In this issue, Joshi and colleagues show that Notch-1 regulates cell cycle by targeting expression of cyclinD3 and Cdk4/6. This article adds to the growing body of literature pointing to aberrant Notch activity as a master controller during T-ALL pathogenesis.
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive leukemia with a poor outcome. The majority of human T-ALL has gain-of-function (activating) mutations in Notch-1,1 implicating Notch-1 as a central component in the pathogenesis of T-ALL. Notch proteins are widely expressed transmembrane receptors involved in many cellular processes such as differentiation, proliferation, and apoptosis. Notch-1 signaling is critical for normal T-cell development. However, aberrant regulation of Notch, leading to persistent activation, leads to unchecked cell growth. All members of the Notch family (Notch 1-4) have been implicated in tumorogenesis.2 Notch ligands are members of Delta-Serrate-Lag2 (DSL) family that are expressed on neighboring cells. Ligand binding causes a 2-step proteolytic cleavage of Notch that releases the active intracellular domain of Notch (N1C) from the cell surface membrane. The last cleavage step is mediated by γ-secretase complex. N1C translocates to the nucleus, where it functions as a transcription factor. In the canonical NOTCH signaling pathway, Notch binds to the transcription factor CSL, displaces transcriptional repressors, recruits activators, and thus increases transcription of target genes.
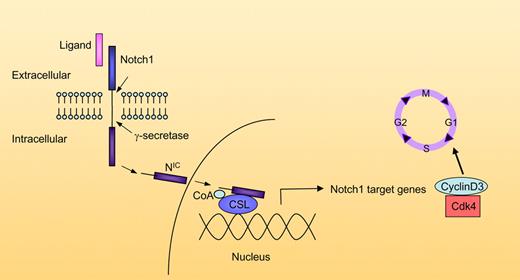
Schematic representation of Notch pathway interactions with the cell cycle. The ligand (DSL) binds to the extracellular domain of the Notch receptor. Following ligand binding, Notch is cleaved at the extracellular domain. The membrane-bound intracellular domain is then cleaved by the γ-secretase complex, releasing the active intracellular domain NIC. NIC moves to the nucleus, where it binds to CSL, recruits coactivators (CoA), and induces gene transcription. Target Notch genes include cyclinD3 and Cdk4, which promote progression through the cell cycle. Additional target genes are not shown.
Schematic representation of Notch pathway interactions with the cell cycle. The ligand (DSL) binds to the extracellular domain of the Notch receptor. Following ligand binding, Notch is cleaved at the extracellular domain. The membrane-bound intracellular domain is then cleaved by the γ-secretase complex, releasing the active intracellular domain NIC. NIC moves to the nucleus, where it binds to CSL, recruits coactivators (CoA), and induces gene transcription. Target Notch genes include cyclinD3 and Cdk4, which promote progression through the cell cycle. Additional target genes are not shown.
In the current study, Joshi et al demonstrate that Notch-1 regulates expression of components of cell-cycle machinery, which contribute to dysregulated cell growth.3 Specifically, they demonstrate that Notch-1 directly binds to the cyclinD3 promoter and increases cyclinD3 expression. In addition, Notch-1 increases expression of Cdk4 and Cdk6, the catalytic partners of cyclin D3. The Cdk4-cyclin D3 complex is essential for progression through G1/S phase of cell cycle. The authors also demonstrate that Cdk4 and cyclin D3 expression is dependent on Notch expression in a mouse model of T-cell leukemia. In Notch-dependent T-cell lymphomas, Cdk4 and cyclinD3 are increased, and the increase can be abrogated by use of inhibitors of γ-secretase (GSI). In further support of the concept that cyclinD3 is downstream of Notch, mice lacking cyclinD3 are protected from the development of leukemias induced by the activated, intracellular form of the Notch.4 Thus, their data support a direct role of Notch in regulating cell-cycle progression in which activating mutations of Notch would be able to enhance transformation by deregulating cell growth.
Because of its central role in T-ALL pathogenesis, Notch is an obvious therapeutic target. Since cleavage and release of N1C by γ-secretase is a critical step in Notch signaling, small molecular inhibitors of γ-secretase (GSI) have been proposed for use in T-ALL. Indeed, in vitro GSI administration causes cell-cycle arrest of T-ALL cell lines with activating mutations in Notch1. However, early clinical trials of GSI in refractory T-ALL have not shown significant clinical response.5 The current study suggests that combinatorial therapy with inhibitors of Notch and inhibitors of cell cycle may be beneficial to a subset of patients with T-ALL. However, targeting Notch signaling has additional complexities. For example, Notch can decrease expression of p27, a Cdk inhibitor; increase expression of Myc; and upregulate of the PI3K-AKT signaling pathway. Thus, identification of additional Notch target genes and molecular characterization of T-ALL mutations will facilitate development of rational therapy for T-ALL patients.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal