genetic evidence of the existence of an LPS receptor
Heppner and Weiss9 and later Sultzer10 found that mice of the C3H/HeJ substrain were highly resistant to LPS. Resistance was determined by cells of hematopoietic origin,11 indicating that the lethal effects of LPS were mediated by derivatives of the blood, eventually identified (at least in a galactosamine-sensitized model of LPS-induced death) as macrophages.12 In due course, all biologic effects of LPS were shown to be suppressed by the C3H/HeJ mutation. This included the immunoadjuvant effect of LPS, for example,13 and its ability to stimulate B-cell division.14
Later another strain, C57BL/10ScCr, was found to be equally resistant to the lethal effect of LPS.15 The mutation in this strain was shown to be allelic to that in the C3H/HeJ strain.16 The C3H/HeJ mutation was said to affect the Lps locus, and was mapped to chromosome 4 in the late 1970s by monitoring its linkage to classical phenotypic markers. It was placed between the Mup (major urinary protein) and Ps (polysyndactyly) loci: an interval now known to span approximately 30 Mb of DNA, encompassing hundreds of genes. At this stage, mapping work ceased for more than a decade. Without a map of genetic markers, and without techniques for cloning large pieces of DNA into overlapping contigs, no further advance was possible.
However, the mere fact that a single mutation could entirely abolish LPS sensing taught many things. It suggested the existence of a single nonredundant pathway for LPS perception and, presumably, a single receptor for LPS. No longer could LPS be considered a toxin that worked by affecting the integrity of plasma membranes of cells throughout the host. Moreover, long before the identity of the Lps locus was known, it was shown that C3H/HeJ mice were unusually susceptible to infection by Gram-negative bacteria. From this, it was correctly concluded that detecting LPS was important to the development of a response to the microbe. Despite its inherent toxicity, LPS did not evolve to be a weapon, but only became one incidentally, in consequence of the powerful response the mammalian host had evolved to deal with Gram-negative microbes.
A marker to follow: tumor necrosis factor
To identify the LPS receptor, it was necessary to follow a biologic activity induced by receptor activation. While LPS causes many changes in the host, the elicitation of cytokine production is perhaps the most convenient change to monitor, and one with clear biologic relevance. In 1985, tumor necrosis factor (TNF) was isolated from mice17,18 and shown to be an important component in the pathogenesis of LPS-induced shock.19 This finding established TNF as a primary mediator of inflammation and led to broad efforts by pharmaceutical companies to neutralize TNF with therapeutic intent. TNF neutralization ultimately proved to be most useful in the therapy of specific chronic inflammatory diseases, including rheumatoid arthritis.
There is no precise explanation for why some disease processes but not others are TNF-dependent. However, where LPS responses were concerned, the measurement of TNF production (normal in macrophages obtained from C3H/HeN mice but absent in macrophages obtained from C3H/HeJ mice) proved a reliable quantitative means of phenotypic assignment during genetic mapping.
The positional cloning of Lps
Several attempts to find the elusive LPS receptor were made using conventional biochemical methods and expression cDNA cloning. A part of the LPS receptor was identified by Wright, et al,20 who found that CD14, a GPI-tethered leucine-rich repeat protein present on mononuclear phagocytes, was required for LPS signaling. But the question remained: how did LPS signal across the membrane of the cell, and what was the product of the Lps locus (unlinked to the CD14 locus on mouse chromosome 18)? Positional cloning gave the answer. The Lps locus was mapped on 2093 meioses to a 2.6-Mb critical region.21
It must be recalled that at the time, there was no published mouse genome sequence, nor even a draft of a sequence corresponding to the Lps critical region, which was terra incognita. Accordingly, the critical region was cloned into bacterial artificial chromosomes (BACs) and a single yeast artificial chromosome (YAC), which were sequentially fragmented and sequenced in a search for genes. Positional cloning methods evolved over the term of the search for the Lps locus (which was accomplished over the period spanning 1993-1998), and in the beginning, exon trapping and hybridization selection were used to search for candidate genes. Increasingly, computational methods (gene prediction programs like GRAIL, developed at the Oak Ridge National Laboratory, and BLAST searches against expressed sequence tag [EST] databases maintained by the National Center for Biotechnology Information and The Institute for Genomic Research) were used. As EST databases grew in complexity, BLAST searches of acquired sequence against EST databases became the preferred search tool.
Many candidates were found, and were taken more seriously than they would have been in later years, because homology (rather than identity) at the nucleotide level could not be discounted, given that EST databases were incomplete. Some false candidates arose from chimerism of BAC or YAC clones. A succession of pseudogenes and fragments of known genes were identified in the critical region, cloned from both C3H/HeN and C3H/HeJ cDNA, and each excluded in turn. The correct gene, Tlr4, was found in the last BAC scheduled for sequencing. Unlike the great majority of candidates, it gave a perfect match with the sequences captured from the BAC. Moreover, it made excellent biologic sense as a candidate because of its homology to the interleukin-1 (IL-1) receptor, and because of the known immunologic function of the Drosophila Toll protein. Moreover, it was a receptor with leucine-rich repeats in the ectodomain, and therefore structurally similar to CD14, known to be a component of the LPS sensor.
The identification of a point mutation in the cytoplasmic domain of the C3H/HeJ (but not the C3H/HeN) cDNA followed a few days later and argued strongly that TLR4 was “the” protein required to transduce LPS signals. The added finding that C57BL/10ScCr mice lacked the locus entirely while the closely related LPS-sensitive strain C57BL/10ScSn retained the locus affirmed beyond any doubt that the Lps locus had been found.22
The Drosophila model
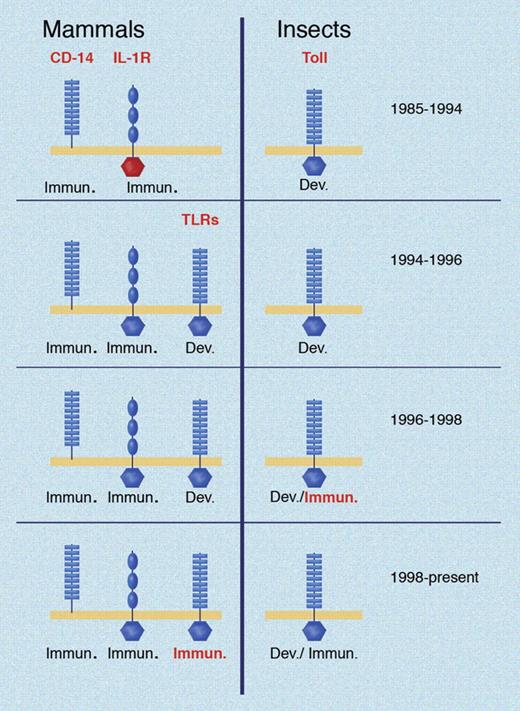
At the time Toll-like receptor 4 (TLR4) was identified as the membrane-spanning component of the LPS receptor, it was one of 5 mammalian proteins with homology to Drosophila Toll. The first of these receptors (now known as TLR1) was recognized in 199423 and mapped to a mouse chromosome in 1996,24 and presumed to function in development. Molecules of the Toll family were known as early as 1991 to have cytoplasmic domain homology to the mammalian IL-1 receptor25 : then seen as a curious fact since IL-1 was well known to elicit inflammatory responses, while Drosophila Toll was well known to have an essential function in development. It could only be assumed that a certain molecular ancestor had been dedicated to development in one species and inflammation in another (Figure 1).
Key developments in the TLR field over time. Conceptual advances shown in red for both insects and mammals. Toll was identified as a developmental protein in 1985. CD14 was identified as a part of the LPS receptor in 1990; the IL-1 receptor was cloned in 1988 and noted to have domain homology to Toll in 1991. In 1994, mammalian TLRs were first identified, but incorrectly assumed to have developmental functions based on what was known in Drosophila at the time. In 1996, the dual immunologic/developmental character of Drosophila Toll was recognized. The immune function of a mammalian TLR was first demonstrated in 1998. Illustration made with assistance of Marie Dauenheimer.
Key developments in the TLR field over time. Conceptual advances shown in red for both insects and mammals. Toll was identified as a developmental protein in 1985. CD14 was identified as a part of the LPS receptor in 1990; the IL-1 receptor was cloned in 1988 and noted to have domain homology to Toll in 1991. In 1994, mammalian TLRs were first identified, but incorrectly assumed to have developmental functions based on what was known in Drosophila at the time. In 1996, the dual immunologic/developmental character of Drosophila Toll was recognized. The immune function of a mammalian TLR was first demonstrated in 1998. Illustration made with assistance of Marie Dauenheimer.
Toll was originally named as one of 12 genes in the “dorsal group” responsible for the differentiation of the embryo into dorsal and ventral structures. A plasma membrane receptor, Toll is activated by Spaetzle, a protein cleaved from an inactive precursor through proteolysis. Toll signaling entails the activation of a serine kinase, Pelle, and ultimately an NF-κB homologue, Dorsal. Hundreds of genes are then induced and govern the differentiation of dorsal and ventral structures.
In 1996, Hoffmann and colleagues showed that the synthesis of Drosomycin, an antifungal peptide required to protect fruit flies against infection by Aspergillus fumigatus, was also activated by Toll signaling, and dependent upon it. It appeared that Toll had a dual life in the fly and was involved in both development and host resistance to infection. In 1997, mammalian TLR4 was shown to activate NF-κB when overexpressed and artificially ligated in mammalian cells.26 However, this left open the question of whether the protein had a developmental or immunologic role in mammals and did not shed light on the ligand for TLR4 or, indeed, reveal whether any ligand existed.
The positional cloning of the Lps locus, which revealed TLR4 as the core of the LPS receptor, indicated that in the mouse, the function of the protein was highly circumscribed. Here, no overt developmental function seemed to be involved, since mice lacking TLR4 developed normally. However, the immune function of Toll and the mammalian TLRs had been conserved for hundreds of millions of years. An immediate proposal, visited both in Drosophila and in mammals, held that the Drosophila Tolls and the mammalian TLRs might each detect different microbial insults, signaling to generate the preponderance of the innate immune response.
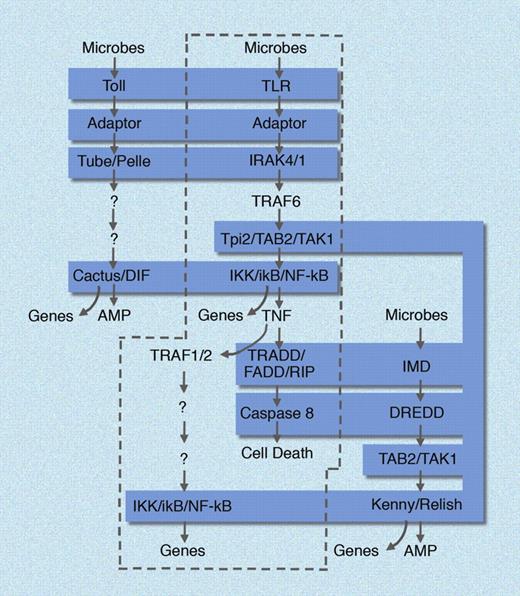
In Drosophila, only Toll (among 9 paralogues that exist in that species) seems to have retained an immunologic function. Moreover, a separate response pathway in Drosophila, known as the Imd pathway, was seen to mimic the TNF signaling pathway in many of its aspects. It is interesting to consider that in mammals, the TLR pathways (which lead to TNF production) and the TNF pathway itself (mediated by 2 signaling receptors for this cytokine) can be viewed as one unbroken pathway with an extracellular component, TNF.27 In the fly, the 2 pathways have been separated (Figure 2).
Homologies between mammalian and insect immune defense pathways. In mammals (enclosed in dashed curve), TLR and TNF signaling pathways are unified, in that TNF is produced in response to all TLR ligands, and then signals via the TNF receptors to induce cell death and NF-κB activation in other cells. In Drosophila, the Toll pathway and the Imd pathway are separated, each responding independently to microbial stimuli. Molecular homologies are shaded in blue. AMP indicates antimicrobial peptide. Genes, implies the induction of many hundreds of NF-κB dependent genes. Figure is not all encompassing and is meant to emphasize core similarities between the pathways. Illustration made with assistance of Marie Dauenheimer.
Homologies between mammalian and insect immune defense pathways. In mammals (enclosed in dashed curve), TLR and TNF signaling pathways are unified, in that TNF is produced in response to all TLR ligands, and then signals via the TNF receptors to induce cell death and NF-κB activation in other cells. In Drosophila, the Toll pathway and the Imd pathway are separated, each responding independently to microbial stimuli. Molecular homologies are shaded in blue. AMP indicates antimicrobial peptide. Genes, implies the induction of many hundreds of NF-κB dependent genes. Figure is not all encompassing and is meant to emphasize core similarities between the pathways. Illustration made with assistance of Marie Dauenheimer.
In mammals, gene targeting established that each TLR recognizes a different set of signature molecules from the microbial world. Moreover, these sensors yield protection that is nonredundant. Blocking all TLR signaling by mutational inactivation of 2 of the TLR adapter proteins, MyD88 and TRIF, causes severe immunocompromise. Usually this leads to fatal opportunistic infections in mice before the development of an experienced adaptive immune system.
It comes as no surprise that specific TLR ligands like LPS, which have long been known to have adjuvant activity, require their cognate TLRs to exert an adjuvant effect. On the other hand, it has variously been stated that TLRs are “essential,”28 “responsible,”29 or “required”30 for an adaptive immune response. It is now clear that this is not the case. No classical adjuvant—not even those that contain TLR ligands—requires TLR signaling. Hence, in TLR signaling deficient mice, normal or near-normal antibody responses can be elicited with a wide variety of classical adjuvants.31 Some adjuvants, such as alum, contain no TLR ligands, or provoke no TLR-mediated responses. Indeed, strong adaptive immune responses (as to allografts) can occur in the absence of any form of adjuvant at all.
The principal ligands for TLRs 1, 2, 3, 5, 6, 7, and 9 (Table 1) were determined by gene targeting between 1999 and 2003, as reviewed elsewhere.32 TLR10, which exists in humans and is most closely related to TLRs 1, 2, and 6, has been lost from the mouse genome. Its ligand cannot be explored in the mouse, and remains uncertain. TLRs 11, 12, and 13 have been lost from the human genome, and of the 3, only one ligand for TLR11 (not necessarily the sole ligand) has been identified.33
Mouse TLRs, their principal ligands, and the adaptors that serve them
| TLR . | Associated proteins . | Ligands . | Adaptors . |
|---|---|---|---|
| TLR1/2 | CD14, CD36, Dectin1 | Ac3LP, Glycolipids | MyD88,Tirap |
| TLR2/6 | CD14, Dectin1 | Ac2LP, LTA, Zymosan | MyD88, Tirap |
| TLR3 | polyI:C, dsRNA | TRIF | |
| TLR4 | CD14, MD-2 | LPS, Taxol, Heparan, Hyaluronate, F-prot, RSV, G-prot, VSV, Env prot, MMTV, others | MyD88,Tirap TRIF,TRAM |
| TLR5 | Flagellin | MyD88 | |
| TLR7 | ssRNA, imiquimod, loxoribine, other | MyD88 | |
| TLR9 | ? Effete | CpG, DNA | MyD88 |
| TLR11 | Profilin | MyD88 | |
| TLR12 | ? | ? | |
| TLR13 | ? | ? |
| TLR . | Associated proteins . | Ligands . | Adaptors . |
|---|---|---|---|
| TLR1/2 | CD14, CD36, Dectin1 | Ac3LP, Glycolipids | MyD88,Tirap |
| TLR2/6 | CD14, Dectin1 | Ac2LP, LTA, Zymosan | MyD88, Tirap |
| TLR3 | polyI:C, dsRNA | TRIF | |
| TLR4 | CD14, MD-2 | LPS, Taxol, Heparan, Hyaluronate, F-prot, RSV, G-prot, VSV, Env prot, MMTV, others | MyD88,Tirap TRIF,TRAM |
| TLR5 | Flagellin | MyD88 | |
| TLR7 | ssRNA, imiquimod, loxoribine, other | MyD88 | |
| TLR9 | ? Effete | CpG, DNA | MyD88 |
| TLR11 | Profilin | MyD88 | |
| TLR12 | ? | ? | |
| TLR13 | ? | ? |
Associated proteins indicate cell-surface molecules that are known to have essential accessory roles for the detection of at least some ligands based on the effects of ENU-induced mutations or gene targeting.
Most or all of the TLRs, like Toll itself, are believed to be functional multimers. Some, like the TLR2 complexes with TLRs 1 or 6, are heteromeric. Some appear to be homomeric, and in some cases, non-TLR subunits are part of the signaling complex. For example, TLR4 seems not to detect LPS directly, but only as a complex with MD2, a small tightly associated LPS binding subunit. Crystallographic analysis has shown the nature of the interaction between specific TLR ligands and the Toll-like receptors. Notably, interactions between LPS and the MD2:TLR4 complex, between poly I:C and TLR3, and between lipopeptides and TLR2/TLR1 complexes have been demonstrated.
Some TLRs, particularly those that sense nucleic acids (TLRs 3, 7, 8, and 9), are expressed within the endosome/lysosome compartment, and transit to this location from the ER with the assistance of UNC93B, a multispanning ER protein identified for its role in TLR signaling through genetic analysis as described below. Other TLRs are expressed primarily on the cell surface, but their ability to transit to the endosome is not excluded, nor is the possibility that in some cells under some conditions, the nucleic acid sensing TLRs might gain access to the cell surface. TLR distribution is complex, in terms of which cells express them. Some seem nearly ubiquitous; others are confined to only a few cell types. A discussion of their pattern of distribution will not be covered here, partly because the facts are not all available yet, given that even low-level expression of specific TLRs may have important signaling consequences.
While many ligands of both endogenous and exogenous origin have been said to engage individual TLRs, it is possible that some of the reports reflect contamination of ligand preparations with LPS, lipopeptides, or DNA. Nonetheless, it is clear that endogenous molecules sometimes do activate TLR signaling. Notably, host DNA can activate TLR9, and is probably relevant to the pathogenesis of autoimmune diseases in which antibodies against chromatin constituents may assist in the internalization of DNA molecules, whereupon they may drive further cell activation by TLR signaling.
While TLR4 is dedicated largely to detecting Gram-negative bacteria, it is also important in combating some viral infections: notably respiratory syncitial virus (RSV)34 and rhabdoviral (vesicular stomatitis virus; VSV) infections.35 TLRs 3 and 9 are important in resistance to herpes viral infections.36 TLR7 is important in resistance to influenza infection.37,38 Gram-positive bacteria are managed by TLR2 complexes. Hence, there is a clear division of labor, but some microbes are detected by multiple TLRs.
Other systems for perception of infection and the damage it causes
As described below, all TLR signaling can be eliminated by 2 mutations (in genes encoding the adaptor proteins MyD88 and TRIF). The severe immunocompromise evident in such mice indicates an extremely important and nonredundant role for TLRs in innate defense. Nonetheless, other systems for microbial perception also exist. RIG-I like helicases (RLHs) have recently been identified as sensors of cytoplasmic nucleic acids (reviewed in39 ), and offer protection against specific viruses. Proteins of the NOD/NALP family of receptors (sometimes called NLRs), detect a wide variety of injurious stimuli and are central to the generation of an IL-1 response.40-42 Some of the stimuli for NLR activation are microbial, and these probably contribute to distant mobilization of immune responses.
NK cell receptors (both activating and inhibitory) also mediate innate immune sensing.43 Some NK cell receptors, as exemplified by Ly49H (which detects the virally encoded m157 protein of mouse cytomegalovirus44 ), respond to foreign proteins including foreign MHC molecules. Another type of NK activating receptor (NKG2D) responds to “stress” proteins such as RaeI or H60, produced as a result of viral infection or irradiation.45 A particularly interesting mechanism of mediating the immune response is presented by NK inhibitory receptors, which normally deliver an inhibitory signal to “calm” the NK cell in the presence of self ligand (class I MHC), but are silenced in the absence of self. These “missing self” receptors respond to virally mediated down-regulation of class I MHC antigen (an evasion mechanism used by many viruses to prevent recognition by cytotoxic T lymphocytes).
Other mechanisms have also been exploited for the detection of microbes. In Drosophila, microbial proteases can directly trigger proteolytic cascades in the hemolymph of the host and initiate a response.46 It is not clear whether strictly homologous mechanisms for cellular awareness of microbes exist in mammals, although the activation of complement by microbial proteases is well known (for example,47 ) and might be seen as an analogous process.
The forward genetic approach
As described in this review, it was a forward genetic approach that led to the understanding that TLRs are a key system for detecting microbes in mammals. Forward genetics also has contributed to the elucidation of the signaling pathways used by the TLRs, and to the more general requirements for a successful innate immune response.
Forward genetics is equivalent to classical genetics empowered by the use of mutagens to produce phenotype and advanced methods for the identification of mutations. It is an approach in which hypotheses are not used in the initial approach to a biologic question. Rather, a phenotype (an alternative state of the phenomenon under study) is the starting point of forward genetic analysis, and its cause determined by positional cloning. By setting up an appropriate phenotypic screen such as measuring TNF production in macrophages in response to TLR signaling,48 the investigator can identify several mutants and thus genes involved in the phenomenon of interest. Once the geneticist has identified several molecules that are essential for a phenomenon to occur as it normally does, he or she may often draw a clear “picture” of the events that must occur to support the phenomenon. Hypotheses are often necessary along the way; for example, to probe the mechanism by which a particular protein works in the process being analyzed.
The power of genetics rests partly in its lack of bias. Often the hypothesis-driven approach to biologic problems creates self-fulfilling prophecies, as experiments are constructed to prove the hypothesis rather than to test it. Genetics is also a source of surprise. If one accepts that a particular biologic system has complexity that extends beyond the bounds of the investigator's imagination, genetics is the tool with which to probe it. Hypotheses are, after all, restricted to what one can imagine. Very commonly, genetics will disclose an essential requirement for a molecule that would never have been “guessed” to be involved in a particular process.
Forward genetics has been pursued in many different model organisms. The mouse has been a latecomer in this respect, but its direct use as a model has been much empowered by the complete sequencing of the mouse genome, and by the ability to resequence more rapidly than ever before. The mutagen of choice for the generation of phenotypes in the mouse is N-ethyl-N-nitrosourea (ENU), an alkylating agent that creates point mutations at random at a frequency of 1 change per 1 to 2 Mb of haploid DNA. There is no convincing evidence for “hot spots” in the genome.
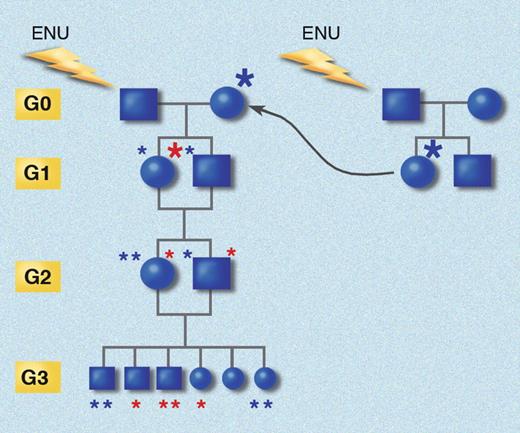
It appears that ENU increases the rate of phenovariance in mice approximately 15- to 20-fold over the background rate. Each ENU-mutagenized G0 male mice will produce approximately 20 G1 offspring heterozygous for approximately 3000 mutations. Homozygosity for some of these mutations can be produced by breeding through 3 generations by backcrossing G1 females with the G0 male and then by inbreeding siblings of the next 2 generations (Figure 3). Substantial attrition is observed in G3 mice, and most probably, a 150- to 200-fold increase in mutation rate would be uniformly lethal in G3 animals, based on the rate of observed lethal mutations within defined genetic intervals covered by balancer chromosomes.49
Inbreeding protocol for generating homozygous mutations. G0 mice are bred to germline mutant G1 females, and siblings from both the G1 and G2 generations are crossed. The chance for homozygosity for each G1 mutation is only 1 in 16 in every G3 mouse. Red asterisks indicate mutations originating from the G0 sire; blue asterisks indicate mutations originating in the G1 dam. Star size indicates derivation from an immediate ancestor (large) or from a more remote ancestor (small). Generations (yellow boxes) are aligned. This strategy has the advantage of introducing X-linked mutations into the pedigree, and causes homozygosity for autosomal mutations at a rate 1.39 times greater than would be the case if additional mutations were not introduced by breeding each G0 with a G1 mutant female. Illustration made with assistance of Marie Dauenheimer.
Inbreeding protocol for generating homozygous mutations. G0 mice are bred to germline mutant G1 females, and siblings from both the G1 and G2 generations are crossed. The chance for homozygosity for each G1 mutation is only 1 in 16 in every G3 mouse. Red asterisks indicate mutations originating from the G0 sire; blue asterisks indicate mutations originating in the G1 dam. Star size indicates derivation from an immediate ancestor (large) or from a more remote ancestor (small). Generations (yellow boxes) are aligned. This strategy has the advantage of introducing X-linked mutations into the pedigree, and causes homozygosity for autosomal mutations at a rate 1.39 times greater than would be the case if additional mutations were not introduced by breeding each G0 with a G1 mutant female. Illustration made with assistance of Marie Dauenheimer.
Almost all phenotypes induced by ENU emanate from changes in coding sense, caused by missense, nonsense, or splicing errors. Table 2 summarizes the characteristics of 83 single base substitutions produced with ENU, detected as phenovariants, and identified in our laboratory to date. Among these 83 mutations, all produced coding change. Fifty-four (65%) of mutations produced missense codons; 12 (14%) produced nonsense codons; 17 (20%) caused aberrant splicing, and of these, all were located within 10 bp of a splice junction.
Nonselected base substitutions (n = 83) that induce phenovariance caused by ENU and identified by positional cloning
| Type . | Change . | No. . | Change . | No. . | Total . | Fraction, % . |
|---|---|---|---|---|---|---|
| Transition | A → G | 12 | T → C | 13 | 25 | 30.1 |
| Transversion | A → T | 12 | T → A | 19 | 31 | 37.3 |
| Transversion | A → C | 1 | T → G | 2 | 3 | 3.6 |
| Transversion | C → A | 6 | G → T | 7 | 13 | 15.7 |
| Transition | C → T | 7 | G → A | 4 | 11 | 13.3 |
| Transversion | C → G | 0 | G → C | 0 | 0 | 0 |
| Type . | Change . | No. . | Change . | No. . | Total . | Fraction, % . |
|---|---|---|---|---|---|---|
| Transition | A → G | 12 | T → C | 13 | 25 | 30.1 |
| Transversion | A → T | 12 | T → A | 19 | 31 | 37.3 |
| Transversion | A → C | 1 | T → G | 2 | 3 | 3.6 |
| Transversion | C → A | 6 | G → T | 7 | 13 | 15.7 |
| Transition | C → T | 7 | G → A | 4 | 11 | 13.3 |
| Transversion | C → G | 0 | G → C | 0 | 0 | 0 |
In addition to single basepair changes, ENU can probably cause single base insertions or deletions, although these are very rare.
The forward strand changes have been tabulated for these mutations. Note that A→T transversions and A→G transitions are the most common ENU induced substitutions (comprising 67.4% of point mutations in total). Seventy-one percent of mutations affect A/T pairs, while 29% of mutations affect C/G pairs. C→G substitutions are rare (none was observed among 83 point mutations that caused phenovariance). Rarely, single base deletions, insertions, or doublet substitutions are observed as well and it is possible that these mutations are also caused by ENU.
Many of these mutations are currently displayed at http://mutagenetix.scripps.edu.
Mutations induced by ENU are often phenotypically distinct from those produced by gene targeting. Some yield hypomorphic but viable alleles of genes for which null alleles are embryonic lethal. Others produce neomorphic alleles (encoding proteins with functions qualitatively different from either the WT or null allele phenotypes). Others produce antimorphic (“dominant negative”) alleles. All can be useful in understanding the phenomenon under study.
TLR signaling as dissected by ENU mutagenesis
We are approaching a time when TLRs and the signaling apparatus they trigger will be viewed as elegant molecular machines, detecting defined molecular structures and reporting them to the responding cell.
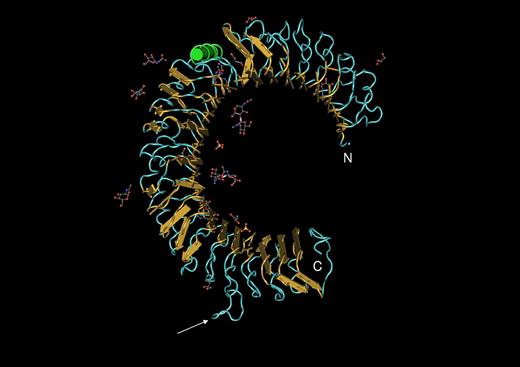
TLRs are homo- or heterodimeric receptor proteins of fairly large size (780-1100 aa in length). Approximately 80% of the polypeptide chain projects above the plasma membrane (for TLRs 1, 2, 4, 5, and 6) or into the endosomal vesicle (for TLRs 3, 7, 8, 9, and probably 13) and is mostly composed of leucine-rich repeats. Without known exceptions such proteins adopt a curved solenoid shape (Figure 4), which has been verified for several of the TLRs by X-ray crystallography.50-53 TLRs sometimes act in complexes with other proteins. For example, TLR4 cannot signal (and perhaps cannot reach the cell surface) in the absence of MD-2, a small secreted protein that is tightly associated with the TLR4 ectodomain. As already mentioned, CD14 also participates in TLR4 signaling, though the precise relationship between the 2 molecules remains unclear. TLR2 signals in conjunction with either TLR1 or TLR6, and never as a homodimer.
The ectodomain of a TLR3 subunit.51 “Worms” rendering with yellow arrows indicating beta sheet and green coil indicating alpha helix, imaged with the program CN3D. From NCBI protein database, PDB 1ZIW. Amino terminal (N) and carboxy terminal (c) ends of the structure are indicated. Loops outside the solenoid (arrow) may give flexibility to the protein. Carbohydrate residues (ball and stick) may influence association between subunits and/or binding of ligand.
The ectodomain of a TLR3 subunit.51 “Worms” rendering with yellow arrows indicating beta sheet and green coil indicating alpha helix, imaged with the program CN3D. From NCBI protein database, PDB 1ZIW. Amino terminal (N) and carboxy terminal (c) ends of the structure are indicated. Loops outside the solenoid (arrow) may give flexibility to the protein. Carbohydrate residues (ball and stick) may influence association between subunits and/or binding of ligand.
The relative complexity of TLR signaling on the cytoplasmic side (with inferred chaining of adaptors and in some cases the incorporation of multiple adaptor types into the active complex) permits qualitative differences in signaling by individual TLRs depending upon the stimulus used. Moreover, at least in some instances, TLR complexes have a “modular” design. For example, TLR4 retains some signaling potential even in the absence of CD14. It is still able to sense rough (poorly glycosylated) LPS or lipid A, but can no longer sense smooth (highly glycosylated) LPS. Moreover, when activated by lipid A in the absence of CD14, TLR4 signals only via the MyD88/TIRAP pathway (and not via the TRIF/TRAM pathway).54 These findings suggest that both the supramolecular organization of the TLR4 complex and the nature of the ligand used determine how the receptor will “fire” when activated. TLR4 may be thought of as a switch with multiple stops.
TLR2 is also partially dependent on CD14 for signaling, whether in complex with TLR1 or TLR6. And CD36 behaves as an accessory component of the TLR2/TLR6 heterodimer.55 TLR2 is also believed to associate with Dectin-1 to cooperate in the detection of certain glycans. Indeed, TLR2 and TLR4 can be activated by several structurally diverse ligands (Table 1). Some of these are microbial signature molecules, but others are endogenous, and some are proteins (discussed below).
While only one component of the TLR9 receptor complex is known, more components are suspected based on unpublished genetic data (the Effete mutation impairs TLR9 signaling but does not reside in TLR9 itself). Here too, different ligands produce different effects, with type A CpG oligonucleotides promoting inflammatory cytokine production more strongly than type I IFN, and type B CpG oligonucleotides doing the opposite. This would imply that the type B oligos stimulate the recruitment of IRF7 more efficiently than the type A oligos, and again, might suggest at least 2 distinguishable active conformations of the receptor.
It is generally agreed that signaling causes recruitment of adapter proteins to the TIR domain of the receptor. The nature of the interaction involves both the so-called BB loop (within which the residue altered by the classical Lps mutation resides, as well as the residue altered by the more recently identified ENU-induced allele Lps3), and the Poc site, mutated in the strain Pococurante. Pococurante is a receptor-selective MyD88 mutation that permits signaling via the TLR2/6 heterodimer but not the other TLRs. It gives an inferential model as to how MyD88 signals.56 MyD88 itself undergoes dimerization, as do the TIR domains of receptors. This most likely entails association between the αE helices of the TIR domains.56,57 It is likely, in fact, that “chains” of adapter proteins form upon receptor engagement leading to the next phase of signaling, which entails recruitment of IRAK isozymes (particularly IRAK1 and IRAK4), via interactions involving homotypic death domains in the adapters and kinases. IRAK1 is phosphorylated by IRAK4,58 and further recruitment of TRAF6 to the complex then occurs. TRAF6, a Zn++/RING finger domain protein, is endowed with E3 ubiquitin ligase activity, and activation of TRAF6 entails the attachment of a polyubiquitin chain to specific sites on the molecule,59 which in turn act as anchorage sites for TAB1, TAB2, and TAK1, the TGFβ activated kinase (also known as MAP3K7), and most probably the MEK kinase Tpl2 (also known as MAP3K8). These kinases each participate in the activation of MEK kinases, as well as the IκB kinase complex. The latter in turn causes degradation of IκB, and subsequent translocation of NF-κB to the nucleus. Cofactors for NF-κB function include the recently discovered Akirins.60 NF-κB causes transcriptional activation of numerous cytokine genes. Other kinase signaling events (still poorly understood) cause posttranscriptional changes in the expression of numerous cytokine mRNAs.61,62 A schematic illustration of the TLR signaling pathways defined by ENU mutagenesis to date is presented in Figure 5. Mutations induced by ENU affect perhaps one-half of the proteins currently known to be required for TLR signaling, and a cohesive picture can be assembled on this basis.
TLR signaling pathways established by mutagenesis. Each red × represents a different mutation. The relationship of proteins within the signaling pathways has been deduced biochemically and by reference to domain structures of the target molecules. Some mutations have yet to be found, but affect proteins critical for signal transduction. Illustration made with assistance of Marie Dauenheimer.
TLR signaling pathways established by mutagenesis. Each red × represents a different mutation. The relationship of proteins within the signaling pathways has been deduced biochemically and by reference to domain structures of the target molecules. Some mutations have yet to be found, but affect proteins critical for signal transduction. Illustration made with assistance of Marie Dauenheimer.
Signal attenuation and the development of autoimmunity
Phosphatases,63-65 de-ubiquitinating enzymes,66-69 and other forms of inhibition impede cell activation via the TIR domain signaling pathways. An important level of control was recently identified through mutagenesis. Mutations affecting the protein tyrosine phosphatase SHP1 have long been known to cause a severe autoinflammatory and autoimmune disease, exemplified by the phenotype Motheaten (Me), which affects the SHP1-encoding Ptpn6 gene.70 A milder hypomorphic allele of Ptpn6, called Spin, was shown to cause inflammatory and autoimmune disease as well. This disease was fully suppressed by introducing the mice into a germ-free environment, or by mutations in MyD88, IRAK4, or the interleukin-1 receptor IL-1R1. It appears that microbes initiate signaling via TLRs, causing IL-1 production, which in turn acts through its own receptor to cause more IL-1 production. The endless loop appears to be the basis of autoimmune disease, normally prevented by the “braking” effect of SHP1.71
Similar endless loops may cause other severe inflammatory diseases, such as hemophagocytic lymphohistiocytosis, modeled in mice by an ENU-induced mutation called Jinx, affecting Unc13d, homologous to the human MUNC13-4 gene.72 In this case, the mutation prevents CTL and NK cell degranulation. A viral stimulus (lymphochoriomeningitis [LCMV] infection) provokes CD8 cell activation, which causes production of IFNγ. However, because there is no degranulation, eradication or control of the viral infection do not occur. IFNγ production, in turn, provokes myeloid cell expansion and increases the burden of virus with which the host must cope. An endless cycle follows in with myeloid cells present antigen to CD8 cells, driving increased expansion and reciprocal stimulation of the myeloid cells.
Finally, in an archetypal autoimmune disease (systemic lupus erythematosus; SLE) it has been shown that within B cells that have engulfed chromatin or ribonucleoproteins through the B-cell receptor, endosomal TLR9 or TLR7/8 signaling drives cell activation and expansion of the autoreactive clone.73 This may be an important positive feedback loop, just like the others mentioned previously.
Conclusions and prospects for future understanding
Forward genetic methods—those that begin with phenotype—have the special virtue of producing surprises. As briefly described here, genetics revealed the key entry points to microbe sensing and the innate immune response, which encompasses inflammation, with its well known (though incompletely understood) ties to the adaptive immune response. Genetics continues to aid in the dissection of the molecular events that attend microbe sensing. It has also given fresh insight into the nature of untoward inflammation and of true autoimmunity, in which the adaptive immune response attacks self. As the field of immunology moves forward, fresh attention will certainly be focused on the TLRs and the signaling pathways to which they are linked. They are among the most important receptors ever discovered and, just as they are tied to the initial phase of immune defense, they are tied to some of the most basic mechanisms of disease.
Acknowledgments
Funding for the research in this paper was provided by the National Institutes of Health (grant number HHSN272200700038C).
National Institutes of Health
Authorship
Contribution: B.A.B. wrote the paper.
Conflict-of-interest disclosure: The author declares no competing financial interests.
Correspondence: Dr Bruce A. Beutler, Department of Genetics, The Scripps Research Institute, 10550 N Torrey Pines Road, La Jolla, CA 92037; e-mail: bruce@scripps.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal