Abstract
Notch signaling plays a role in normal lymphocyte development and function. Activating Notch1-mutations, leading to aberrant downstream signaling, have been identified in human T-cell acute lymphoblastic leukemia (T-ALL). While this highlights the contribution of Notch signaling to T-ALL pathogenesis, the mechanisms by which Notch regulates proliferation and survival in normal and leukemic T cells are not fully understood. Our findings identify a role for Notch signaling in G1-S progression of cell cycle in T cells. Here we show that expression of the G1 proteins, cyclin D3, CDK4, and CDK6, is Notch-dependent both in vitro and in vivo, and we outline a possible mechanism for the regulated expression of cyclin D3 in activated T cells via CSL (CBF-1, mammals; suppressor of hairless, Drosophila melanogaster; Lag-1, Caenorhabditis elegans), as well as a noncanonical Notch signaling pathway. While cyclin D3 expression contributes to cell-cycle progression in Notch-dependent human T-ALL cell lines, ectopic expression of CDK4 or CDK6 together with cyclin D3 shows partial rescue from γ-secretase inhibitor (GSI)-induced G1 arrest in these cell lines. Importantly, cyclin D3 and CDK4 are highly overexpressed in Notch-dependent T-cell lymphomas, justifying the combined use of cell-cycle inhibitors and GSI in treating human T-cell malignancies.
Introduction
Notch proteins are a family of ligand-activated large (300 kDa) single-pass transmembrane heterodimeric receptors.1 Notch controls multiple cell fate decisions and differentiation processes during lymphocyte development and function and is required at various stages of T-cell development.2,3 Deregulated Notch signaling during T-cell development leads to malignant transformation, including the cancer most closely associated with aberrant Notch expression in humans, acute T-cell acute lymphoblastic leukemia (T-ALL), which constitutes approximately 15% to 20% of ALLs seen in adults and children.4,5 The oncogenic potential of Notch was first identified in (t7;9) chromosomal rearrangement in approximately 2% of human T-ALL, whereby intracelluar Notch1 is translocated to the T-cell receptor (TCR) β gene.6 More than 50% of human T-ALLs bear mutations in Notch1, indicating a prominent role for Notch in this T-cell malignancy.7 Inhibitors of Notch signaling abrogate the growth of human and murine T-ALL cell lines bearing Notch1 gain-of-function mutations, indicating Notch is required in established tumors.8,9
In vertebrates, 4 notch receptors (Notch 1-4) are activated by 5 different Notch ligands expressed on various cell types: Jagged1, Jagged2, and Delta-like (DL)1, DL3, and DL4.2,3 After ligand-binding, proteolytic cleavage by γ-secretase releases the signaling-competent intracellular domain of Notch (NIC).10-12 NIC is composed of a RAM domain, ankyrin repeats (ANK) that mediate protein-protein interactions, nuclear localization sequences, a transactivation domain (TAD), and a C-terminal PEST domain regulating protein turnover. Human T-ALL cases frequently bear activating mutations in the extracellular heterodimerization domain and/or the C-terminal PEST domain of Notch1, resulting in ligand-independent activation.7 During canonical Notch signaling, NIC translocates to the nucleus, engages its nuclear binding protein CSL (CBF-1, mammals; suppressor of hairless, Drosophila melanogaster; Lag-1, Caenorhabditis elegans) and transcribes downstream target genes, including the HES family of transcriptional repressors.13,14 In the absence of NIC, CSL recruits repressor complexes to the regulatory regions of Notch/CSL target genes, inhibiting transcription. NIC interaction with CSL acts as a switch that promotes the assembly of CSL coactivator complexes.15-17 γ-Secretase inhibitors (GSIs) block proteolytic cleavage of Notch receptors, thereby preventing activation of Notch. Use of GSI in activated T cells results in down-regulation of nuclear factor (NF)-κB activity, cytokine (interleukin-2 [IL-2] and interferon-γ [IFN-γ]) production, and cell proliferation.18
In T-cell lymphomas, context-specific putative target genes have been identified through which Notch1 may promote transformation by altering cell-growth kinetics.19-21 The D-type cyclins (cyclins D1, D2, and D3) are the first cyclins to be induced as cells enter the G1 phase of the cell cycle,22-24 and, thus, if regulated by Notch signaling, are highly relevant to the mechanism of Notch-induced lymphomagenesis. D-type cyclins associate with and activate cyclin-dependent kinases (CDK), CDK4 and CDK6.23 Activation of CDKs at specific time points during the cell cycle regulates phosphorylation and inactivation of the retinoblastoma protein (Rb) and derepression of E2F transcription factors, driving the cell into S phase.22,23 Notch activation up-regulates cyclin D1 and CDK2 activity in rat kidney epithelial cells increasing cell proliferation.25 Notch induces transcription of S phase kinase-associated protein 2 (SKP2), which targets CDK-inhibitors p27 and p16 for proteosomal degradation, resulting in premature entry into S phase.26 In pancreatic cancer cells, down-regulating Notch1 induces a G0/G1 cell-cycle arrest associated with reduced levels of cyclin D1 expression and increased p21CIP and p27KIP1 expression.27 Notch signaling is also a potent regulator of cell cycle in human T-ALL cell lines, because treating them with GSI induces G0/G1 cell-cycle arrest.7,9
Activated human primary T lymphocytes up-regulate cyclin D3,28 correlating with entry of these cells into G1.29 Cyclin D3−/− mice show defective thymocyte development, characterized by a marked deficit of CD4+CD8+ (double positive [DP]) T cells, and lack the proliferative burst seen during the DN-3 to DN-4 transition.30 This developmental requirement for cyclin D3 extends to experimental models of T-ALL, because cyclin D3−/− mice are resistant to T-ALL induction by activated forms of Notch1.30 In addition, cyclin D3 is frequently overexpressed in cancers of the lymphoid system.31-33
Based on these data, we sought to determine how Notch regulates cell-cycle progression in T cells and whether cyclin D3 is a direct target of Notch. Here we demonstrate that Notch regulates cyclin D3 expression in mature T cells and transformed T lymphoblasts, and propose a possible mechanism for regulated cyclin D3 expression in activated T cells. D-type cyclins function together with their catalytic partners CDK4 and CDK6 to facilitate cell-cycle progression, and we also show CDK4 and CDK6 are Notch target genes in peripheral T cells and in lymphomas arising in transgenic animals overexpressing Notch1. Our study reveals the oncogenic potential of aberrant Notch signaling in T cells through its deregulation of the cell cycle. It further suggests that cell-cycle regulation by Notch may represent a target for therapeutic intervention using small molecule inhibitors/modifiers in Notch-driven T-cell leukemias.
Methods
Mice, CD4+ T-cell preparation, and GSI treatment
C57BL/6 mice were housed in the University of Massachusetts animal care facility in accordance with Institutional Animal Care and Use Committee (IACUC) guidelines. CD4+ T cells were isolated from 2- to 3-month-old mice using the IMag magnetic bead system (BD Pharmingen, BD Biosciences, San Jose, CA), and 2.25 × 106/mL were pretreated in vitro at 37°C for 20 minutes with 0.1% dimethyl sulfoxide (DMSO) or with 50 μm zIL-CHO in DMSO, a small peptide inhibitor of γ-secretase, then plated onto 12-well plates precoated with 1 μg/mL αmouseCD3ϵ + αmouseCD28. For in vivo analysis, GSI (LY-411,575) in chow, formulated to deliver 5 mg/kg per day, was given to mice for 13 days; control animals received normal chow.
Lysate preparation, immunoblotting, and antibodies
Whole cell lysates were prepared with radio immunoprecipitation assay (RIPA) buffer. Preparation of nuclear extracts was described previously.34 Immunoblotting was performed using anti–cyclin D3, anti-CDK4, anti-CDK6 (Santa Cruz Biotechnology, Santa Cruz, CA); anti-GAPDH (Chemicon International, Billerica, MA); anti–cleaved Notch1 (eBiosciences, San Diego, CA); anti–phospho-Ser780-pRb, and anti–phospho-Ser795-pRb (Cell Signaling Technologies, Danvers, MA).
Dual luciferase assay and reverse transcription–polymerase chain reaction
293T cells (106) were transfected with indicated Notch expression vectors or empty vector using Fugene6 (Roche, Indianapolis, IN). We used 0.4 μg of D3-Luc reporter plasmid in each experiment and 0.1 μg PRL-CMV (pRL-CMV Vector contains the CMV enhancer and early promoter elements to provide expression of Renilla luciferase in cotransfected mammalian cells). Luciferase assays were performed at 48 hours, following the manufacturer's instruction (Dual Lucifersae Assay System; Promega, Madison, WI). For reverse transcription–polymerase chain reaction (RT-PCR), total RNA was isolated using RNA-BEE (Tel-test, Friendswood, TX), following the manufacturer's protocol. Reverse transcription was performed with oligo-dT primers (Invitrogen, Carlsbad, CA); cDNA amplified by PCR using cyclin D3 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers and conditions as described.35
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) analysis was performed on 106 CD4+ T cells using ChIP assay kit (Cell Signaling Solutions; Upstate, Millipore, Billerica, MA) following the manufacturer's protocol. Eluted material was reverse cross-linked and DNA-purified with PCR purification kit (QIAGEN, Valencia, CA) for PCR. Primer Set1 (227 bp, annealing temperature: 52°C) forward: 5′-GAGAATTCGGATCTCACTGCTTA-3′, reverse: 5′-CCGGCAGATTTCTTTGAGTTC-3′; Primer Set2 (297 bp, annealing temperature: 54°C) forward: 5′-TAGCCCAGGCTACTCCTGAA-3′ reverse: 5′-TCTCAGACCCAGGGAAGTGT-3′. PCR conditions: 94°C for 5 minutes, 94°C for 1 minute, 52°C for 1 minute, 72°C for 1 minute for 35 cycles, then 72°C for 7 minutes. Antibodies used: rabbit αNotch1, rabbit αRBP-Jκ, goat αp50, normal rabbit immunoglobulin G (IgG), normal goat IgG (Santa Cruz Biotechnology).
Human T-ALL cell lines, GSI treatment
Human T-ALL cell lines DND-41, HPB-ALL, and T-ALL1 were maintained in media as described.7 For GSI treatment, 3 μm zIL-CHO were used for 7 days with 0.1% DMSO used as vehicle control.
Cell-cycle analysis
DND-41, HPB-ALL, and T-ALL1 were analyzed for cell-cycle progression using DRAQ5 (AXXORA, San Diego, CA). At time of analysis, cells were resuspended in 5 × 105 cells/mL phosphate-buffered saline (PBS), and 5 μm DRAQ5 were added directly to the tube and gently mixed. Cells were incubated at room temperature for 20 minutes and acquired on LSRII using FacsDiva software (both from Becton Dickinson, BD, Franklin Lakes, NJ). Data were analyzed using FlowJo (TreeStar, Ashland, OR).
Plasmids and retroviral infection
pMSCV-IRES-YFP-cyclin D3 expression plasmid was a kind gift from Dr Suzanne J. Baker (St Jude Children's Research Hospital, Memphis, TN). pMSCV-IRES-DsRed-CDK4 or -CDK6 retroviral construct was generated by subcloning CDK4, CDK6 (pCMV-CDK4 or CDK6 expression plasmid, kind gift of Dr Phil W. Hinds (Tufts-New England Medical Center, Boston, MA) into pMSCV-IRES-DsRed vector (gift from Dr Dario A.A. Vignali, St Jude Children's Research Hospital). Expressed proteins of expected size were confirmed by transient expression studies in 293T cells. For retroviral supernatants, 1.5 × 105 293T cells were plated onto 60-mm dishes, 1 day before transfection. The next day, 4 μg of the expression plasmid and 2 μg of the retroviral packaging plasmid pCL-Ampho (Imgenex) were transfected using 18 μL of Fugene6. Forty-eight hours after transfection, viral supernatant was harvested and cleared by spinning at 290g (1500 rpm) for 5 minutes. Two milliliters supernatant were mixed with 16 μL Fugene6 and added to cell suspension (human T-ALL cell line, 106 cells/mL/well) in 6-well culture dishes. Cells were spin-infected at 804g (2500 rpm) for 1 hour at 30°C, then placed into 100-mm culture dishes with fresh supernatant. Cells were cultured at 37°C with 5% CO2 for 48 hours, then placed in fresh media. Twenty-four hours after replacement, infection efficiency was analyzed on an LSRII. After 7 days' culture, cells were sorted for enrichment by gating for fluorescent protein expression (yellow fluorescent protein [YFP], cyclin D3; YFP and DsRed, cyclin D3 and CDK4 or CDK6) using a FACSVantage (Becton Dickinson).
Calculating relative G1 rescue index
The relative G1 rescue index (RI) was calculated to represent the rescue from GSI-induced G1 arrest observed in transduced human T-ALL cell lines relative to G1 arrest observed in wild-type human T-ALL cell lines (Tables S1,2, available on the Blood website; see the Supplemental Materials link at the top of the online article). Difference in percentage of cells in G1 (ΔG1) was calculated by subtracting percentage of cells in G1 with DMSO treatment from percentage of cells in G1 after 7 days' GSI treatment: ΔG1 = percentage of cells in G1GSI − percentage of cells in G1DMSO. δG1 was calculated for all wild-type cell lines (uninfected) and transduced cell lines infected either with vector only (pMSCV-IRES-DsRed II or pMSCV-IRES-YFP) or expression construct (pMSCV-IRES-DsRed II-CDK4 or CDK6 and/or pMSCV-IRES-YFP-cyclin D3). δG1 of transduced cells (vector and cyclin D3 and/or Cdk-4/6) was normalized to wild-type cells: nδG1 = δG1transduced /δG1wild-type × 100. The relative G1 RI was calculated by subtracting nδG1 from 100 for human T-ALL cell lines and represented graphically as: RIT-ALL = 100 − nδG1. Complete G1 rescue is defined as RI = 100.
Analysis of TOP-Notch, MIG-Nic leukemic mice
To generate TOP-Notch/MIT-RX leukemic mice, bone marrow from TOP-Notch animals was transduced with the tetracycline transactivator expression construct, MIT-RX as described previously.36,37 To generate MIG-Nic leukemic mice, wild-type FVB bone marrow was infected with MIG-NicδPEST vector as described above.37 Spleens from diseased animals or wild-type FVB controls were used. Leukemia transplant experiments for tumor regression studies were performed using TOP-Notch/Eμ-tTA mice as described in Table S1. At indicated time points, animals were harvested to generate samples for RT-PCR and Western blot analysis.
Results
Notch signaling regulates cyclin D3 expression in CD4+ T cells in vitro and in vivo
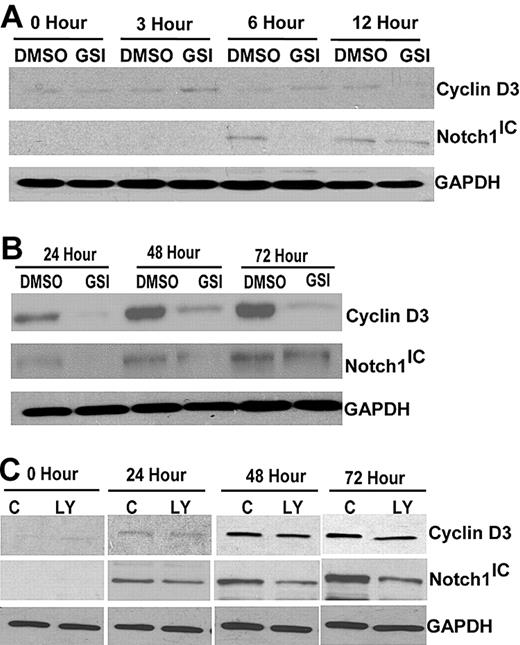
Previous experiments in our laboratory demonstrated Notch expression is induced by TCR signaling.18 To examine whether Notch signaling is required for expression of cyclin D3 in primary T cells, CD4+ T cells isolated from spleens of C57BL/6 mice were stimulated with anti-CD3ϵ and anti-CD28 for different length of times. There was no significant increase in cyclin D3 expression up to 12 hours after stimulation (Figure 1A). We also stimulated CD4+ T cells in the presence of the GSI, 50 μM IL-CHO, after determining that this concentration reduced Notch-1 levels (Figure S1), and found there was little or no effect on cyclin D3 expression compared with vehicle control, DMSO, during the first 12 hours (Figure 1A). However, by 24 hours after stimulation, there was an increase in cyclin D3 protein that continued to rise up to 72 hours after stimulation (Figure 1B). Abrogating Notch signaling with GSI-treatment decreased cyclin D3 expression at 24, 48, and 72 hours after stimulation (Figure 1B). We confirmed our observations using another GSI, DAPT (N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester), and found similar effects, confirming decreased cyclin D3 expression was a result of blocking Notch activity. Next, C57BL/6 mice were treated with GSI (LY-411,575) formulated in rodent chow to deliver 5 mg/kg per day for 2 weeks (Figure S2) to determine how attenuating Notch signaling in vivo affects cyclin D3 expression. CD4+ T cells from GSI-treated and control chow-fed mice were isolated and stimulated with anti-CD3ϵ and anti-CD28 up to 72 hours. Consistent with in vitro effects of GSI treatment on cyclin D3 expression, treating mice in vivo with GSI also reduced cyclin D3 expression, compared with control animals, and this correlated with reduced Notch1 expression (Figure 1C). Together, these data suggest, in stimulated CD4+ T cells, cyclin D3 expression is a downstream target of Notch signaling.
Cyclin D3 expression is dependent on Notch signaling in CD4+ T cells. (A) Whole cell lysates of CD4+ T cells were analyzed by immunoblotting for cyclin D3 or Notch1IC expression. CD4+ T cells were stimulated with 1 μg/mL anti-CD3ϵ and 1 μg/mL anti-CD28 for indicated time periods (0, 3, 6, and 12 hours) in the presence of GSI (IL-CHO, 50 μm) or DMSO (0.1%). (B) Later time points (24, 48, and 72 hours) after 1 μg/mL anti-CD3ϵ and 1 μg/mL anti-CD28 stimulation were analyzed for differences in cyclin D3 and Notch1IC expression with or without GSI treatment. (C) Whole cell lysates from stimulated CD4+ T cells (for 24, 48, and 72 hours) that were isolated from C57BL/6 mice treated in vivo with GSI for 14 days were analyzed for cyclin D3 and Notch1IC. GAPDH was used as a loading control. LY indicates GSI in rodent chow; and C, control rodent chow.
Cyclin D3 expression is dependent on Notch signaling in CD4+ T cells. (A) Whole cell lysates of CD4+ T cells were analyzed by immunoblotting for cyclin D3 or Notch1IC expression. CD4+ T cells were stimulated with 1 μg/mL anti-CD3ϵ and 1 μg/mL anti-CD28 for indicated time periods (0, 3, 6, and 12 hours) in the presence of GSI (IL-CHO, 50 μm) or DMSO (0.1%). (B) Later time points (24, 48, and 72 hours) after 1 μg/mL anti-CD3ϵ and 1 μg/mL anti-CD28 stimulation were analyzed for differences in cyclin D3 and Notch1IC expression with or without GSI treatment. (C) Whole cell lysates from stimulated CD4+ T cells (for 24, 48, and 72 hours) that were isolated from C57BL/6 mice treated in vivo with GSI for 14 days were analyzed for cyclin D3 and Notch1IC. GAPDH was used as a loading control. LY indicates GSI in rodent chow; and C, control rodent chow.
Notch1IC directly regulates cyclin D3 promoter activity
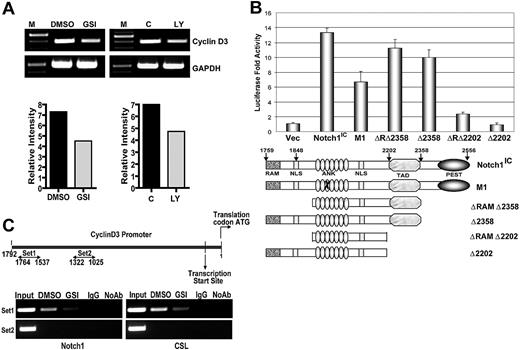
Next, we asked whether Notch signaling influences cyclin D3 expression at the transcript level. CD4+ T cells isolated from spleens of C57BL/6 mice were pretreated with GSI or DMSO then stimulated with anti-CD3ϵ and anti-CD28. After 24 hours, we assessed cyclin D3 transcript levels by RT-PCR. After stimulation, transcription of cyclin D3 was decreased in cells treated with GSI, compared with controls (Figure 2A). In vivo abrogation of Notch signaling also resulted in reduced transcript levels (Figure 2A). We used the dual luciferase assay to determine whether Notch1 regulates expression from the cyclin D3 promoter. Ectopic expression of Notch1IC increased cyclin D3 reporter activity greater than 10-fold, compared with the control plasmid (Figure 2B). To characterize the domain of Notch1IC that is required for regulating the cyclin D3 promoter, we again performed dual luciferase reporter gene assays. Cotransfection of various Notch1IC constructs with the cyclin D3 reporter revealed that the C-terminal TAD is required for cyclin D3 promoter activity (Figure 2B). To determine whether Notch binds directly to the cyclin D3 promoter, we performed ChIP using CD4+ T cells isolated from spleens of C57BL/6 mice, and stimulated with anti-CD3ϵ and anti-CD28 for 24 hours. We immunoprecipitated protein-bound DNA using anti-Notch1, followed by PCR using different primer sets specific for the cyclin D3 promoter region. We were able to amplify a 227-bp region (−1764 to −1537) of the cyclin D3 promoter, indicating direct binding of Notch1 to the cyclin D3 promoter (Figure 2C). Inhibiting Notch activation with GSI treatment abrogated the recruitment of Notch1 to the cyclin D3 promoter (Figure 2C). To further investigate whether CSL is involved in Notch-mediated regulation of the cyclin D3 promoter, we performed ChIP with an antibody that immunoprecipitates CSL. We were able to amplify the same 227-bp region (−1764 to −1534) using anti-CSL, as when using anti-Notch1 and, again, GSI treatment diminished the recruitment of CSL to the promoter region (Figure 2C). We also examined other regions within the cyclin D3 promoter for putative Notch/CSL binding. We observed that a 297-bp region (−1322 to −1025) was not amplified by PCR after immunoprecipitation with either antibody (Figure 2C), suggesting that Notch/CSL interaction on the cyclin D3 promoter is specific, and both binding partners are recruited to a unique location within the promoter region. Together these data indicate that cyclin D3 is a direct target of the Notch/CSL signaling pathway.
Expression from cyclin D3 promoter is regulated by Notch signaling. (A) CD4+ T cells after in vitro GSI pretreatment and from mice treated in vivo with GSI were stimulated with anti-CD3ϵ + anti-CD28 for 24 hours. Expression of cyclin D3 transcript was determined by RT-PCR after isolation of RNA. (B) Cyclin D3 luciferase reporter plasmid was transiently cotransfected with either 0.1 μg Notch1IC expression plasmid or 0.1 μg of indicated Notch1IC mutants53 into 293T cells. Transfected cells were harvested 48 hours later for dual luciferase assay. The relative luciferase activity was calculated by normalizing against Renilla luciferase (pRL-CMV) activity that was used as an internal control. Values shown are representative of 3 separate experiments performed in triplicate. (C) ChIP of stimulated CD4+ T cells with or without GSI treatment. Anti-Notch1 (left panels) and anti-CSL (right panels) were used to precipitate protein complexes bound to DNA. Rabbit isotype control IgG was used as a negative control. Primers sets 1 (top panels) and 2 (bottom panels) were used in PCR to determine whether Notch1 and CSL are recruited to a specific region on cyclin D3 promoter.
Expression from cyclin D3 promoter is regulated by Notch signaling. (A) CD4+ T cells after in vitro GSI pretreatment and from mice treated in vivo with GSI were stimulated with anti-CD3ϵ + anti-CD28 for 24 hours. Expression of cyclin D3 transcript was determined by RT-PCR after isolation of RNA. (B) Cyclin D3 luciferase reporter plasmid was transiently cotransfected with either 0.1 μg Notch1IC expression plasmid or 0.1 μg of indicated Notch1IC mutants53 into 293T cells. Transfected cells were harvested 48 hours later for dual luciferase assay. The relative luciferase activity was calculated by normalizing against Renilla luciferase (pRL-CMV) activity that was used as an internal control. Values shown are representative of 3 separate experiments performed in triplicate. (C) ChIP of stimulated CD4+ T cells with or without GSI treatment. Anti-Notch1 (left panels) and anti-CSL (right panels) were used to precipitate protein complexes bound to DNA. Rabbit isotype control IgG was used as a negative control. Primers sets 1 (top panels) and 2 (bottom panels) were used in PCR to determine whether Notch1 and CSL are recruited to a specific region on cyclin D3 promoter.
NF-κB binds to the cyclin D3 promoter in primary T cells augmenting Notch-dependent cyclin D3 promoter activity
NF-κB has recently emerged as a nuclear binding partner for Notch, and both signaling pathways have been shown to be interconnected.34,38,39 Studies by our laboratory and others have documented that signaling through the TCR activates NF-κB, and various NF-κB family members have been shown to modulate cell cycle by regulating the cyclin D1 promoter.34,40-42 Our laboratory has also shown that Notch directly regulates the IFN-γ promoter, through complexes with p50. Therefore, we asked whether NF-κB regulates cyclin D3 in T cells, in conjunction with Notch signaling. To address this, we cotransfected 293T cells with the NF-κB subunit p50, along with Notch1IC and a cyclin D3 luciferase reporter construct. Our results show that coexpression of p50 induces a greater than 2-fold increase in Notch1IC-dependent transactivation of the cyclin D3 reporter (Figure 3A). Although initial cotransfection of p50 with equal amounts of Notch1IC correlates with increases in reporter activity, Notch1IC was the limiting factor in p50-mediated regulation. Furthermore, these data show that D3 reporter activity is Notch-dependent and p50 cooperates with Notch for this effect.
NF-κB binds to and augments Notch-dependent cyclin D3 promoter activity. (A) Cyclin D3 luciferase reporter plasmid (0.4 μg) and 0.1 μg pRL-CMV plasmid of internal control were transiently transfected into 293T cells. p50 and Notch1IC expression plasmids were cotransfected in ratios, as indicated, together with the reporter plasmids. Notch1IC was transfected in increasing amounts (0.1, 0.2, 0.3, and 0.4 μg) while the amount of p50 expression plasmid was kept constant (0.1 μg), and vice versa. Transfected cells were harvested 48 hours later for dual luciferase assay as previously described. Transfected cells were also harvested 48 hours later for whole cell lysates and immunoblotted for Notch-1 and p50. GAPDH was used as a loading control. (B) Cyclin D3 luciferase reporter plasmid was transiently cotransfected with either Notch1IC expression plasmid or Notch1IC mutants, as indicated, along with the p50 expression plasmid in 4:1 ratios (0.4 μg Notch1IC:0.1 μg p50) into 293T cells. Transfected cells were harvested 48 hours later for dual luciferase assay. Values shown for both luciferase assays are representative of 3 separate experiments carried out in triplicate. (C) ChIP of stimulated CD4+ T cells with or without GSI treatment (as described in “Methods”). Anti-p50 was used to precipitate protein complexes bound to DNA. Goat isotype control IgG was used as a negative control. Two different primer sets (1 and 2) were used to assess specificity of binding of p50 to the cyclin D3 promoter via PCR of immunoprecipitated DNA fractions. Results shown are representative of at least 3 separate experiments.
NF-κB binds to and augments Notch-dependent cyclin D3 promoter activity. (A) Cyclin D3 luciferase reporter plasmid (0.4 μg) and 0.1 μg pRL-CMV plasmid of internal control were transiently transfected into 293T cells. p50 and Notch1IC expression plasmids were cotransfected in ratios, as indicated, together with the reporter plasmids. Notch1IC was transfected in increasing amounts (0.1, 0.2, 0.3, and 0.4 μg) while the amount of p50 expression plasmid was kept constant (0.1 μg), and vice versa. Transfected cells were harvested 48 hours later for dual luciferase assay as previously described. Transfected cells were also harvested 48 hours later for whole cell lysates and immunoblotted for Notch-1 and p50. GAPDH was used as a loading control. (B) Cyclin D3 luciferase reporter plasmid was transiently cotransfected with either Notch1IC expression plasmid or Notch1IC mutants, as indicated, along with the p50 expression plasmid in 4:1 ratios (0.4 μg Notch1IC:0.1 μg p50) into 293T cells. Transfected cells were harvested 48 hours later for dual luciferase assay. Values shown for both luciferase assays are representative of 3 separate experiments carried out in triplicate. (C) ChIP of stimulated CD4+ T cells with or without GSI treatment (as described in “Methods”). Anti-p50 was used to precipitate protein complexes bound to DNA. Goat isotype control IgG was used as a negative control. Two different primer sets (1 and 2) were used to assess specificity of binding of p50 to the cyclin D3 promoter via PCR of immunoprecipitated DNA fractions. Results shown are representative of at least 3 separate experiments.
We next sought to determine which functional domains of Notch1IC are critical for its interaction with p50 in regulating the cyclin D3 promoter. Mutant Notch1IC constructs lacking the C-terminal region showed approximately 50% reduction in reporter activity compared with full-length Notch1IC. Consistent with our previous results, we observed that TAD was required for facilitating p50-mediated cyclin D3 promoter activity (Figure 3B). We then analyzed the cyclin D3 promoter for consensus NF-κB binding sites and found 2 putative binding sites within the 227-bp region (−1764 to −1537) that we previously immunoprecipitated for ChIP using anti-Notch and anti-CSL (Figure 2C). To further investigate whether NF-κB binds directly to the cyclin D3 promoter, we used anti-p50 antibody to perform ChIP assays. We isolated CD4+ T cells from spleens of C57BL/6 mice, pretreated them with GSI or DMSO, and stimulated them with anti-CD3ϵ and and anti-CD28 for 24 hours. After de-cross-linking DNA, we PCR-amplified the −1764 to −1534 region of the cyclin D3 promoter. We successfully amplified this region in DMSO-treated samples, indicating the p50 subunit of NF-κB binds directly to the cyclin D3 promoter and GSI treatment abrogates this interaction (Figure 3C). As a negative control, we used an additional primer set to amplify −1322 to −1025 of the cyclin D3 promoter that did not immunoprecipitate using antibodies against Notch1 or CSL. We were unable to amplify this region after retrieving de–cross-linked DNA fragments, confirming the specificity with which p50 binds to the cyclin D3 promoter (Figure 3C). Together, with our previous ChIP data (Figure 2C), these results indicate p50, CSL, and Notch1 are recruited to the cyclin D3 promoter to directly regulate cyclin D3 expression in primary T cells.
Cyclin D3 is a target of Notch signaling in human T-ALL cell lines, and its overexpression provides partial rescue from GSI-induced G1 arrest
Notch signaling regulates various genes critical to the molecular pathogenesis of T-ALL.5 Therefore, we asked whether cyclin D3 was a target of Notch signaling in human T-ALL cell lines and if overexpression of cyclin D3 could rescue these cell lines from the effects of GSI. As reported previously, DND-41, HPB-ALL, and T-ALL1 treated with GSI show G1 arrest,7,21 reiterating that transit through the cell cycle is Notch-dependent in these cell lines (Figure 4A). Next, we examined whether G1 arrest in DND-41, HPB-ALL, and T-ALL1 cell lines correlated with changes in cyclin D3 expression. Analysis of protein extracts from cell lines treated with GSI revealed reduced cyclin D3 expression when Notch signaling is abrogated compared with vehicle control, DMSO (Figure 4B). Decreased Notch1 expression in all cell lines confirmed the inhibitory effects of GSI treatment on Notch processing (Figure 4B). We observed similar results with DAPT, confirming cyclin D3 is a target of Notch signaling in human T-ALL cell lines (Figure S3). These results indicate cyclin D3 expression is temporally associated with cell-cycle progression in Notch-dependent human lymphomas. We then asked whether overexpressing cyclin D3 could rescue cell lines from the effects of GSI- treatment (Figure S4). DND-41, HPB-ALL, and T-ALL1 cell lines were retrovirally transduced with MSCV-cyclin D3-IRES-YFP or MSCV-IRES-YFP empty vector and sorted to enrich for the YFP-expressing population. Cyclin D3-YFP–expressing, as well as the YFP vector control, cells were divided and treated with either GSI or DMSO. We then compared differences in percentages of cells in G1 under the 2 treatment conditions (GSI or DMSO) in the transduced cell lines (cyclin D3-YFP or YFP-vector) with those of nontransduced cell lines to calculate G1 rescue indices (Table S1). Compared with vector alone, in the DND-41 and T-ALL1 cell lines, overexpressing cyclin D3 resulted in partial rescue from G1 arrest due to abrogated Notch signaling (Figures 4C, S5). However, cyclin D3 overexpression was not sufficient to rescue HPB-ALL from GSI treatment (Figures 4C, S5). Together, these data suggest, in human T-cell lymphomas, that cyclin D3 is an important target of the cell-cycle machinery and is regulated by Notch signaling.
Cyclin D3 is a target of Notch signaling in human T-ALL cell lines. (A) Cell-cycle analysis was performed on human T-ALL cell lines DND-41, HPB-ALL, and T-ALL1 that were treated with 0.1% DMSO (top panels) or GSI, 3 μm IL-CHO (bottom panels) for 7 days. FACS plots are representative of results of 3 independent replicates. Graphical representation of the mean (± SE) of percentage of cells in G1 phase of cell cycle under the 2 treatment conditions for the 3 experiments conducted. Two-tailed Student t tests were performed using GraphPad Prism version 4.0 for OS X (GraphPad Software, San Diego, CA) **P < .05.. (B) Whole cell extracts were prepared from human T-ALL cell lines after 7 days of GSI or DMSO treatment to analyze difference in cyclin D3 (top) and Notch1 expression (middle). GAPDH (bottom) was used as a loading control. (C) Graphical representation of relative G1 rescue indices. Human T-ALL cell lines were retrovirally infected with either MSCV-IRES-YFP (vector, open bar) or MSCV-cyclin D3-IRES-YFP (cyclin D3, solid bar). Infected cells were sorted for enrichment and treated with DMSO or GSI for 7 days. Cell-cycle analysis was performed to determine the percentage of cells arrested in G1 with GSI-treatment, compared with DMSO. Relative G1 rescue indices were calculated for each cell line after normalizing against GSI-induced G1 arrest in the uninfected cell line.
Cyclin D3 is a target of Notch signaling in human T-ALL cell lines. (A) Cell-cycle analysis was performed on human T-ALL cell lines DND-41, HPB-ALL, and T-ALL1 that were treated with 0.1% DMSO (top panels) or GSI, 3 μm IL-CHO (bottom panels) for 7 days. FACS plots are representative of results of 3 independent replicates. Graphical representation of the mean (± SE) of percentage of cells in G1 phase of cell cycle under the 2 treatment conditions for the 3 experiments conducted. Two-tailed Student t tests were performed using GraphPad Prism version 4.0 for OS X (GraphPad Software, San Diego, CA) **P < .05.. (B) Whole cell extracts were prepared from human T-ALL cell lines after 7 days of GSI or DMSO treatment to analyze difference in cyclin D3 (top) and Notch1 expression (middle). GAPDH (bottom) was used as a loading control. (C) Graphical representation of relative G1 rescue indices. Human T-ALL cell lines were retrovirally infected with either MSCV-IRES-YFP (vector, open bar) or MSCV-cyclin D3-IRES-YFP (cyclin D3, solid bar). Infected cells were sorted for enrichment and treated with DMSO or GSI for 7 days. Cell-cycle analysis was performed to determine the percentage of cells arrested in G1 with GSI-treatment, compared with DMSO. Relative G1 rescue indices were calculated for each cell line after normalizing against GSI-induced G1 arrest in the uninfected cell line.
Notch regulates CDK4 and CDK6 expression and pRb phosphorylation in primary T cells
The fact that we could only partially restore cell-cycle progression in some human T-ALL cell lines after ectopic expression of cyclin D3 prompted us to investigate other components of the cell cycle that might be influenced by Notch signaling. Cyclins function through their catalytic partners, CDK4 and CDK6, to facilitate cell-cycle progression. Therefore, we considered CDK4 and CDK6 as 2 putative targets of Notch regulation in primary T cells. As described above, we isolated CD4+ T cells from spleens of C57BL/6 mice, then stimulated the cells with anti-CD3ϵ and anti-CD28 in the presence or absence of GSI. At 6 hours after stimulation, we found increased expression both of CDK4 and CDK6 in DMSO-treated cells, but GSI treatment abrogated this up-regulation, signifying that even early induction of CDK4 and CDK6 is Notch-dependent (Figure 5A). Expression of both proteins continued to increase up to 72 hours, with maximal expression observed 48 hours after stimulation (Figure 5B). Interestingly, CDK6 expression was more robust than CDK4 expression at these later time points. Consistent with effects at early time points, inhibiting Notch signaling reduced expression of CDK4 and CDK6 at 24, 48, and 72 hours after stimulation (Figure 5B). These observations led us to ask whether differences in CDK4 or CDK6 expression altered phosphorylation of their target protein, pRb. D-cyclins, along with CDK4 and CDK6, are known to phosphorylate pRb at Ser780 and Ser795.43-46 We examined phosphorylation of pRb at these residues in CD4+ T cells stimulated for 24, 48, and 72 hours with anti-CD3ϵ and anti-CD28, with or without GSI treatment. We found Ser780 was phosphorylated by 24 hours and remained phosphorylated up to 72 hours after stimulation (Figure 5C). Phosphorylation of pRB Ser795 was also observed at 24 hours, although to a lesser extent than that of Ser780, and also increased up to 72 hours (Figure 5C). In contrast, GSI treatment abrogated phosphorylation of both pRb residues (Figure 5C), indicating Notch signaling regulates temporal expression of CDK4 and CDK6 and subsequent pRb phosphorylation, thus facilitating cell-cycle progression in primary T cells.
CDK4 and CDK6 expression and pRb phosphorylation is dependent on Notch signaling in CD4+ T cells. (A) Whole cell lysate of CD4+ T cells that were stimulated as described before for indicated time points (0, 3, 6, and 12 hours) in the presence (in vitro) of GSI or DMSO were analyzed by immunoblotting for differences in CDK4 and CDK6 expression. (B) Whole cell lysates of CD4+ T cells that had been stimulated, as described, for indicated time points (24, 48, and 72 hours) in the presence (in vitro) of GSI or DMSO were analyzed by immunoblotting for differences in CDK4 and CDK6 expression. Changes in temporal expression of CDK4 and CDK6 after stimulation are represented graphically in lower panels. (C) Nuclear lysates from in vitro GSI- or DMSO-treated CD4+ T cells that were stimulated for 24, 48, and 72 hours were analyzed by immunoblotting for differences in pRb phosphorylation at residues Ser780 (top panel) and Ser795 (bottom panel). The nuclear protein PARP was used as a loading control.
CDK4 and CDK6 expression and pRb phosphorylation is dependent on Notch signaling in CD4+ T cells. (A) Whole cell lysate of CD4+ T cells that were stimulated as described before for indicated time points (0, 3, 6, and 12 hours) in the presence (in vitro) of GSI or DMSO were analyzed by immunoblotting for differences in CDK4 and CDK6 expression. (B) Whole cell lysates of CD4+ T cells that had been stimulated, as described, for indicated time points (24, 48, and 72 hours) in the presence (in vitro) of GSI or DMSO were analyzed by immunoblotting for differences in CDK4 and CDK6 expression. Changes in temporal expression of CDK4 and CDK6 after stimulation are represented graphically in lower panels. (C) Nuclear lysates from in vitro GSI- or DMSO-treated CD4+ T cells that were stimulated for 24, 48, and 72 hours were analyzed by immunoblotting for differences in pRb phosphorylation at residues Ser780 (top panel) and Ser795 (bottom panel). The nuclear protein PARP was used as a loading control.
Cotransducing CDK4 or CDK6 with cyclin D3 provides variable rescue of human T-ALL cell lines from GSI-induced G1 arrest
Having shown CDK4 and CDK6 expression, as well as pRb phosphorylation, are targets of Notch signaling in primary T cells, we wanted to determine whether this was also true for human T-ALL cell lines. We treated the 3 T-ALL cell lines with GSI to abrogate Notch signaling and assessed CDK4 and CDK6 expression by immunoblotting. Our results show that GSI treatment reduces CDK4 and CDK6 expression to variable degrees in these cell lines (Figure 6A), with 25% to 30% reduction in CDK4 and CDK6 in GSI-treated DND-41 and HPB-ALL cells. The T-ALL1 cell line showed maximal reduction (almost 50%) in CDK4 and CDK6 expression (Figure 6A). We next asked whether decreased expression of CDK4 and CDK6 correlated with reduced pRb phosphorylation, as it did in primary T cells. Consistent with those data, we observed phosphorylation of pRb was altered by abrogating Notch signaling using GSI. As expected, reduced phosphorylation was variable and cell line-dependent; however, both Ser780 and Ser795 pRb residues showed reduced phosphorylation when the cell lines were treated with GSI (Figure 6B). These results indicate, in addition to cyclin D3, its catalytic partners CDK4 and CDK6, may also be targets of Notch signaling that cooperate to contribute to unchecked cell-cycle progression in T-cell lymphomas. These data prompted us to ask whether we could rescue human T-ALL lines from GSI-induced G1 arrest by cotransducing CDK4 or CDK6 together with cyclin D3. We used vector-YFP control cell lines and 3 cell lines stably expressing cyclin D3-YFP (Figure 5C and Figure S6) to coexpress DsRed-CDK4 or DsRed-CDK6. Cells expressing both YFP and DsRed were sorted to enrich for cells ectopically expressing cyclin D3 with CDK4 or CDK6. Sorted cells were treated with GSI or DMSO, and cell-cycle analysis was performed. We compared the differences in percentages of cells in G1 under the 2 treatment conditions (GSI or DMSO) to calculate G1 RIs, as before (Table S2). In all 3 cell lines, overexpressing cyclin D3 either with CDK4 or CDK6 could rescue cells from G1 arrest, as evidenced by the increase in RI (Figures 6C, S6). Interestingly, in all 3 cell lines, the G1 rescue effects of coexpressing cyclin D3 together with CDK4 or CDK6 were additive, compared with those of expressing cyclin D3 alone (Figures 4C, S1). This was particularly true in HPB-ALL cells, in which we did not observe any rescue with ectopic expression of cyclin D3 alone (Figure 6C, Table S1). Taken together, our data indicate cyclin D3, CDK4, and CDK6 are Notch-regulated targets that promote transit through the cell cycle in T-cell lymphomas.
CDK4 and CDK6 expression and pRb phosphorylation are targets of Notch signaling in human T-ALL cell lines. (A) Whole cell extracts were prepared from human T-ALL cell lines after 7 days of in vitro GSI or DMSO treatment to analyze differences in CDK4 (top) and CDK6 expression (middle). GAPDH (bottom) was used as a loading control. Band intensities of protein expression were normalized to GAPDH and represented graphically using ImageJ software, version 1.31, supported by Wayne Rasband (NIH). (B) Immunoblot of whole-cell lysates prepared from human T-ALL cell lines that were treated with GSI or DMSO for 7 days to analyze differences in pRb phosphorylation, pRB Ser780 (top panels), pRb Ser795 (middle panels), and GAPDH (bottom panels; loading control). (C) Graphical representation of relative G1 rescue indices. Human T-ALL cell lines DND-41, HPB-ALL, and T-ALL1 were retrovirally infected either with MSCV-IRES-YFP and MSCV-IRES-DsRed II (vector, black bar), MSCV-cyclin D3-IRES-YFP and MSCV-CDK4-IRES-DSRed (CDK4 + D3, gray bar), or MSCV-cyclin D3-IRES-YFP and MSCV-CDK6-IRES-DSRed (CDK6 + D3, white bar). The infected cells were sorted for enrichment and treated with DMSO or GSI for 7 days. Cell-cycle analysis was performed to determine the percentage of cells arrested in G1 with GSI treatment compared with DMSO. Relative G1 rescue indices were calculated for each cell line after normalizing against the GSI-induced G1 arrest in the uninfected cell line.
CDK4 and CDK6 expression and pRb phosphorylation are targets of Notch signaling in human T-ALL cell lines. (A) Whole cell extracts were prepared from human T-ALL cell lines after 7 days of in vitro GSI or DMSO treatment to analyze differences in CDK4 (top) and CDK6 expression (middle). GAPDH (bottom) was used as a loading control. Band intensities of protein expression were normalized to GAPDH and represented graphically using ImageJ software, version 1.31, supported by Wayne Rasband (NIH). (B) Immunoblot of whole-cell lysates prepared from human T-ALL cell lines that were treated with GSI or DMSO for 7 days to analyze differences in pRb phosphorylation, pRB Ser780 (top panels), pRb Ser795 (middle panels), and GAPDH (bottom panels; loading control). (C) Graphical representation of relative G1 rescue indices. Human T-ALL cell lines DND-41, HPB-ALL, and T-ALL1 were retrovirally infected either with MSCV-IRES-YFP and MSCV-IRES-DsRed II (vector, black bar), MSCV-cyclin D3-IRES-YFP and MSCV-CDK4-IRES-DSRed (CDK4 + D3, gray bar), or MSCV-cyclin D3-IRES-YFP and MSCV-CDK6-IRES-DSRed (CDK6 + D3, white bar). The infected cells were sorted for enrichment and treated with DMSO or GSI for 7 days. Cell-cycle analysis was performed to determine the percentage of cells arrested in G1 with GSI treatment compared with DMSO. Relative G1 rescue indices were calculated for each cell line after normalizing against the GSI-induced G1 arrest in the uninfected cell line.
Cyclin D3 and CDK4 are highly expressed in Notch-dependent T-cell lymphomas in vivo
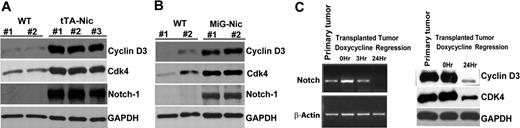
Finally, we utilized the Top-Notch mouse model of Notch-induced lymphoma to address whether Notch1 regulates expression of cell-cycle proteins in T-cell leukemia in vivo.37 We killed Top-Notch leukemic mice showing overt signs of lymphoma and prepared protein extracts from spleens. We confirmed high levels of NIC expression in diseased animals, as well as high levels of cyclin D3 and CDK4 compared with controls (Figure 7A). Interestingly, we did not find detectable CDK6 expression in these animals (data not shown), suggesting functional redundancy of CDK4 and CDK6. We obtained similar results from MIG-Nic leukemic mice (Figure 7B). Next, we performed leukemia transplantation experiments to show that, in the leukemic cells derived from tTA-Nic mice, cyclin D3 and CDK4 expression is dependent on the continuous expression of the Top-Notchic transgene. tTA-Nic mice that were moribund due to lymphoma were euthanized, and single-cell suspensions prepared from lymphoid organs were transplanted IV into the tail vein of nontransgenic mice. After these mice developed tumors, they were treated with doxycycline to shut off Notch expression. We performed RT-PCR analysis on these regression samples to determine the time point at which Notch was turned off (Figure 7C). We then prepared protein lysates from this time point and observed that abrogation of Notch results in reduced cyclin D3 as well as CDK4 expression. These results further strengthen the link between Notch signaling and cell-cycle regulation. Together, these findings indicate that, in Notch-dependent lymphomas, cyclin D3, and CDK expression is regulated by Notch1 and may contribute to Notch-driven T-cell leukemogenesis.
Regulation of cyclin D3 and CDK4 expression by Notch1 in T-cell lymphomas. (A) Protein extract from spleens of 2 wild-type FVB mice (WT) and 3 Top-Notch leukemic (tTA-Nic) mice were used to analyze the expression of cyclin D3, CDK4, and Notch-1. GAPDH was used as a loading control. (B) Protein extracts from spleens of 2 wild-type FVB mice (WT) and 2 MIG-Nic leukemic mice were used to determine cyclin D3, CDK4, and Notch-1 expression, as indicated. GAPDH was used as a loading control. (C) E μ-tTA/TOP-Notch mice were generated as previously described.36,37 Samples prepared for regression analysis were prepared as described in Table S1. Left panel shows the RT-PCR performed on primary tumor samples and on transplanted samples from mice undergoing doxycycline regression for different lengths of time (0, 3, and 24 hours). Notch transcripts were analyzed, and β-actin was used as a positive control. Right panel shows Western blot analysis performed using total cell lysate for detection of cyclin D3 and CDK4 from primary tumor samples and transplanted samples from mice undergoing doxycycline regression for various lengths of time (0 and 24 hours).
Regulation of cyclin D3 and CDK4 expression by Notch1 in T-cell lymphomas. (A) Protein extract from spleens of 2 wild-type FVB mice (WT) and 3 Top-Notch leukemic (tTA-Nic) mice were used to analyze the expression of cyclin D3, CDK4, and Notch-1. GAPDH was used as a loading control. (B) Protein extracts from spleens of 2 wild-type FVB mice (WT) and 2 MIG-Nic leukemic mice were used to determine cyclin D3, CDK4, and Notch-1 expression, as indicated. GAPDH was used as a loading control. (C) E μ-tTA/TOP-Notch mice were generated as previously described.36,37 Samples prepared for regression analysis were prepared as described in Table S1. Left panel shows the RT-PCR performed on primary tumor samples and on transplanted samples from mice undergoing doxycycline regression for different lengths of time (0, 3, and 24 hours). Notch transcripts were analyzed, and β-actin was used as a positive control. Right panel shows Western blot analysis performed using total cell lysate for detection of cyclin D3 and CDK4 from primary tumor samples and transplanted samples from mice undergoing doxycycline regression for various lengths of time (0 and 24 hours).
Discussion
We have identified cyclin D3 as a direct target of Notch1 that promotes cell-cycle progression and proliferation both in peripheral and leukemic T cells. We show cyclin D3 expression in T cells is dependent on Notch1 signaling both in vitro and in vivo. Our data demonstrate that Notch1, along with its canonical binding partner CSL, associates with the cyclin D3 promoter. In addition, we show the NF-κB subunit p50 binds the cyclin D3 promoter and enhances Notch1-dependent promoter activity. Taken together, our data using Notch-dependent human T-ALL lines identify cyclin D3 as a target of constitutively active Notch signaling, since ectopic expression of cyclin D3 can partially rescue these cell lines from GSI-induced G1 arrest. Finally, we show that cyclin-dependent kinases CDK4 and CDK6 are also targets of Notch signaling and, along with cyclin D3, contribute to deregulated cell-cycle progression in leukemic T cells.
We demonstrated that 24 hours after activation, primary T cells show a robust increase in cyclin D3 expression that is abrogated with GSI treatment. We have shown that Notch1 associates with the cyclin D3 promoter and the TAD of Notch1IC is required for regulating cyclin D3 promoter, because its removal almost completely abrogates promoter activity. We also found the ANK region of Notch1IC is critical for expression from the cyclin D3 promoter. These results confirm the importance of the ANK and TAD domains of Notch1 in regulating cyclin D3 and are known to be required for Notch-driven T-cell leukemogenesis.47
Interestingly, in addition to the canonical Notch binding partner, CSL, we also found the NF-κB subunit p50 binds to the cyclin D3 promoter. It was recently demonstrated that the Notch transcriptional complex (NTC), consisting of CSL/ICN1/MAML, dimerizes on properly spaced pairs of CSL binding sites and that certain target genes require this architecture for Notch responsiveness.48,49 However, Notch-mediated effects are exquisitely context-dependent, a property that likely stems from the existence of response elements containing combinations of binding sites for CSL and other transcription factors.46,47 In the case of cyclin D3 promoter, we were able to identify 2 putative NF-κB binding sites in a 227-bp region amplified after ChIP. Consensus NF-κB binding sites may incorporate a nested CSL binding site.16 This raises the question of whether there is competition between NF-κB and CSL for overlapping binding sites on a promoter or for DNA binding factors, such as p50, that cooperate with NTC to transactivate Notch-regulated genes. While we show NF-κB and CSL both are recruited to the cyclin D3 promoter, directly regulating cyclin D3 expression, more work is required to elucidate the mechanism involved in this regulation. Our observations underscore an important role for NF-κB in peripheral T-cell function and, very recently, NF-κB has been implicated as a major downstream target of Notch1 in human T-ALL.50
Our results in primary T cells reveal CSL recruitment to the promoter region is diminished by abrogating Notch activity. The Drosophila homolog of CSL, Su(H), is dynamic in its association with DNA and shows significant increase in target-gene occupancy after Notch activation, highlighting the importance of cooperative binding of higher-order NTCs and the influence of other DNA binding cofactors to stabilize the Su(H) activation-complex on DNA.51 It remains to be determined whether regulation of the cyclin D3 promoter involves a similar mechanism.
Several targets of Notch signaling in human T-ALLs have been identified. Transcriptional regulators c-Myc, Lef-1, and the m-TOR pathway have been shown to be associated with Notch-induced transformations.19-21 Notch-dependent human T-ALL cell lines have previously been used to identify targets of deregulated Notch signaling.19,21 Our results have identified cyclin D3 as a target of Notch signaling in the human T-ALL cell lines DND-41, HPB-ALL, and T-ALL1. However, ectopic expression of cyclin D3 alone could not completely rescue these cell lines from GSI-induced G1 arrest. We have demonstrated that while cyclin D3 is an important direct target of Notch signaling in Notch-dependent cell lines, there are other cell-cycle regulators that may be influenced by GSI treatment.
Cyclin-dependent kinases are important for cell-cycle progression in normal T cells and during leukemic transformation.52 We show that CDK4 and CDK6 both are important for Notch-dependent cell-cycle progression. In activated T cells, there is increased expression of CDK4 and CDK6 that is abrogated by GSI treatment. Unlike cyclin D3, their expression is induced at early time points, with robust increase 24 hours after stimulation. We observed an additive rescue from G1 arrest with ectopically expressed CDK4 or CDK6, together with cyclin D3, in all 3 Notch-dependent cell lines, suggesting cyclin D3 and its catalytic CDKs may be co-opted as part of the pro-oncogenic Notch pathway. CDK4 levels have been shown to be reduced in T-ALL lines after treatment with the GSI, compound E.21 Failure to effect a complete rescue from GSI-induced G1 arrest could be attributed to inhibition by endogenous cyclin-dependent kinase inhibitors (CKIs) that are negatively regulated by Notch activation.26 CKI p27 was up-regulated after GSI treatment in Notch-dependent human T-ALL cell lines.21 The exact mechanism by which Notch regulates D cyclins and their kinases remains to be explored.
Data presented here reveal a possible mechanism, downstream of deregulated Notch signaling, that may facilitate leukemic transformation. These observations identify cell-cycle components as adjunct therapeutic targets for designing combination treatment strategies using γ-secretase inhibitors for T-ALL treatment.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the members of the Osborne laboratory for useful discussions and technical support, especially Rebecca G. Lawlor for help with the tissue culture work. We thank Sripriya Subramanian for help with cell sorting. We also thank Dr Rachit Bakshi for helpful discussions and critical reading of the manuscript. Lastly, we thank Drs Phil Hinds, Dario Vignali, and Suzanne Baker for their generosity in providing plasmids.
This work was supported by National Institutes of Health (NIH) grants PO1 AG025531 and RO1 AI049361.
National Institutes of Health
Authorship
Contribution: I.J. designed and performed experiments and wrote the manuscript; L.M.M. and J.T. contributed to the design of experiments and interpretation of data; R.D. provided experimental samples for the analysis of TOP-Notch and MIG-Nic mice; A.J.C., J.C.A., and P.S. provided essential reagents for experiments and were also involved in review of the manuscript; A.F. and T.E.G. provided GSI for in vitro and in vivo work; and B.A.O. provided support for the work, analyzed and interpreted the data, and reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Barbara A. Osborne, Department of Veterinary & Animal Sciences, 311 Paige Lab, University of Massachusetts, Amherst, MA 01003; e-mail: osborne@vasci.umass.edu.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal