Abstract
The simian virus 40 (SV40) T antigen is a potent oncogene able to transform many cell types and has been implicated in leukemia and lymphoma. In this report, we have achieved sporadic SV40 T-antigen expression in mature B cells in mice, by insertion of a SV40 T antigen gene in opposite transcriptional orientation in the immunoglobulin (Ig) heavy (H) chain locus between the D and JH segments. SV40 T-antigen expression appeared to result from retention of the targeted germline allele and concomitant antisense transcription of SV40 large T in mature B cells, leading to chronic lymphocytic leukemia (CLL). Although B-cell development was unperturbed in young mice, aging mice showed accumulation of a monoclonal B-cell population in which the targeted IgH allele was in germline configuration and the wild-type IgH allele had a productive V(D)J recombination. These leukemic B cells were IgDlowCD5+ and manifested nonrandom usage of V, D, and J segments. VH regions were either unmutated, with preferential usage of the VH11 family, or manifested extensive somatic hypermutation. Our findings provide an animal model for B-CLL and show that pathways activated by SV40 T antigen play important roles in the pathogenesis of B-CLL.
Introduction
B-cell chronic lymphocytic leukemia (B-CLL),1,2 the most common leukemia in the Western world, is characterized by the accumulation of a monoclonal population of mature B cells that aberrantly express CD5.3,4 The clinical course of CLL is extremely heterogeneous: whereas some patients survive more than a decade with stable disease, others die of the disease within months despite aggressive treatment. This heterogeneity is associated with variability in the expression pattern of a number of different proteins, and also with the presence of different chromosomal aberrations.5 Approximately half of the CLL cases have 13q14deletions, apparently involving the linked microRNA molecules miR15 and miR16, which are thought to be negative regulators of the antiapoptotic gene Bcl2.2 Deletions on chromosome 17p, affecting the p53 protein, are less frequent and are associated with poor prognosis.6,7 CLLs manifest a unique gene-expression signature that differs from other lymphoid cancers, suggesting a common mechanism of transformation or a homogeneous cell population of origin.2
CLL has been subdivided into 2 prognostic subsets, based on the presence of somatic mutations of the immunoglobulin (Ig) heavy (H) chain variable (VH) genes. A total of 50% to 70% of CLL patients have mutated VH genes, probably reflecting antigen-driven postgerminal center (GC) selection, and have a more favorable prognosis than those with unmutated B-cell receptors. CLL Ig VH regions also exhibit unique complementarity determining region 3 (CDR3) features that characterize and differentiate aggressive unmutated from more indolent mutated CLLs. In particular, unmutated poor outcome cases frequently contain long CDR3s with amino acid residues that favor polyreactivity. Interestingly, both unmutated and mutated CLLs are thought to derive from self-reactive B-cell precursors.8 In this context, it has been found that approximately 3% to 4% of healthy persons older than 40 years of age have a population of monoclonal lymphocytes in their blood with immunophenotypic characteristics of CLL cells.2
The simian virus 40 (SV40) T antigen is a potent oncogene able to transform many cell types9,10 and has been implicated in the etiology of various cancers.11,12 The SV40 T antigen has transforming activity by inactivating p53 and Rb proteins and inducing genomic instability.13 Several studies point to a causative role of SV40 in the formation of human B-cell malignancies, including non-Hodgkin lymphoma.14-16 Transgenic expression of the SV40 T gene under the control of the IgH enhancer induced hyperproliferation of multilineage hematopoiesis, reminiscent of myelodysplastic syndromes in humans.17
In this report, we aimed to accomplish sporadic SV40 T gene expression in the B-cell lineage in mice. We introduced the SV40 T gene, without its promoter and in opposite transcriptional orientation between the IgH chain D and JH segments. As antisense transcription takes place across the D-JH region in pro-B cells,18 it is possible that in our mouse model SV40 T is expressed as part of a large antisense transcript in early pro-B cells that have not yet performed IgH D to JH recombination. But in almost all pro-B cells, the SV40 T gene will be excised during normal IgH D to JH rearrangement. Only in those rare B cells, in which the nonproductive targeted allele has a germline configuration and thus still contains the SV40 T gene, it may possibly be expressed in more mature B-cell stages. In addition, given the ability of the recombination activating gene (Rag)-1 and -2 proteins to catalyze DNA translocation and transposition reactions both in vitro19,20 and in vivo,21-23 it is conceivable that V(D)J-mediated reinsertion of D-JH circles containing the SV40 T oncogene compromises genomic integrity, leading to tumor formation.
In our model, we found that expression of SV40 T antigen is associated with the development of B-cell malignancies with striking similarities to human B-CLL. Tumor formation appears to result from retention of the targeted germline allele and concomitant antisense transcription of SV40 large T in mature B cells.
Methods
Generation of mouse models
To generate the IgH.T (and IgH.TEμ, with an extra copy of the IgH intronic enhancer) targeting constructs, containing the thymidine kinase gene, neomycin resistance gene, SV40 T antigen, internal ribosome entry site, splice accepter and flanking homology arms (created by long-range polymerase chain reaction [PCR] from 129 mouse genomic DNA), we performed multiple cloning steps (strategy available on request). Constructs were linearized and electroporated into E14 embryonic stem (ES) cells, which were subsequently cultured with G418 and gancyclovir. Expanded ES cell clones were screened for homologous recombination by Southern blotting. Chimeric mice were generated by injection of ES cells into blastocysts and bred to C57BL/6 mice. Germline transmission of the targeted allele was verified by Southern blotting, and offspring was genotyped by PCR analysis of DNA from tail snips using SV40 T-specific primers. Excision of the neocassette was achieved by crosses with transgenic mice expressing Cre-recombinase ubiquitously under the control of the cytomegalovirus immediate early enhancer-chicken beta-actin hybrid promoter.
IgH.T, IgH.TEμ, and p53−/− mice (The Jackson Laboratory) were bred and maintained in the Erasmus MC animal care facility under specific pathogen–free conditions. Experimental procedures were reviewed and approved by the Erasmus MC Animal Experiments committee.
Southern blotting
Genomic DNA samples were digested with SacI or XbaI overnight at 37°C and processed using standard procedures. A 760-bp neomycin probe was generated by PCR. The DQ52up probe is an 840-bp BspHI-PciI fragment, located just upstream of DQ52 (Figure 1B). The CH3 probe is a 1131-bp NspI-XcmI fragment spanning the μ CH3 exon. A 1.2-kb XhoI-XbaI fragment (XX1.2; Figure 4D) was used as a probe for the H chain DQ52-Eμ region. Probes were labeled using the Prime-it II kit (Stratagene) and hybridized overnight at 65°C. Fragments were visualized using a phospho-Imager and analyzed with ImageQuant (GE Healthcare).
Flow cytometry, cell purification, and blood smears
Blood was collected from the tail vein, and erythrocytes were lysed using NH4Cl. Preparation of single-cell suspensions and monoclonal antibody (BD Biosciences) incubations for 4-color cytometry have been described.24 Viable cells were counted using a Coulter counter (Beckman Coulter). B cells were purified by magnetic separation using CD19-microbeads and AutoMACS (Miltenyi Biotec). To produce blood thin films, 10 μL of blood was smeared on a glass slide, dried, and fixed in methanol for 3 minutes and stained in 5% Giemsa.
Western blot
Total cell lysates were obtained by addition of lysis buffer (20 mM Tris, 137 mM NaCl, 10 mM ethylenediaminetetraacetic acid, 100 mM NaF, 1% NP-40, 10% glycerol, 1 mM Pefabloc, 1 mM Na3VO4) to cells and incubation on ice for 20 minutes. Lysates were centrifuged (13 000g), and supernatants were boiled with loading buffer, separated on sodium dodecyl sulfate–polyacrylamide gels, and blotted using standard procedures. Membranes were stained with biotinylated anti-SV40T antibodies (Pab108; BD Biosciences), using streptavidin–horseradish peroxide as a second step and developed using enhanced chemiluminescence.
RNA isolation and RT-PCR
Total RNA was isolated by GenElute Mammalian RNA purification (Sigma-Aldrich), DNAse digested, and reverse transcribed with Superscript II and random hexamer primers. PCR primers spanning at least one intron were designed using ProbeFinder software (Roche Diagnostics), and probes were chosen from the universal probe library (Roche Diagnostics). For quantitative real-time PCR, cDNA was amplified in universal Mastermix containing 200 nM of each primer and 100 nM probe, using the ABI Prism 7700 sequence detection system (Applied Biosystems). To confirm specificity of amplification products, samples were analyzed by standard agarose gel electrophoresis. The obtained Ct values were normalized to those of hypoxanthine-guanine phosphoribosyl-transferase (HPRT).25
Heteroduplex PCR and DNA sequencing
Heteroduplex analyses were performed as described.26 For DNA sequence analyses, cDNA samples were amplified using 7 primers located in the framework 1 region: 2 high degeneracy primers MH1–2 and 5 low degeneracy primers MH3–7,27 in combination with a primer located in the 5′ cμ region.28 PCR products were directly sequenced using the BigDye terminator cycle sequencing kit with AmpliTaq DNA polymerase on an ABI PRISM 377 automated sequencer (Applied Biosystems). All H chain regions were sequenced in 2 directions from at least 2 independent PCR products and analyzed by IMGT/V-Quest (IMGT, the international ImMunoGeneTics information system).
Results
Generation of IgH.T and IgH.TEμ mice
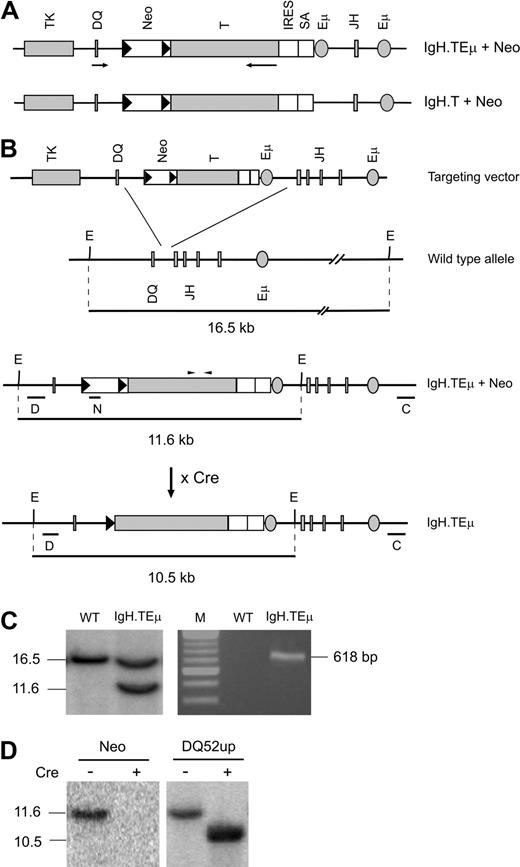
To achieve sporadic SV40 T antigen expression, we generated 2 different mouse models by homologous recombination in ES cells. The SV40 large and small T antigen–coding unit10 was inserted between DQ52 and JH1 in the IgH locus, either with (IgH.TEμ) or without (IgH.T) an extra copy of the IgH intronic enhancer Eμ (Figure 1). ES cell clones with homologous recombination events were identified by Southern blotting, using the 5′-flanking DQ52up probe and the 3′-flanking CH3 probe (Figure 1BC left panel; and data not shown). ES clones were injected into blastocysts to generate chimeric mice that transmitted the targeted IgH allele through the germline, as identified by SV40 T gene–specific PCR (Figure 1C right panel). Excision of the neo-cassette was achieved by crossing mice with transgenic mice ubiquitously expressing Cre recombinase under the control of the cytomegalovirus immediate early enhancer-chicken beta-actin hybrid promoter. Successful excision was verified by Southern blotting using neomycin, DQ52up, and CH3 probes (Figure 1D; and data not shown). Mice harboring a single targeted allele were further bred to C57BL/6 mice. Heterozygous offspring, referred to as IgH.TEμ or IgH.T mice, were born at the expected Mendelian frequencies, did not manifest any developmental defects, appeared normal, and were fertile.
Generation of IgH.TEμ and IgH.T mouse models. (A) Schematic representation of the targeting constructs with (IgH.TEμ + Neo) and without (IgH.T + Neo) an additional Eμ copy. LoxP sites are represented by black triangles; arrows represent transcriptional orientation. Neo indicates neomycin gene; T, T antigen; IRES, internal ribosome entry site; and SA, splice accepter. (B) The targeting vectors included the SV40 T antigen gene in opposite transcriptional orientation without its promoter, flanking regions of 5′ (DQ52) and 3′ (JH) Ig H chain homology, the herpes simplex thymidine kinase gene for negative selection, and the neomycin resistance gene for positive selection. Below the targeting vector: wild-type Ig H chain allele, the locus after homologous recombination, and after Cre-mediated Neo-excision. The position of the PCR primers used to genotype offspring is indicated by arrowheads above the T antigen. Probes used in Southern blots are indicated by horizontal lines. E indicates EcoRI site; D, DQ52up; N, neomycin; and C, CH3. (C) Southern blot of EcoRV-digested ES cell DNA (left panel) and PCR analysis of tail DNA from the indicated mice. The IgH.TEμ ES cell clone analysis shows a 16.5-kb wild-type fragment and an 11.6-kb fragment recombined fragment containing the Neo insertion. (D) Southern blot of EcoRV-digested tail DNA from IgH.TEμ mice before (−) and after (+) Cre-mediated recombination. Blots were hybridized with either the Neo (left panel) or the DQ52up probe (right panel).
Generation of IgH.TEμ and IgH.T mouse models. (A) Schematic representation of the targeting constructs with (IgH.TEμ + Neo) and without (IgH.T + Neo) an additional Eμ copy. LoxP sites are represented by black triangles; arrows represent transcriptional orientation. Neo indicates neomycin gene; T, T antigen; IRES, internal ribosome entry site; and SA, splice accepter. (B) The targeting vectors included the SV40 T antigen gene in opposite transcriptional orientation without its promoter, flanking regions of 5′ (DQ52) and 3′ (JH) Ig H chain homology, the herpes simplex thymidine kinase gene for negative selection, and the neomycin resistance gene for positive selection. Below the targeting vector: wild-type Ig H chain allele, the locus after homologous recombination, and after Cre-mediated Neo-excision. The position of the PCR primers used to genotype offspring is indicated by arrowheads above the T antigen. Probes used in Southern blots are indicated by horizontal lines. E indicates EcoRI site; D, DQ52up; N, neomycin; and C, CH3. (C) Southern blot of EcoRV-digested ES cell DNA (left panel) and PCR analysis of tail DNA from the indicated mice. The IgH.TEμ ES cell clone analysis shows a 16.5-kb wild-type fragment and an 11.6-kb fragment recombined fragment containing the Neo insertion. (D) Southern blot of EcoRV-digested tail DNA from IgH.TEμ mice before (−) and after (+) Cre-mediated recombination. Blots were hybridized with either the Neo (left panel) or the DQ52up probe (right panel).
B-cell development in IgH.T and IgH.TEμ mice
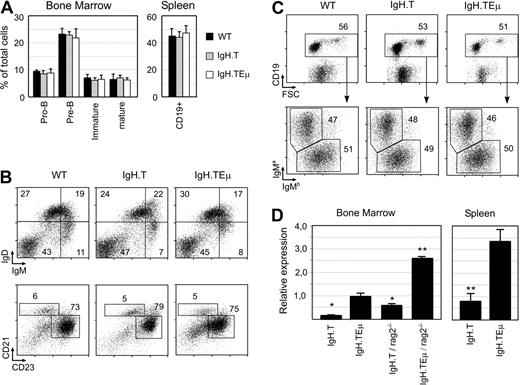
First, we investigated whether SV40 T antigen gene insertion in the IgH locus affected B-cell development or H chain allele usage. Single-cell suspensions from bone marrow (BM) and spleen from approximately 8-week-old IgH.T and IgH.TEμ heterozygous mice and wild-type littermates were analyzed by flow cytometry. The presence of the targeted IgH alleles did not affect BM or spleen cellularity. In addition, the proportions of the various stages of B-cell development in the BM or the proportions of CD19+ B cells in the spleen were unaffected (Figure 2A). The 3 groups of mice had similar IgM/IgD profiles or proportions of immature (CD21lowCD23low), follicular (CD21+CD23+), and marginal zone (CD21highCD23low) B cells in the spleen (Figure 2B).
Normal B-cell development in young IgH.T and IgH.TEμ mice. (A) Proportions of pro-B (CD19+/CD43+/μ−), pre-B (CD19+/μ+), immature (IgM+/IgD−), and mature (IgM+/IgD+) B cells in BM and total CD19+ B cells in the spleen of the indicated mice, as measured by flow cytrometric analysis. Mean values and SD are given for 4 mice per group. (B) Fluorescence-activated cell sorter profiles for IgM and IgD in total spleen cells from the indicated mouse groups (top panel). Total splenic CD19+ B cells were gated and analyzed for CD21 and CD23 expression. The proportions of CD21highCD23high follicular B cells and CD21hiCD23lo marginal zone B cells within the specified gates are given. (C) Flow cytrometric analysis of CD19 expression (top panel) on peripheral white blood cells of WT, IgH.T, and IgH.TEμ mice at 2 months of age. The CD19+ fraction was gated and analyzed for the expression of the IgMa and IgMb allele (bottom panel). Numbers in the plots indicate the percentage of cells within the specified gates. (D) Quantitative PCR analysis of large T mRNA expression in CD19+ BM cells and spleen cells of the indicated mice. Expression was normalized with HPRT, and expression in IgH.TEμ BM was set to 1. Significant differences compared with IgH.TEμ mice (* P < .01, ** P < .001, Mann-Whitney U test). Mean values and SD are given for 3 mice (8-10 weeks old) per group.
Normal B-cell development in young IgH.T and IgH.TEμ mice. (A) Proportions of pro-B (CD19+/CD43+/μ−), pre-B (CD19+/μ+), immature (IgM+/IgD−), and mature (IgM+/IgD+) B cells in BM and total CD19+ B cells in the spleen of the indicated mice, as measured by flow cytrometric analysis. Mean values and SD are given for 4 mice per group. (B) Fluorescence-activated cell sorter profiles for IgM and IgD in total spleen cells from the indicated mouse groups (top panel). Total splenic CD19+ B cells were gated and analyzed for CD21 and CD23 expression. The proportions of CD21highCD23high follicular B cells and CD21hiCD23lo marginal zone B cells within the specified gates are given. (C) Flow cytrometric analysis of CD19 expression (top panel) on peripheral white blood cells of WT, IgH.T, and IgH.TEμ mice at 2 months of age. The CD19+ fraction was gated and analyzed for the expression of the IgMa and IgMb allele (bottom panel). Numbers in the plots indicate the percentage of cells within the specified gates. (D) Quantitative PCR analysis of large T mRNA expression in CD19+ BM cells and spleen cells of the indicated mice. Expression was normalized with HPRT, and expression in IgH.TEμ BM was set to 1. Significant differences compared with IgH.TEμ mice (* P < .01, ** P < .001, Mann-Whitney U test). Mean values and SD are given for 3 mice (8-10 weeks old) per group.
To determine whether SV40 T gene insertion resulted in an altered frequency of IgH allele usage, we took advantage of IgM allotype differences between the targeted IgH allele (129 strain-derived IgMa) and the wild-type C57BL/6 allele (IgMb) in F1 mice. By flow cytometry using allotype-specific antibodies, we found equal usage of the targeted and the wild-type IgH allele in mature splenic CD19+ B cells (Figure 2C).
Quantitative reverse-transcribed (RT) PCR analyses of purified BM CD19+ B-cell lineage fractions from IgH.TEμ/IgH.T mice revealed the presence of SV40 T gene transcripts (Figure 2D). Significantly higher levels of SV40 T transcripts were detected in IgH.TEμ than in IgH.T B-cell fractions, consistent with the capacity of the Eμ element to increase transcription. Because the SV40 T gene will be excised during D to JH segment rearrangement, we reasoned that inclusion of SV40 T sequences in IgH transcripts is expected to be largely limited to the early pro-B cells stage in the BM, before the initiation of D to JH gene recombination. Consistent with this, we found that purified CD19+ B-cell lineage fractions from IgH.T and IgH.TEμ on the Rag-2−/− background had increased levels of SV40 T transcription, compared with IgH.T and IgH.TEμ mice on the wild-type background (Figure 2D). Because B-cell development is blocked at the pro-B cell stage in IgH.T;Rag-2−/− and IgH.TEμ;Rag-2−/− mice, this finding indicated that SV40 T antigen gene transcription was indeed particularly present in pro-B cells that have a germline IgH configuration.
Importantly, we detected significant SV40 T antigen transcription in mature splenic CD19+ B cells from IgH.T and IgH.TEμ mice (Figure 2D). Taken together, these findings show that SV40 T gene insertion in the IgH chain locus did not affect B-cell development or H chain allele usage and that in a fraction of mature B cells SV40 T is retained and transcribed.
Unilateral IgH allele usage in aged IgH.T and IgH.TEμ heterozygous mice
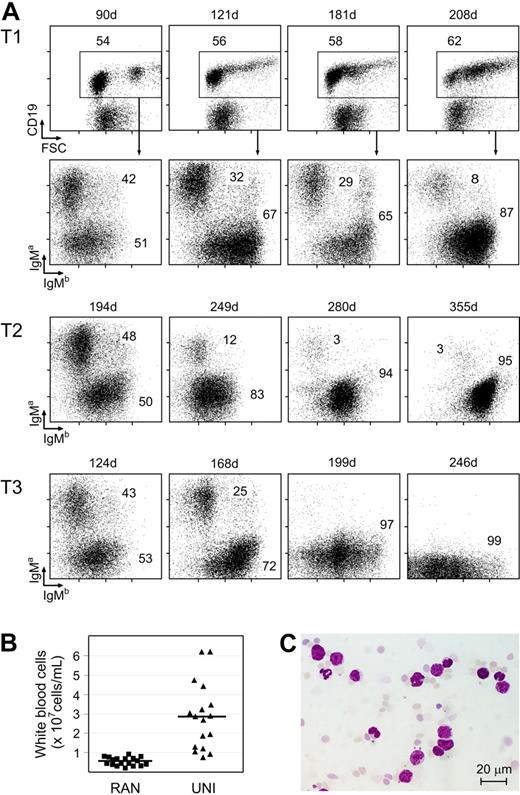
To monitor selective outgrowth of monoclonal B-cell populations over time, we periodically determined IgMa/IgMb profiles in peripheral blood B cells from panels of IgH.T (n = 15) and IgH.TEμ (n = 31) mice (Figure 3A). Young animals consistently had close to equal proportions of IgMa- and IgMb-expressing B cells. In contrast, we observed in all aging IgH.TEμ mice and in 2 of 15 IgH.T mice a gradual appearance of a CD19+ population with preferential expression of the nontargeted IgMb allele. These IgMb cells were also characterized by large forward scatter values, indicating increased average cell size. In mice with unilateral IgMb expression (> 90% IgMb, designated UNI mice), significantly higher numbers of peripheral white blood cells were found, compared with mice with random IgH allele usage (45%-55% IgMb, RAN mice; Figure 3B). Examination of blood smears showed the presence of enlarged cells with clumped chromatin and little cytoplasm (Figure 3C). Nevertheless, animals showed no outward signs of distress in this period.
Unilateral Ig H chain usage in aged IgH.TEμ heterozygous mice. (A) Flow cytrometric analysis of surface CD19, IgMa, and IgMb expression on peripheral blood mononuclear cells from 3 individual IgH.TEμ mice, T1, T2, and T3, representative for mice analyzed that developed unilateral Ig H chain usage over time. The CD19+ fraction was gated (shown only for mouse T1), and the IgMa/IgMb profile was analyzed (shown for all 3 mice). Data are shown as dot plots. Numbers indicate the percentages of cells within the specified populations. Age of the animal at the time of analysis is indicated by the numbers above the plots. (B) Total white blood cell count in IgH.TEμ mice showing random usage (RAN, n = 17) of IgMa and IgMb alleles, or unilateral usage of IgMb (UNI, with < 10% IgMa and > 90% IgMb B cells, n = 18; P < .001, Mann-Whitney U test). The RAN mouse group consists of IgH.TEμ (< 5 months of age) as well as IgH.T mice (< 9 months). The age of onset of UNI group is indicated in Table S1. Each symbol indicates an individual mouse (■, RAN; and ▴, UNI). Bars show average with SD of 17 (RAN) and 18 (UNI) animals. (C) Giemsa staining of blood smear of a UNI mouse, showing the presence of large cells with little chromatin. Size marker represents 20 μm.
Unilateral Ig H chain usage in aged IgH.TEμ heterozygous mice. (A) Flow cytrometric analysis of surface CD19, IgMa, and IgMb expression on peripheral blood mononuclear cells from 3 individual IgH.TEμ mice, T1, T2, and T3, representative for mice analyzed that developed unilateral Ig H chain usage over time. The CD19+ fraction was gated (shown only for mouse T1), and the IgMa/IgMb profile was analyzed (shown for all 3 mice). Data are shown as dot plots. Numbers indicate the percentages of cells within the specified populations. Age of the animal at the time of analysis is indicated by the numbers above the plots. (B) Total white blood cell count in IgH.TEμ mice showing random usage (RAN, n = 17) of IgMa and IgMb alleles, or unilateral usage of IgMb (UNI, with < 10% IgMa and > 90% IgMb B cells, n = 18; P < .001, Mann-Whitney U test). The RAN mouse group consists of IgH.TEμ (< 5 months of age) as well as IgH.T mice (< 9 months). The age of onset of UNI group is indicated in Table S1. Each symbol indicates an individual mouse (■, RAN; and ▴, UNI). Bars show average with SD of 17 (RAN) and 18 (UNI) animals. (C) Giemsa staining of blood smear of a UNI mouse, showing the presence of large cells with little chromatin. Size marker represents 20 μm.
Collectively, these data show the accumulation over time of atypical, large B cells exclusively expressing the IgMb allele from the nontargeted IgH locus.
Accumulation of monoclonal B-cell populations in aged IgH.T and IgH.TEμ mice
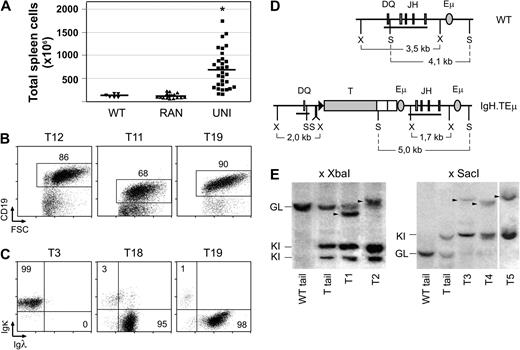
IgH.T and IgH.TEμ mice in the UNI group had normal B-lineage cell numbers in BM, but spleen sizes were significantly increased, compared with wild-type littermates or IgH.T/IgH.TEμ RAN mice (Figure 4A; supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). A majority (∼ 60%) of the IgH.T and IgH.TEμ UNI mice had enlarged lymph nodes with cell numbers up to approximately 15 times those of healthy animals. As shown for 3 examples of UNI mice in Figure 4B, flow cytometric analyses of the spleen revealed the presence of a population of mature CD19+ B cells with increased cell size, compared with wild-type control cells (CD19/FSC profiles, Figure 2C).
Accumulation of monoclonal B-cell populations in aged IgH.T and IgH.TEμ mice. (A) Spleen size of WT (▾, n = 6), RAN (▴, n = 18), and UNI (■, n = 33) animals. Each symbol indicates an individual mouse; mean values are indicated by a line. * Significant difference with WT (P < .001; Mann-Whitney U test). (Ages of mice are given in Table S1). (B) Flow cytometric analysis of total spleen cells in the 3 UNI mice indicated. Numbers indicate the percentage of CD19+ B cells. (C) Expression of Igκ or Igλ L chain in splenic IgM+ B cells of 3 UNI mice. Numbers indicate the percentage of cells within the specified quadrants. (D) Schematic representation of WT and targeted alleles, and position of restriction sites used for Southern analysis of Ig H chain recombination. Horizontal line represents the position of the XX1.2 probe, spanning the DQ-JH region. S indicates SacI; X, XbaI. (E) DNA rearrangements in total spleen cells of UNI animals. DNA was digested with XbaI or SacI and hybridized to the XX1.2 probe. Tail DNA from WT and UNI animals was used as reference for the WT germline fragment (GL) and the knockin germline fragment (KI). Note that digestion with XbaI results in 2 germline KI fragments. A vertical line has been inserted between samples T4 and T5 (SacI digests) to indicate a repositioned gel lane.
Accumulation of monoclonal B-cell populations in aged IgH.T and IgH.TEμ mice. (A) Spleen size of WT (▾, n = 6), RAN (▴, n = 18), and UNI (■, n = 33) animals. Each symbol indicates an individual mouse; mean values are indicated by a line. * Significant difference with WT (P < .001; Mann-Whitney U test). (Ages of mice are given in Table S1). (B) Flow cytometric analysis of total spleen cells in the 3 UNI mice indicated. Numbers indicate the percentage of CD19+ B cells. (C) Expression of Igκ or Igλ L chain in splenic IgM+ B cells of 3 UNI mice. Numbers indicate the percentage of cells within the specified quadrants. (D) Schematic representation of WT and targeted alleles, and position of restriction sites used for Southern analysis of Ig H chain recombination. Horizontal line represents the position of the XX1.2 probe, spanning the DQ-JH region. S indicates SacI; X, XbaI. (E) DNA rearrangements in total spleen cells of UNI animals. DNA was digested with XbaI or SacI and hybridized to the XX1.2 probe. Tail DNA from WT and UNI animals was used as reference for the WT germline fragment (GL) and the knockin germline fragment (KI). Note that digestion with XbaI results in 2 germline KI fragments. A vertical line has been inserted between samples T4 and T5 (SacI digests) to indicate a repositioned gel lane.
Accumulation of B cells expressing the nontargeted IgMb H chain allele could either reflect a selective polyclonal hyperplasia or a monoclonal expansion of IgMb-expressing B cells. To distinguish these 2 possibilities, we did an initial screening for Ig L chain usage in the expanding B-cell populations in IgH.T and IgH.TEμ UNI mice and found that the CD19+ B-cell population was almost entirely Ig κ positive (with < 0.5% of λ+ B cells) or in 2 cases largely Ig λ+ (Figure 4C). Usage of only a single L chain indicated monoclonality of the accumulating B-cell populations in IgH.T and IgH.TEμ UNI mice. This was confirmed by detailed analysis of the IgH rearrangement status by Southern blotting of XbaI digests of genomic DNA from purified CD19+ splenic B-cell fractions from 9 heterozygous IgH.TEμ UNI mice. We used a JH region probe, which recognizes a 3.5-kb XbaI fragment in a germline wild-type IgH allele and a 2.0/1.7-kb doublet fragment in a germline knockin IgH.TEμ allele, as present in genomic DNA from non-B cells (Figure 4D-E). All 9 samples contained prominent approximately 2.0/1.7-kb fragments of the IgH.TEμ allele in the germline configuration. In addition, approximately 3.5-kb fragments of the germline wild-type IgH allele were detected (generally of low density), most probably reflecting non-B cells present in the spleen samples. In addition, uniquely rearranged wild-type IgH alleles were present (in the 2 cases shown in Figure 4E, these fragments are ∼ 3 and ∼ 4 kb).
Similar results were obtained when we tested DNA from 12 additional heterozygous IgH.TEμ UNI mice using SacI digests. All 12 splenic B-cell fractions showed an approximately 5-kb fragment of the IgH.TEμ allele in the germline configuration, a weak approximately 4.1-kb germline wild-type IgH allele, as well as uniquely rearranged wild-type alleles in the range of approximately 7 to 10 kb (shown for 3 cases in Figure 4E). The identification of unique, singly rearranged JH fragments confirmed the expansion of single B-cell clones in UNI animals. Furthermore, the presence of germline knockin IgH.TEμ alleles indicated that the targeted IgH locus with the SV40 T gene was still in germline configuration. This was confirmed by the finding that a SV40 large T antigen–specific probe hybridized to these germline knockin IgH.TEμ restriction fragments, without revealing any other unique fragments (in 9 of 9 samples tested; data not shown).
In summary, aged IgH.T and IgH.TEμ UNI mice show accumulation of monoclonal B-cell populations that have retained the targeted IgH allele in the germline configuration. We did not find evidence for the occurrence of Rag-mediated transposition, as no SV40 T sequences were detected on new unique restriction fragments.
Monoclonal B-cell populations in IgH.TEμ mice have a CLL-like phenotype
Splenic B-cell fractions from aged IgH.T/IgH.TEμ UNI mice were found to express significantly higher levels of SV40 T antigen transcripts than B cells from RAN mice, as determined by quantitative RT-PCR (Figure 5A). SV40 large T protein was identified in all tumor samples by Western blotting (Figure 5B). As expected, no SV40 T transcription or protein was detected in wild-type (WT) splenic B cells (not shown).
Monoclonal B-cell populations in aged IgH.TEμ mice express T antigen and CLL markers. (A) Quantitative RT-PCR analysis of large T antigen expression in RAN (▾, n = 5) and UNI (■, n = 16) purified splenic CD19+ B-cell fractions. Expression was normalized with HPRT, and relative expression in RAN B-cell fractions was set to 1. Each symbol represents an individual mouse; mean values are indicated by a line (*Significant difference with WT; P < .01; Mann-Whitney U test). (B) Western blot showing expression of large T protein (LT, arrow) in purified B-cell fractions from IgH.TEμ UNI mice. Numbers on the left indicate size in kDa. Examples are representative for 20 CLL samples analyzed. (C) Expression of CD19, IgM, IgD, CD5, and CD43 on BM, spleen, and lymph node cells from a IgH.TEμ UNI mouse. CD19+ cells were gated and analyzed for their IgM/IgD and CD5/CD43 profiles. Data are representative of 33 animals analyzed (for age of mice, see Table S1).
Monoclonal B-cell populations in aged IgH.TEμ mice express T antigen and CLL markers. (A) Quantitative RT-PCR analysis of large T antigen expression in RAN (▾, n = 5) and UNI (■, n = 16) purified splenic CD19+ B-cell fractions. Expression was normalized with HPRT, and relative expression in RAN B-cell fractions was set to 1. Each symbol represents an individual mouse; mean values are indicated by a line (*Significant difference with WT; P < .01; Mann-Whitney U test). (B) Western blot showing expression of large T protein (LT, arrow) in purified B-cell fractions from IgH.TEμ UNI mice. Numbers on the left indicate size in kDa. Examples are representative for 20 CLL samples analyzed. (C) Expression of CD19, IgM, IgD, CD5, and CD43 on BM, spleen, and lymph node cells from a IgH.TEμ UNI mouse. CD19+ cells were gated and analyzed for their IgM/IgD and CD5/CD43 profiles. Data are representative of 33 animals analyzed (for age of mice, see Table S1).
We characterized the phenotype of the monoclonal B-cell populations in IgH.T and IgH.TEμ mice by flow cytometry and quantitative RT-PCR. In these analyses, we did not detect phenotypic differences between clonal B-cell populations from IgH.T or IgH.TEμ mice. The clonal B-cell populations expressed CD19, B220, CD2, and substantial levels of surface IgM, but surface IgD was generally low (Figure 5C). They were negative for the early differentiation markers surrogate light chain, IL-7R or AA4.1 and the mature B-cell markers CD21, CD23, and CD138 (data not shown). Importantly, all B-cell leukemia samples expressed significant levels of CD43 and CD5 on the cell surface (Figure 5C), both of which are associated with B-CLL in human. In all UNI mice substantial populations of CD19+CD5+CD43+ cells were identified in blood, lymph nodes, spleen, and bone marrow (Table S1).
p53-deficiency increases CLL incidence in IgH.TEμ mice
All mice in the IgH.TEμ panel manifested leukemia formation in blood, spleen, and BM before the age of 10 months (Figure 6A), but in the IgH.T panel only 2 of 15 (Figure 6B). At one year of age, the remaining mice in the IgH.T group were killed and found to have no B-cell abnormalities.
Effect of p53 on CLL development in IgH.TEμ and IgH.T mice. (A) Kaplan-Meier survival curve of IgH.TEμ (black, n = 31), IgH.TEμ/p53−/− (red, n = 6), and p53−/− (blue, n = 9) mice. (B) Kaplan-Meier survival curve in IgH.T (black, n = 15), IgH.T/p53−/− (red, n = 10), and p53−/− (blue, n = 9) mice. Tumor-free survival is plotted over time.
Effect of p53 on CLL development in IgH.TEμ and IgH.T mice. (A) Kaplan-Meier survival curve of IgH.TEμ (black, n = 31), IgH.TEμ/p53−/− (red, n = 6), and p53−/− (blue, n = 9) mice. (B) Kaplan-Meier survival curve in IgH.T (black, n = 15), IgH.T/p53−/− (red, n = 10), and p53−/− (blue, n = 9) mice. Tumor-free survival is plotted over time.
In human B-CLL, loss of p53 function is associated with accelerated disease progression and poor prognosis.6,7 To study the effect of p53 loss in our mouse model, IgH.TEμ and IgH.T mice were crossed onto a p53-deficient background. Relative to p53−/− mice, the groups of IgH.TEμ;p53−/− and IgH.T;p53−/− mice did not show a significant increase in tumor formation (Figure 6). However, whereas p53−/− littermate controls all developed T-cell tumors, 5 of 6 animals in the IgH.TEμ;p53−/− panel developed B-cell tumors, with the remaining animal developing a T-cell tumor. Likewise, 4 of 10 mice in the IgH.T;p53−/− panel had B-cell tumors. The age at which malignancies were evident decreased in IgH.TEμ;p53−/− mice, compared with IgH.TEμ mice (median survival of 138 and 161 days, respectively; P < .01, the log-rank test). This was also the case for IgH.T;p53−/− mice, compared with IgH.T mice (median survival of 143 days and > 365 days, respectively; P < .001). The phenotypes of B-cell tumors in IgH.TEμ;p53−/− and IgH.T;p53−/− were similar to those present in IgH.TEμ or IgH.T animals on the wild-type background, as characterized by flow cytometry and Southern blotting analysis of IgH alleles (not shown).
Taken together, these results demonstrate that, although SV40 large T antigen acts as an inhibitor of the p53 pathway, complete loss of p53 increases B-CLL incidence in IgH.TEμ and IgH.T mice.
Ig VH usage in IgH.TEμ leukemias resembles unmutated human CLL
Based on the presence or absence of Ig VH gene mutations, human CLL patients are categorized into 2 subgroups with different prognosis. We therefore decided to characterize the V regions from 8 IgH.TEμ and one IgH.T mouse. We purified CD19+ splenic B-cell fractions and performed Ig VH family specific RT-PCR analyses. First, we confirmed clonality of the leukemias by heteroduplex analysis, in which homoduplexes and heteroduplexes resulting from denaturation and renaturation of IgH V region RT-PCR products were separated in nondenaturing polyacrylamide gels based on their conformation.29 For most samples, we observed clear single homoduplexes, confirming the amplification of clonally rearranged V regions, but one case showed the presence of oligoclonal bands. DNA sequence analysis revealed that 4 leukemias expressed VH11.2, another 4 expressed VH segments from the VHJ558 family, and one expressed both VH11.2 and VHJ558 segments, indicating biclonality.
In the leukemias expressing VH11.2, which is the only member of the VH11 family, the sequence was identical to the germline gene present in the IMTG database (www.imgt.org; Table 1). The VH11.2 gene is preferentially expressed in B-1 lymphocytes and is associated with autoreactivity and oligoclonal expansions of CD5+ B cells in old mice.30 Three of the J558 family genes expressed were (nearly) identical to their most homologous published germline genes. By contrast, 2 VHJ558+ leukemias manifested extensive somatic hypermutation (SHM). One IgH.TEμ B-CLL expressed VH1-69, which is often found in human CLL,31 and contained 15 mutations (10 in CDRs) and one IgH.T B-CLL expressed VH1-74 and contained 13 mutations (5 in CDRs). These 2 CLL samples probably originated from GC-experienced B cells with high activity of activation-induced cytidine deaminase (Aid), which induces SHM.
Characterization of IgH chains of B-CLL
| Tumor no. . | IMGT VH segment . | VH family . | % difference from GL . | N, P . | DH segment . | N, P . | JH segment . | DH RF . | CDR3 . | CDR3 length . | pI . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| T1 | VH11-2 | VH11 | 0 | — | DST4.2 | — | JH1 | 3 | MRYSNYWYFDV | 11 | 6.4 |
| T5 | VH11-2 | VH11 | 0 | — | DST4.2 | — | JH1 | 3 | MRYSNYWYFDV | 11 | 6.4 |
| T20 | VH11-2 | VH11 | 0 | — | DFL16.1 | — | JH1 | 3 | MRYGSSYWYFDV | 12 | 6.4 |
| T23 | VH11-2 | VH11 | 0 | — | DSP2.1 | — | JH1 | 3 | MRYGNYWYFDV | 11 | 6.4 |
| T4 | VH11-2 | VH11 | 0 | — | DFL16.1 | — | JH1 | 3 | MRYGSSYWYFDV | 12 | 6.4 |
| T9 | VH1-18 | VHJ558 | 0 | aggg | DFL16.1 | gtagg | JH3 | 3 | ARRDYGSSYVGWFAY | 15 | 9.0 |
| T16 | VH1-55 | VHJ558 | 1 | ggg | - | — | JH2 | ARGFDY | 6 | 6.4 | |
| T3 | VH1-58 | VHJ558 | 0 | gagagagtg | DSP2.2 | ctgg | JH3 | 3 | ARERVYDYDLAWFAY | 15 | 4.7 |
| T23 | VH1-69 | VHJ558 | 6.8 | t | DFL16.2 | t | JH2 | 3 | ARYDYYYYYC | 9 | 6.3 |
| T32 | VH1-74 | VHJ558 | 5.8 | gggat | D3–3 | — | JH4 | 3 | ARDDPLGYAMDY | 12 | 4.1 |
| Tumor no. . | IMGT VH segment . | VH family . | % difference from GL . | N, P . | DH segment . | N, P . | JH segment . | DH RF . | CDR3 . | CDR3 length . | pI . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| T1 | VH11-2 | VH11 | 0 | — | DST4.2 | — | JH1 | 3 | MRYSNYWYFDV | 11 | 6.4 |
| T5 | VH11-2 | VH11 | 0 | — | DST4.2 | — | JH1 | 3 | MRYSNYWYFDV | 11 | 6.4 |
| T20 | VH11-2 | VH11 | 0 | — | DFL16.1 | — | JH1 | 3 | MRYGSSYWYFDV | 12 | 6.4 |
| T23 | VH11-2 | VH11 | 0 | — | DSP2.1 | — | JH1 | 3 | MRYGNYWYFDV | 11 | 6.4 |
| T4 | VH11-2 | VH11 | 0 | — | DFL16.1 | — | JH1 | 3 | MRYGSSYWYFDV | 12 | 6.4 |
| T9 | VH1-18 | VHJ558 | 0 | aggg | DFL16.1 | gtagg | JH3 | 3 | ARRDYGSSYVGWFAY | 15 | 9.0 |
| T16 | VH1-55 | VHJ558 | 1 | ggg | - | — | JH2 | ARGFDY | 6 | 6.4 | |
| T3 | VH1-58 | VHJ558 | 0 | gagagagtg | DSP2.2 | ctgg | JH3 | 3 | ARERVYDYDLAWFAY | 15 | 4.7 |
| T23 | VH1-69 | VHJ558 | 6.8 | t | DFL16.2 | t | JH2 | 3 | ARYDYYYYYC | 9 | 6.3 |
| T32 | VH1-74 | VHJ558 | 5.8 | gggat | D3–3 | — | JH4 | 3 | ARDDPLGYAMDY | 12 | 4.1 |
IMGT indicates the international ImMunoGeneTics information system (www.imgt.org); GL, germline; pI, isoelectric point; —, not applicable; RF, reading frame; VH, Ig H chain V segment; N, P, N,P nucleotides; JH, Ig H chain J segment; and DH, Ig H chain D segment.
Nonstochastic usage was not only observed for VH segments but also for D and JH segments: eg, DST4.2 was found twice (normally DST segment usage is < 2%) and all VH11–2 VH segments were joined to JH1 (50%; normally, JH1 usage is < 16%). Unmutated poor outcome human B-CLL cases often express IgH chains with long CDR3s, containing multiple neutral tyrosine and serine residues that may confer CDR3 flexibility and favor polyreactivity. The CDR3 length, which is on average approximately 12 amino acids in murine B cells,32 was 11 to 12 amino acids in 6 leukemias, whereas 2 leukemias had a short (6 and 9 amino acids) and 2 leukemias had a long CDR3 length (15 amino acids). Eight of 10 CDR3s were enriched for tyrosines and serines (3-6 residues), and all CDR3s contained multiple charged amino acids, whereby the pI values were relatively neutral in 7 of 10 cases. As a result of the biased use of certain VH, D, and JH gene segments, the 5 VH11.2+ leukemias had identical or strikingly homologous IgH V regions, similar to the stereotypic antigen-binding sites observed in human B-CLL patients.
Similar transcription factor expression in Aidhigh and Aidlow B-CLL cells
SHM of Ig VH genes provides prognostic information for CLL patients,1,2 whereas hypermutation is predictive of a more favorable prognosis, lack of mutation predicts a poor prognosis. However, both cases identified with somatic hypermutation manifested high splenic cellularity (0.4 and 1.4 × 109 cells; Table S1).
We investigated Aid expression in purified splenic CD19+ B-cell fractions from leukemic mice by RT-PCR. We found that Aid expression was increased in a subset of these CLLs, compared with B-cell populations from wild-type or nonleukemic IgH.T/IgH.TEμ mice (Figure 7A). This Aidhigh subset included leukemic samples expressing unmutated VH11, whereas Aid expression in the 2 samples with high levels of SHM was low. The Aidhigh B-CLL subset did not show late onset or slow disease progression, compared with Aidlow B-CLL.
Similar transcription factor expression in AIDhigh and AIDlow CLL cells. (A) Quantitative RT-PCR analysis of AID expression in RAN (▾, n = 7) and UNI (■, n = 16) purified CD19+ B-cell fractions. Expression was normalized with HPRT, and relative expression in wild-type B-cell fractions was set to 1. (B) Expression of indicated transcription factors as measured by quantitative RT-PCR in AIDhigh (▨, n = 5; relative expression > 2, panel A) and AIDlow (□, n = 11; relative expression < 2). Expression was normalized with HPRT, and relative expression in RAN B-cell fractions was set to 1. Mean values and SEM are given. * Significant differences between AIDlow and AIDhigh CLL cells (P < .001, t test).
Similar transcription factor expression in AIDhigh and AIDlow CLL cells. (A) Quantitative RT-PCR analysis of AID expression in RAN (▾, n = 7) and UNI (■, n = 16) purified CD19+ B-cell fractions. Expression was normalized with HPRT, and relative expression in wild-type B-cell fractions was set to 1. (B) Expression of indicated transcription factors as measured by quantitative RT-PCR in AIDhigh (▨, n = 5; relative expression > 2, panel A) and AIDlow (□, n = 11; relative expression < 2). Expression was normalized with HPRT, and relative expression in RAN B-cell fractions was set to 1. Mean values and SEM are given. * Significant differences between AIDlow and AIDhigh CLL cells (P < .001, t test).
Finally, we used quantitative RT-PCR to analyze the Aidlow and Aidhigh subsets for the expression of transcription factors implicated in B-cell activation and terminal differentiation. Compared with untransformed B cells, leukemic IgH.T/IgH.TEμ B cells showed markedly increased expression of the transcription factors Obf and Eza (consistent with the reported increased Eza expression in human CLL33 ), whereas expression of Id2, an inhibitor of Eza, was reduced. Expression of Irf4, which correlates with clinical outcome in B-CLL patients,34 was approximately 2 to 3 times increased. Overall, Aidlow and Aidhigh leukemia samples did not manifest significant differences in the expression of the transcription factors analyzed. Only expression of Pax-5, a B-cell-specific factor that is down-regulated on terminal differentiation, was significantly higher in Aidhigh leukemia samples. This finding does not necessarily indicate that Aidlow have differentiated further toward plasma cells, as they did not show higher levels of Blimp-1, which is highly expressed in plasma cells.
Taken together, B-CLL cells with unmutated V regions express substantial levels of Aid transcripts, which parallels findings in human B-CLL. Moreover, Aidlow and Aidhigh B-CLL manifest similar transcription factor expression patterns, supporting a common origin of the 2 B-CLL subsets.
Discussion
We describe the generation of a B-CLL mouse model based on sporadic SV40 large T antigen expression in mature B cells. Leukemic cells present in these mice displayed many characteristics also found in human B-CLL, in particular in the subgroup of patients with germline IgH V regions associated with poor prognosis. These features include (1) a mature CD19+CD5+ phenotype, (2) VH regions with predominantly germline-encoded sequences, (3) nonstochastic VH-family usage, and (4) CDR3 regions with high serine/tyrosine content.
The predominant use of VH11 in our IgH.TEμ leukemic cells cannot result from random transformation of CD5+ B-1 cells because only approximately 10% of this B-cell population use VH11.35 It rather suggests the involvement of (auto)antigen-driven selection. Increased VH11 usage was also found in clonal expansions of splenic CD5+ B lymphocytes in aging mice.30 Moreover, the VH11- DFL16.1- JH1 H chain present in leukemias T4 and T20 was identical to the published CLL clone TCL1-005 from a Tcl1 transgenic mouse.36 Thus, the B-CLL cells express a restricted B-cell receptor (BCR) repertoire with several cases of (nearly) identical CDR3s. In this respect, our model replicates the human B-CLL BCR sequences that closely resemble those of known autoreactive or polyreactive antibodies. We found that VH regions were either unmutated, with preferential usage of the VH11 family, or manifested extensive somatic hypermutation and used VHJ558. The finding of high Aid expression in unmutated VH11-expressing CLL, together with the similar transcription factor expression patterns in Aidlow and Aidhigh leukemia samples, would support a common post-GC origin of both CLL subsets.
In our IgH.T/IgH.TEμ models, expression of SV40 large T antigen was achieved by insertion of a promoterless SV40 T gene in opposite transcriptional orientation between the D and JH segments. We detected SV40 T transcripts in mature B cells of IgH.T/IgH.TEμ mice, most probably from those rare targeted IgH alleles that have retained their germline configuration. The SV40 large T antigen is probably expressed as part of an antisense transcript within the D-JH region of the IgH locus, similar to the antisense transcripts described in pro-B cells.18 The apparent absence of pro-B cell leukemias in IgH.T/IgH.TEμ mice indicates that transient expression of the SV40 T gene (until it is lost as a result of D to JH recombination) did not result in malignant transformation of pro-B cells. Likewise, we did not detect early T-cell lineage leukemias, despite the presence of IgH chain antisense transcription in early thymocytes.18 From the finding of equal usage of the targeted and the wild-type IgH allele, we conclude that insertion of the T gene or the additional Eμ element did not affect V(D)J recombination. However, as IgH.TEμ mice manifested a substantially higher tumor incidence than IgH.T mice, the presence of an extra copy of Eμ within the D-JH region may increase the level of antisense transcription. Thus, efficient transformation might be dependent on synergistic effects of Eμ and the IgH 3′ enhancers37 that lead to up-regulation of SV40 T expression in more mature B-cell stages. Conversely, the absence of transformation of pre-B cells or immature B cells that have retained the SV40 T gene could be explained by a level of T expression that is below the threshold for transformation.38,39 It is also possible that transformation requires secondary events that only take place in mature B cells.
The phenotypes of the leukemias in our mouse models resemble those found in Eμ-TCL1 mice, which express the T-cell leukemia/lymphoma-1 proto-oncogene under the control of the Eμ element and an Ig VH promoter.40 TCL1 has been functionally linked to enhanced Pkb/Akt-mediated signaling pathways involved in cell proliferation and survival. In contrast, SV40 large T protein interacts with numerous cellular proteins and pathways, most notably the Rb and p53 pathways,10,13,41,42 but may also induce cell survival via Akt activation.43 In our mouse model, animals that lack p53 show accelerated tumor formation, even though SV40 large T inactivation of p53 is thought to be necessary to generate lymphoid tumors.44 This implies that, in the presence of p53, some of the potential transforming properties of large T are inhibited or diminished, as was shown in a mouse model of pancreatic islet carcinogenesis.38,45 Together with the existing communication and coordination between the p53 and Akt pathways, it is very well possible that the mechanisms of malignant transformation by SV40 T antigen and TCL1 are related. Compared with the Eμ-TCL1 mice, in which VH regions of leukemias consistently differed only marginally (< 2.0%) from the germline, our IgH.TEμ/IgH.T mouse models are unique in that, next to leukemias with unmutated BCR, we also observed leukemias with extensive SHM (Table 1). It is presently not clear whether this difference between the Eμ-TCL1 and IgH.TEμ mice would point to differences in the mechanism of malignant transformation or would be related to secondary events in leukemia formation.
Although evidence accumulated that the V(D)J recombinase system is capable of mediating transposition of cleaved signal ends into nonspecific sites in the genome, both in vitro and in vivo,19,21 we did not find evidence for transposition events: all CLLs analyzed had retained the T gene in the IgH locus, which still had the germline configuration. Obviously, we cannot exclude that in some B cells transposition did take place, but with tranposition into transcriptionally inactive regions and therefore not resulting in leukemia. Even though transposition has been shown to occur in vivo,21,23,46 its frequency is very low. Therefore, our mouse model would argue for the existence of mechanisms that very effectively protect against transposition in vivo.47
To be able to investigate the molecular mechanisms involved in CLL pathogenesis, next to the New Zealand Black mouse strain, which is a naturally occurring model of late-onset CLL, several transgenic mouse models have been generated.48 Because in human B-CLL is a heterogeneous disease, each of the current models will provide invaluable knowledge about the molecular mechanisms of transformation in B-CLL. As a preclinical model to test novel treatment strategies for CLL, our leukemia model has the advantage that tumor progression can be monitored by an IgM allotype-specific flow cytometry assay. Finally, gene profiling studies have shown that the SV40 T antigen genetic signature (1) is composed primarily of genes regulating cell replication, proliferation, DNA repair, and apoptosis, (2) does not reflect a general cancer signature, and (3) is uniquely activated primarily in tumors with aberrant expression of p53, Rb, or Brca1 and not in tumors initiated through Myc or ras overexpression.49 As human breast, lung, and prostate tumors expressing this set of genes represent subsets of tumors with the most aggressive phenotype,49 our SV40 large T-dependent CLL model should allow the identification of genes that are associated with treatment resistance and poor prognosis.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the Dutch Cancer Foundation (KWF/NKB; D.C.v.G., R.W.H.) and the Netherlands Organization for Scientific Research /Mozaiek (V.B.T.T.).
Authorship
Contribution: P.J.t.B., V.B.T.T., M.J.W.d.B., A.M., and G.K. performed experiments; P.J.t.B. and V.B.T.T. analyzed results; P.J.t.B. made the figures; and P.J.t.B., D.C.v.G., and R.W.H. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rudi W. Hendriks, Department of Pulmonary Medicine, Erasmus MC Rotterdam, PO Box 2040, 3000 CA Rotterdam, The Netherlands; e-mail: r.hendriks@erasmusmc.nl.