Abstract
In addition to orchestrating the expression of all erythroid-specific genes, GATA-1 controls the growth, differentiation, and survival of the erythroid lineage through the regulation of genes that manipulate the cell cycle and apoptosis. The stages of mammalian erythropoiesis include global gene inactivation, nuclear condensation, and enucleation to yield circulating erythrocytes, and some of the genes whose expression are altered by GATA-1 during this process are members of the p53 pathway. In this study, we demonstrate a specific in vitro interaction between the transactivation domain of p53 (p53TAD) and a segment of the GATA-1 DNA-binding domain that includes the carboxyl-terminal zinc-finger domain. We also show by immunoprecipitation that the native GATA-1 and p53 interact in erythroid cells and that activation of p53-responsive promoters in an erythroid cell line can be inhibited by the overexpression of GATA-1. Mutational analysis reveals that GATA-1 inhibition of p53 minimally requires the segment of the GATA-1 DNA-binding domain that interacts with p53TAD. This inhibition is reciprocal, as the activation of a GATA-1–responsive promoter can be inhibited by p53. Based on these findings, we conclude that inhibition of the p53 pathway by GATA-1 may be essential for erythroid cell development and survival.
Introduction
GATA-1 is the founding member of the GATA factor family of DNA-binding proteins,1 and is critical to the development of the erythroid,2,3 megakaryocyte,4 mast,5 and eosinophil6 cell lineages. It is multifunctional and acts as a global regulator of erythroid-specific genes.7 It is both an activator and repressor of transcription8,9 and participates in long-range gene regulation through the ß-globin locus control region10,11 and the c-kit locus.12 GATA-1 alters chromatin structure; it is a substrate for acetylation,13,14 sumolation,15 and ubiquitination,16 and recruits the corresponding activities to chromatin.
The GATA-1 DNA-binding domain (DBD) consists of 2 zinc-finger domains (C-X2-C-X17 and C-X2-C type) and is highly conserved among the 6 vertebrate GATA factors. The carboxyl-terminal zinc finger (GATA-1 CF) constitutes the primary DBD and is capable of independent high-affinity binding9 to the consensus motif A/TGATAA/G.17 The amino-terminal zinc-finger domain (GATA-1 NF) binds independently to a related sequence containing a GATC core with lower affinity,18,19 and stabilizes binding to DNA with multiple GATA sites.9,20,21 In addition to their role in DNA binding, both zinc fingers participate in protein-protein interactions22,23 with multiple partners, including friend-of-GATA-1 (FOG-1)23 and PU.1.24
GATA-1 is required for generation of erythroid cells,2,3 and GATA-1-negative embryonic stem cells have been used to demonstrate that definitive erythroid cells lacking GATA-1 are blocked at the proerythroblast stage of development.25 Unlike wild-type cells, GATA-1 null definitive erythroid progenitors die of apoptosis, leading to the conclusion that GATA-1 controls the survival of erythroid cells in addition to contributing to growth and differentiation. GATA-1 stimulates the expression of Bcl-xL,26 an antiapoptotic Bcl-2 family member that is critical to the erythroid lineage,27 and Bcl-xL most likely contributes to increased cell survival seen in the presence of GATA-1. This was confirmed in studies with the GATA-1.05 mouse,28 in which it was shown that 5% of the normal level of GATA-1 is sufficient to stop the apoptosis of day 11 definitive erythroid precursors and to increase the expression of Bcl-xL.29 The observed apoptosis in the absence of GATA-1 occurs without the accumulation of the tumor suppressor protein p53, through an undetermined mechanism.25,29
The process of terminal erythroid differentiation is one that might induce the p53 pathway because DNA breakage accompanies nuclear condensation and enucleation. p53 is activated by chromosome instability and by DNA damage; however, the p53 pathway remains largely inactive during erythropoiesis even though apoptosis is an integral part of the process.30 p53 accumulates at low levels during nuclear apoptosis in the normoblast stage of erythropoiesis just before enucleation, but surprisingly, the cells survive.31 A modified p53 response rather than the classical pathway is observed, concomitant with a reduction of caspase 3/7, the ultimate agent of apoptosis. Recently, erythroid conditional knockouts of the inhibitory p53 partner protein genes murine double minute (Mdm)–2 and Mdm-4 in wild-type and in p53−/− mice have shown unequivocally that a competent p53 pathway is present and is induced in erythroid cells in the absence of these proteins.32 Mdm-2 and -4 are clearly involved in preventing the p53 response, but there are most likely other factors because a specialized p53 response is observed (in normoblasts).
The last few cell divisions are critical in erythroid differentiation, because a controlled exit from the cell cycle without induction of apoptosis is essential. GATA-1 regulates cell cycle progression in a concentration- and context-dependent manner, and can both block and accelerate the cell cycle.33 The restoration of GATA-1 to the GATA-1 null erythroid cell line, G1E, altered the expression of many cell cycle regulators in a manner consistent with the induction of cell cycle arrest33 ; at least one of these, c-myc, was shown to be directly inhibited by GATA-1.33 The expression of the p21 gene, which causes cell cycle withdrawal, was stimulated by GATA-1 in a murine erythroleukemia cell line (MEL).34 p53 also induces cell cycle arrest in part by regulating the same genes. It directly inhibits the transcription of the proliferation-inducing oncogene, c-myc,35 and activates the p21 gene, a primary component of the p53 response. In other instances, GATA-1 and p53 appear to have opposing functions, as GATA-1 stimulates the antiapoptotic Bcl-xL gene, but p53 induces several proapoptotic Bcl-2 family members such as Bax36 and Noxa,37 and all function in the same intrinsic apoptotic pathway. In addition, the tumor suppressor RB interacts with both the p53-Mdm-2 complex38 and GATA-1.39 Thus, several proteins that are prominent in GATA-1 function are also participants in the p53 pathway. The observation that the p53 transactivation domain (TAD) has similar amino acid composition (high in acidic and hydrophobic residues) to the TAD of PU.1, which interacts with the GATA-1 DBD, resulting in inhibition of GATA-1,24 caused us to consider that a similar mechanism might explain the interplay between the GATA-1 and p53 pathways.
In this study, we demonstrate a specific interaction between the TAD of p53 (p53TAD) and a segment of GATA-1 that includes the linker between the zinc fingers and the carboxyl-terminal zinc-finger domain (GATA-1 L plus CF). We also show that the native proteins interact and can be coimmunoprecipitated from MEL cell extract. Furthermore, the activation of p53-responsive promoters in an erythroid cell line is inhibited by overexpression of GATA-1, but not by the expression of a GATA-1 mutant that lacks a region required for binding p53. The inhibition is reciprocal, because the activation of a GATA-1–responsive promoter can be inhibited by p53.
Methods
Clones
The p53 glutathione-S-transferase (GST) fusion constructs were provided by Dr Rong Li (University of Virginia, Charlottesville, VA). The GATA-1 GST fusion constructs were prepared by inserting hGATA-1 fragments into the pGEX-5X (GE Healthcare) vector. The GATA-1199-317 and GATA-1252-317 expression constructs were prepared by inserting the fragments of hGATA-1 into the pet11d (Novagen) vector. Mutations were made by site-directed mutagenesis and sequenced.
GST fusion proteins
The GST-GATA-1 fragments were expressed in Escherichia coli strain BL21(DE3) (Novagen). The cells were lysed, centrifuged at 100 000g, and incubated with glutathione (GSH)–Sepharose resin (GE Healthcare). GST-GATA-1 fragments were eluted off the resin with GSH (Sigma-Aldrich). The proteins were dialyzed into storage buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, and 20% glycerol), and rebound to fresh GSH-Sepharose for binding studies.
Protein purification
Binding assays
GST pulldowns.
A total of 1 μM GST or GST–GATA-1 fragment was incubated with the resin in 0.5× Superdex. Purified p53 fragments (0.01-1 μM) were then incubated for 1 hour. After washing, the bound proteins were eluted and loaded onto a 15% acrylamide gel and separated by electrophoresis in Tris-glycine. The proteins were then transferred to an Immobilon-P membrane (Millipore), and the membrane was incubated overnight in Tris-buffered saline/Blotto. The membrane was then incubated with an anti-p53 primary antibody (p53 DO-1 or Pab 1801; Santa Cruz Biotechnology) and with a secondary antibody (goat anti–mouse immunoglobulin [Ig]G horseradish peroxidase [HRP]; Santa Cruz Biotechnology). The proteins were detected through chemiluminescence using ECL Plus (GE Healthcare). The concentrations of GST fusion proteins were determined from A280, and the membrane was Coomassie stained to ensure equal inputs.
Isothermal titration calorimetry-binding experiments.
Isothermal titration calorimetry (ITC) titrations were performed essentially as described.40 (For more details, see supplemental materials and methods, available on the Blood website; see the Supplemental Materials link at the top of the online article.)
Immunoprecipitations
MEL cell nuclei were isolated41 and nuclear extract was made, as described.41,42 A total of 320 μg of extract and 40 μL of protein G Plus agarose precleared with anti-mouse IgG (Pierce) was used for each immunoprecipitation. Samples were rotated at 4°C for 4 hours, followed by centrifugation at 1000g at 4°C. The agarose pellets were washed in phosphate-buffered saline (PBS) and suspended in sodium dodecyl sulfate–polyacrylamide gel electrophoresis loading buffer with 0.1% 2-mercaptoethanol (2-ME) and heated to 100°C for 10 minutes. GATA-1 antibodies (N6) sc265 and (M20) sc1234; p53 antibodies pab 240, 246, and FL393; cyclin B1 (GNS1) antibody, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) sc47724, anti–rat IgG HRP sc2032, and the protein G plus agarose were obtained from Santa Cruz Biotechnology. The anti–rat IgG2a clone R2A-2 antibody was obtained from Sigma-Aldrich. Denatured protein samples were electrophoresed and blotted with Tris glycine gels using polyvinylidene difluoride membrane. Three percent milk in PBS was used for blocking and antibody binding, and blots were washed with PBS with 0.1% Tween 20. Blots were developed with SuperSignal West Dura (Pierce).
DNA constructs
FR 7Luc, a GATA-responsive reporter gene, was made by inserting a blunt-end 470-bp HpaII to XmnI restriction fragment from the chicken folate receptor gene promoter into the SmaI site of pGL3 basic. This fragment contains 2 GATA sites.43 The GATA-1 mutant CM was made by site-directed mutagenesis. Amino acids KKR(245-247) and K(314-316) of human GATA-1 were converted to alanine and sequenced.
Cell culture and transfections
MEL cells were grown in Iscove modified Dulbecco medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (PS). The 6C2 cells were grown in α minimum essential medium with 10% FBS and 2% chicken serum, 0.5 mM 2-ME, 1 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), and 1% PS. Cells were maintained at 37°C and 5% CO2. The 6C2 cells were transfected with Effectene (QIAGEN), according to manufacturer's instructions, and harvested 18 to 24 hours later. A total of 5 × 106 cells per sample was used with 150 ng of control plasmid (cytomegalovirus [CMV] Renilla luciferase), 250 to 500 ng of reporter plasmid p53 Luc (Stratagene), or pBAX Luc and 250 to 500 ng of CMV p53 (in pLPC), whereas 0.5 to 3 μg of GATA-1 expression vectors or control vectors was used. The amount of CMV p53 was titrated to a range where a difference between the control plasmids or GATA-1 mutants and wild-type GATA-1 was observed. The Dual Luciferase Assay Kit (Promega) was used to process and analyze the samples, according to manufacturer's instructions. Luciferase activity was determined with a Victor Multilabel Counter from PerkinElmer. QT6 cells were transfected with Lipofectamine or Lipofectamine 2000 (Invitrogen). The Dual Luciferase Assay was used. All transfections were normalized to CMV Renilla luciferase, by setting the highest value for the positive control to 100% and adjusting other values in proportion.
Nuclear magnetic resonance studies
Nuclear magnetic resonance (NMR) spectra were collected at 300 K on a Varian 500 MHz spectrometer. The GATA-1 plus CF/DNA complex consisted of a 1:1 molar ratio of 15N-labeled GATA-1 plus CF and unlabeled double-stranded DNA (5′-GTTGCAGATAAACATT) at 0.5 mM, as previously described.44 For studies of the p53TAD, the sample consisted of 15N-labeled p53 at 0.5 mM (pH = 6.5, 12.5 mM NaCl, 90% H2O/10% D2O). For binding of the GATA-1 plus CF/DNA complex with p53TAD, we performed 2 titrations. The first titration consisted of a 0.5 mM GATA-1 plus CF (15N-labeled)/DNA complex, and unlabeled p53TAD was added in incremental amounts to a 1:1 ratio. The second titration consisted of 0.5 mM 15N-labeled p53TAD, and the unlabeled GATA-1 plus CF/DNA complex was added in incremental amounts to a 1:1 ratio.
Results
The zinc-finger domains of GATA-1 bind the full-length TAD of p53 in vitro
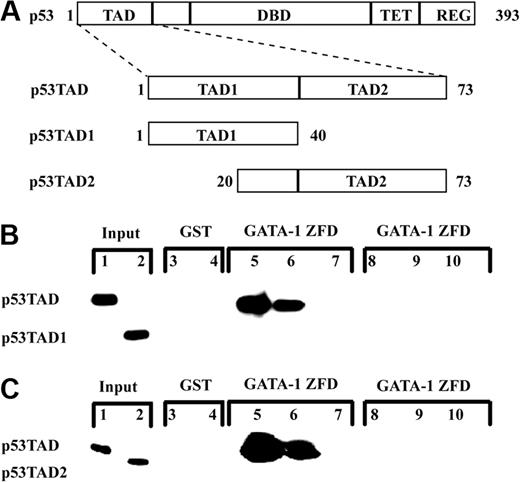
The TAD of p53 (p53TAD) is located within its amino terminus (residues 1-73). p53TAD contains 2 subdomains, p53TAD1 (residues 1-40) and p53TAD2 (residues 40-73), which are each capable of independently activating transcription (Figure 1A). Given the similarity between the TADs of p53 and PU.1, we tested whether the zinc-finger domains of GATA-1 interacted with the TAD of p53. A GST fusion protein containing the 2 zinc-finger domains of GATA-1 (GATA-1 ZFD; GATA-1199-317) was incubated with increasing concentrations of purified fragments of p53 corresponding to p53TAD (p531-73) and p53TAD1 (p531-40) in a GST pulldown assay. GST-GATA-1 ZFD bound to p53TAD, but not to p53TAD1, and GST alone did not interact with either p53TAD or p53TAD1 (Figure 1B). Next, we tested whether a fragment of p53 containing p53TAD2 (p5320-73) interacted with the GST-GATA-1 ZFD. Again, GST-GATA-1 ZFD bound to p53TAD, but not to the p53TAD2 subdomain (Figure 1C). Based on the GST pulldown assay, both subdomains of the TAD of p53 (p53TAD) are necessary and sufficient for interaction with the ZFDs of GATA-1 (GATA-1 ZFD).
The full-length p53TAD binds to the zinc-finger domains of GATA-1 in an in vitro pulldown assay. (A) Schematic of the TAD of p53 demonstrating position of the full-length p53TAD and the location of the 2 independent subdomains: p53TAD1 and p53TAD2. (B) Binding of p53TAD and p53TAD1 to GST-GATA-1 ZFD (GATA-1199-317). Various concentrations (1.0, 0.1, and 0.01 μM) of p53TAD (lanes 5-7) and p53TAD1 (lanes 8-10) were incubated with 1 μM GST-GATA-1 ZFD. In the GST lanes, 1 μM purified p53TAD (lane 3) and p53TAD1 (lane 4) was incubated with 1 μM GST as a control. The input lanes are p53TAD (lane 1) and p53TAD1 (lane 2) at 0.5%. Bound protein was detected with an anti-p53 antibody DO-1. (C) Binding of p53TAD and p53TAD2 to GATA-1 ZFD. Various concentrations (1.0, 0.1, and 0.01 μM) of p53TAD (lanes 5-7) and p53TAD2 (lanes 8-10) were incubated with 1 μM GST-GATA-1 ZFD. In the GST lanes, 1 μM purified p53TAD (lane 3) and p53TAD2 (lane 4) was incubated with 1 μM GST as a control. The input lanes are p53TAD (lane 1) and p53TAD2 (lane 2) at 0.5%. Bound protein was detected with an anti-p53 antibody Pab 1801.
The full-length p53TAD binds to the zinc-finger domains of GATA-1 in an in vitro pulldown assay. (A) Schematic of the TAD of p53 demonstrating position of the full-length p53TAD and the location of the 2 independent subdomains: p53TAD1 and p53TAD2. (B) Binding of p53TAD and p53TAD1 to GST-GATA-1 ZFD (GATA-1199-317). Various concentrations (1.0, 0.1, and 0.01 μM) of p53TAD (lanes 5-7) and p53TAD1 (lanes 8-10) were incubated with 1 μM GST-GATA-1 ZFD. In the GST lanes, 1 μM purified p53TAD (lane 3) and p53TAD1 (lane 4) was incubated with 1 μM GST as a control. The input lanes are p53TAD (lane 1) and p53TAD1 (lane 2) at 0.5%. Bound protein was detected with an anti-p53 antibody DO-1. (C) Binding of p53TAD and p53TAD2 to GATA-1 ZFD. Various concentrations (1.0, 0.1, and 0.01 μM) of p53TAD (lanes 5-7) and p53TAD2 (lanes 8-10) were incubated with 1 μM GST-GATA-1 ZFD. In the GST lanes, 1 μM purified p53TAD (lane 3) and p53TAD2 (lane 4) was incubated with 1 μM GST as a control. The input lanes are p53TAD (lane 1) and p53TAD2 (lane 2) at 0.5%. Bound protein was detected with an anti-p53 antibody Pab 1801.
Identification of the minimal p53-interacting region of GATA-1
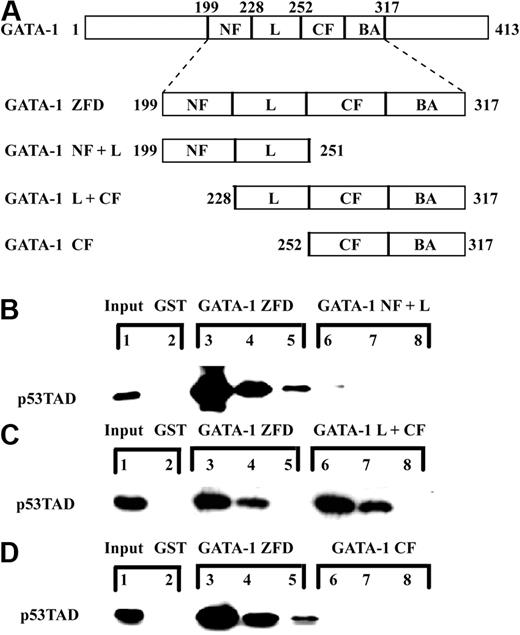
After establishing that both of the subdomains of the TAD of p53 (p53TAD) were required for interaction with the zinc-finger domains of GATA-1 (GATA-1 ZFD), we tested the ability of the individual zinc-finger domains of GATA-1 to interact with p53TAD. Three GST fusion proteins containing different combinations of the zinc-finger domains of GATA-1 were tested with p53TAD (Figure 2A). The 3 constructs included (1) the amino-terminal zinc finger plus the linker region (GATA-1 NF plus L; GATA-1200-251); (2) the linker region plus the complete carboxyl-terminal zinc finger domain (GATA-1 L plus CF; GATA-1228-317); and (3) the carboxyl-terminal zinc finger (GATA-CF; GATA-1252-317). First, GST-GATA-1 NF plus L was incubated with p53TAD, and the binding was compared with GST-GATA-1 ZFD. This experiment revealed that the GATA-1 NF plus L was not sufficient for the interaction with p53TAD (Figure 2B). Next, we tested the GATA-1 L plus CF fragment. Incubation of p53TAD with GST-GATA-1 L plus CF resulted in similar binding to p53TAD as observed for GATA-1 ZFD (Figure 2C). Lastly, GATA-CF (GATA-1252-317) was tested for its ability to bind to p53TAD. The results demonstrated that GATA-1 CF was not sufficient for binding p53TAD (Figure 2D). Based on the results in the pulldown assay, the binding of p53TAD to GATA-1 minimally requires the linker region and the carboxyl-terminal zinc finger of GATA-1 (GATA-1 L plus CF).
The linker region, the carboxyl-terminal zinc finger, and the basic arm of GATA-1 are sufficient for binding p53TAD in an in vitro pulldown assay. (A) Schematic of GATA-1 highlighting the zinc-finger domains (GATA-1 ZFD) and the various deletion mutants of GATA-1 ZFD used as GST fusion proteins. (B) Comparative binding of GST-GATA-1 ZFD and GST-GATA-1 NF plus L (GST-GATA-1200-251) to p53TAD. Various concentrations (1.0, 0.1, and 0.01 μM) of p53TAD were incubated with either 1 μM GST-GATA-1 ZFD (lanes 3-5) or GST-GATA-1 NF plus L (lanes 6-8). In the GST lane, 1 μM purified p53TAD was incubated with 1 μM GST as a control (lane 2). The input lane is 0.5% p53TAD (lane 1). (C) Comparative binding of GST-GATA-1 ZFD and GST-GATA-1 L plus CF (GST-GATA-1228-317) to p53TAD. Various concentrations (1.0, 0.1, and 0.01 μM) of p53TAD were incubated with either 1 μM GST-GATA-1 ZFD (lanes 3-5) or GATA-1 L plus CF (lanes 6-8). In the GST lane, 1 μM purified p53TAD was incubated with 1 μM GST as a control (lane 2). The input lane is 0.5% p53TAD (lane 1). (D) Comparative binding of GST-GATA-1 ZFD and GST-GATA-1 CF (GST-GATA-1252-317) to p53TAD. Various concentrations (1.0, 0.1, and 0.01 μM) of p53TAD were incubated with either 1 μM GST-GATA-1 ZFD (lanes 3-5) or GATA-1 CF plus BA (lanes 6-8). In the GST lane, 1 μM purified p53TAD was incubated with 1 μM GST as a control (lane 2). The input lane is 0.5% p53TAD (lane 1). In all experiments, bound protein was detected with an anti-p53 antibody DO-1.
The linker region, the carboxyl-terminal zinc finger, and the basic arm of GATA-1 are sufficient for binding p53TAD in an in vitro pulldown assay. (A) Schematic of GATA-1 highlighting the zinc-finger domains (GATA-1 ZFD) and the various deletion mutants of GATA-1 ZFD used as GST fusion proteins. (B) Comparative binding of GST-GATA-1 ZFD and GST-GATA-1 NF plus L (GST-GATA-1200-251) to p53TAD. Various concentrations (1.0, 0.1, and 0.01 μM) of p53TAD were incubated with either 1 μM GST-GATA-1 ZFD (lanes 3-5) or GST-GATA-1 NF plus L (lanes 6-8). In the GST lane, 1 μM purified p53TAD was incubated with 1 μM GST as a control (lane 2). The input lane is 0.5% p53TAD (lane 1). (C) Comparative binding of GST-GATA-1 ZFD and GST-GATA-1 L plus CF (GST-GATA-1228-317) to p53TAD. Various concentrations (1.0, 0.1, and 0.01 μM) of p53TAD were incubated with either 1 μM GST-GATA-1 ZFD (lanes 3-5) or GATA-1 L plus CF (lanes 6-8). In the GST lane, 1 μM purified p53TAD was incubated with 1 μM GST as a control (lane 2). The input lane is 0.5% p53TAD (lane 1). (D) Comparative binding of GST-GATA-1 ZFD and GST-GATA-1 CF (GST-GATA-1252-317) to p53TAD. Various concentrations (1.0, 0.1, and 0.01 μM) of p53TAD were incubated with either 1 μM GST-GATA-1 ZFD (lanes 3-5) or GATA-1 CF plus BA (lanes 6-8). In the GST lane, 1 μM purified p53TAD was incubated with 1 μM GST as a control (lane 2). The input lane is 0.5% p53TAD (lane 1). In all experiments, bound protein was detected with an anti-p53 antibody DO-1.
GATA-1 and p53 interact in erythroid cells
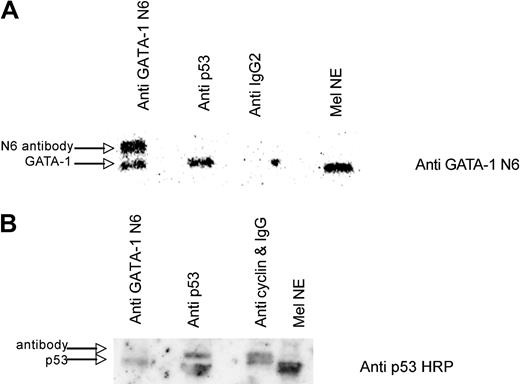
To determine whether the native p53 and GATA-1 proteins physically associate with one another in erythroid cells, immunoprecipitations were carried out with MEL cell nuclear extract and antibodies against GATA-1, p53, cyclin B1, and IgG. The immunoprecipitates were analyzed by Western blotting, and the antibodies themselves were run in adjacent lanes of the gels in addition to molecular weight markers. There was a single GATA-1 band in the MEL cell extract that migrated at 50 kilodaltons with both GATA-1 antibodies (N6 and M20), and the antibodies ran at slightly higher molecular weights. GATA-1 is precipitated with both the GATA-1 and p53 antibodies, but not with anti IgG2 (Figure 3A). The immunoprecipitations were repeated using anti-cyclin B1 antibody, followed by anti-rat IgG2 as the negative control, and probing the blots with an antibody reactive to a different epitope of GATA-1 (M20 Santa Cruz Biotechnology; data not shown) or an HRP anti-p53 antibody (Figure 3B). GATA-1 and p53 again coimmunoprecipitate, strongly suggesting that they associate with one another in MEL cells.
GATA-1 and p53 interact in vitro in MEL cell extract. (A) A total of 320 μg of MEL nuclear extract was incubated with the following: lane 1, 4 μL of anti-GATA-1 N6 and 1 μL of anti-rat IgG2a; lane 2, 4 μL of anti-p53 (PAB 240) and 1 μL of anti-mouse IgG; lane 3, 5 μL of anti-rat IgG2 for 1.5 hours at 4°C with rotation and 40 μL of protein G plus agarose was added with 4 additional hours of rotation at 4°C; lane 4, MEL nuclear extract. A Western blot is shown using GATA-1 N6 antibody. (B) Lane 1, 8 μL of anti-GATA-1 N6 and 1 μL of anti-rat IgG2a; lane 2, 8 μL of anti-p53 (PAB 246) and 1 μL of anti-mouse IgG; lane 3, 8 μL of anti-cyclin B1 (GNS1) and 1 μL of anti-mouse IgG for 1.5 hours at 4°C with rotation, and 40 μL of protein G plus agarose was added with 4 more hours of rotation at 4°C; lane 4, MEL nuclear extract. A Western blot with HRP-conjugated anti-p53 is shown.
GATA-1 and p53 interact in vitro in MEL cell extract. (A) A total of 320 μg of MEL nuclear extract was incubated with the following: lane 1, 4 μL of anti-GATA-1 N6 and 1 μL of anti-rat IgG2a; lane 2, 4 μL of anti-p53 (PAB 240) and 1 μL of anti-mouse IgG; lane 3, 5 μL of anti-rat IgG2 for 1.5 hours at 4°C with rotation and 40 μL of protein G plus agarose was added with 4 additional hours of rotation at 4°C; lane 4, MEL nuclear extract. A Western blot is shown using GATA-1 N6 antibody. (B) Lane 1, 8 μL of anti-GATA-1 N6 and 1 μL of anti-rat IgG2a; lane 2, 8 μL of anti-p53 (PAB 246) and 1 μL of anti-mouse IgG; lane 3, 8 μL of anti-cyclin B1 (GNS1) and 1 μL of anti-mouse IgG for 1.5 hours at 4°C with rotation, and 40 μL of protein G plus agarose was added with 4 more hours of rotation at 4°C; lane 4, MEL nuclear extract. A Western blot with HRP-conjugated anti-p53 is shown.
GATA-1, p53, and GATA DNA do not form a ternary complex in vitro
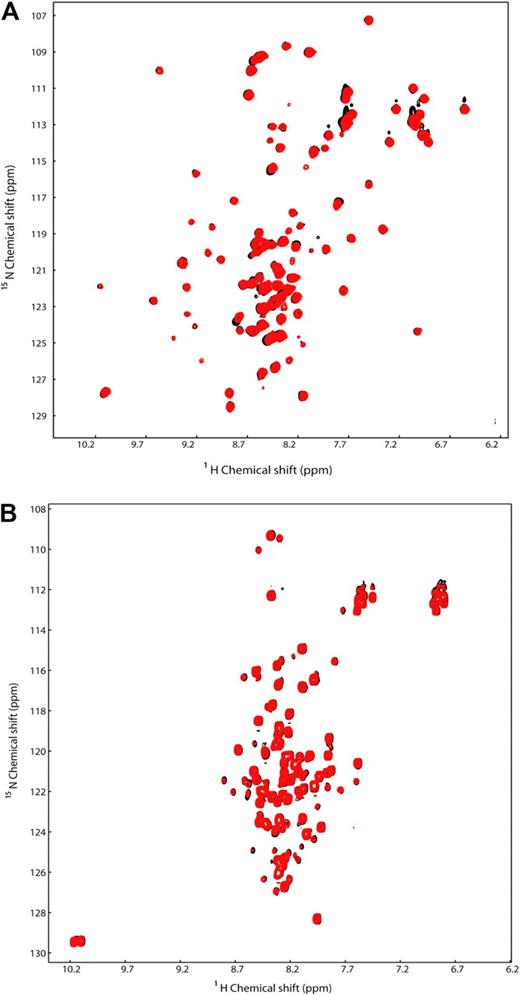
The binding site for the p53TAD on GATA-1 overlaps the minimal DBD of GATA-1. To test whether p53, DNA, and GATA-1 L plus CF are capable of forming a ternary complex, we performed 2 sets of NMR experiments. To a sample containing 15N-labeled GATA-1 L plus CF (0.5 mM), we added a stoichiometric amount of unlabeled AGATAA containing DNA (0.5 mM) and recorded a 2-dimensional (2D) 1H-15N heteronuclear single quantum coherence (HSQC) spectrum (Figure 4A black signals). To the sample containing the 15N-GATA-1 L plus CF/DNA complex, we then added unlabeled p53TAD (0.5 mM), and we observed that there were no differences in the 1H and 15N resonances of 15N-GATA-1 L plus CF (Figure 4A red signals). This result indicates that under these experimental conditions, there does not appear to be a ternary complex involving GATA-1, DNA, and p53. To further test for the presence of a ternary complex, we performed a second set of NMR experiments. First, we recorded a 2D 1H-15N HSQC spectrum of a sample containing 15N-labeled p53TAD (0.5 mM; Figure 4B black signals). To the sample containing the 15N-p53TAD, we then added the unlabeled GATA-1 L plus CF/DNA complex (0.5 mM), and again we did not observe any differences in the 1H and 15N resonances of 15N-p53TAD (Figure 4B red signals). These results further support the conclusion that under the NMR conditions, there does not appear to be a ternary complex involving GATA-1, DNA, and p53. These experiments suggest that p53TAD and the AGATAA DNA compete for binding to GATA-1 L plus CF, and that GATA-1 L plus CF binds tighter to the DNA than to the p53TAD. This is consistent with the fact that GATA-1 L plus CF binding to DNA is approximately 10-fold tighter than to the p53TAD (see “GATA-1 L plus CF and p53TAD interact strongly and specifically in vivo”).
GATA-1, p53TAD, and a GATA-1 DNA-binding site do not form a ternary complex. (A) Overlay of the 2D 1H-15N HSQC spectra for a 0.5 mM sample of 15N-labeled GATA-1 L plus CF/DNA complex (black) and in the presence of 0.5 mM unlabeled p53TAD (red). (B) Overlay of the 2D 1H-15N HSQC spectra for a 0.5 mM sample of 15N-labeled p53TAD in the free form (black) and in the presence of 0.5 mM unlabeled GATA-1 L plus CF/DNA complex (red).
GATA-1, p53TAD, and a GATA-1 DNA-binding site do not form a ternary complex. (A) Overlay of the 2D 1H-15N HSQC spectra for a 0.5 mM sample of 15N-labeled GATA-1 L plus CF/DNA complex (black) and in the presence of 0.5 mM unlabeled p53TAD (red). (B) Overlay of the 2D 1H-15N HSQC spectra for a 0.5 mM sample of 15N-labeled p53TAD in the free form (black) and in the presence of 0.5 mM unlabeled GATA-1 L plus CF/DNA complex (red).
GATA-1 and p53 interfere with one another
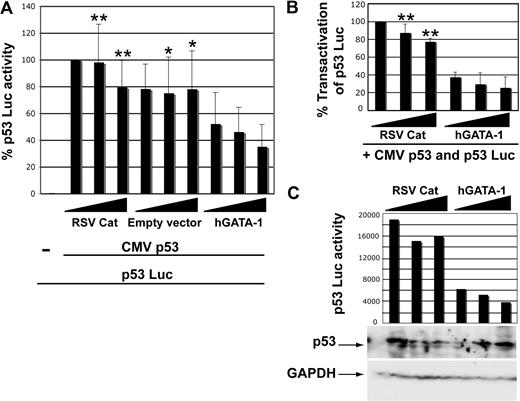
To determine whether the p53 and GATA-1 proteins can influence the activity of one another, transactivation assays were performed. A p53-responsive reporter gene (p53 Luc) can be activated in 6C2 cells by the expression of CMV p53 (Figure 5A bars 1 and 2). The coexpression of Rous sarcoma viral promoter upstream from the chloramphenicol acetyltransferase gene (RSV-Cat) or the inclusion of empty expression vector has little effect on p53 activation, but the addition of RSV hGATA-1 inhibits this activation in a dose-dependent manner (Figure 5A-B). Protein was prepared from a portion of the transfected cells and subjected to Western blotting with anti-p53 antibody to show that the inhibition of p53 activity seen in these experiments is not due to repression of p53 expression by GATA-1 (Figure 5C). We observed high levels of p53 even with the highest concentration of GATA-1 expression vector, and GAPDH levels are comparable among all samples. Similar results were obtained in K562 cells, and p53 can also inhibit the activation of a GATA-1–responsive reporter gene in transactivation assays (data not shown). Under similar conditions, GATA-1 did not inhibit the dexamethasone-induced activation of a glucocorticoid response element (GRE) reporter gene by the glucocorticoid receptor, indicating that the inhibition is specific (data not shown). Furthermore, addition of erythropoietin to K562 cells caused a slight decrease in p53Luc activation that correlated with a slight increase in endogenous GATA-1 levels (supplemental Figure 1), and this result is consistent with the results seen after the addition of RSV hGATA-1.
GATA-1 can inhibit transactivation by p53 in 6C2 cells. (A) All samples were transfected with 150 ng of CMV Renilla luciferase and 0.5 μg of p53 Luc reporter plasmid; and 0.25 μg of CMV p53 was added to all but the first sample. Increasing amounts of RSV-Cat, empty RSV expression vector, or RSV hGATA-1 were added (1, 2, and 3 μg of DNA). The dual luciferase reporter assay was performed on aliquots of the samples, and firefly luciferase activity normalized to Renilla luciferase is plotted. The error bars indicate the standard deviations and n = 3. Means of the 2 highest DNA concentrations of RSV-Cat and empty vector were compared with means for equal weights of hGATA-1 by Student t test; *P < .05 and **P < .005. (B) RSV-Cat and hGATA-1 were compared in 6 transfections. Results were normalized to Renilla luciferase activity. The firefly luciferase value for 1 μg of RSV-Cat was then set to 100%, and all other values were normalized to this. The error bars indicate the standard deviation and n = 6. We compared means for equal weights of the 2 highest DNA concentrations of the RSV-Cat and hGATA-1 plasmids to test for statistical significance by Student t test; **P < .005. (C) A comparable transfection was performed adding CMV Renilla luciferase, p53 Luc reporter plasmid, and CMV p53 to all samples and cotransfecting increasing amounts of RSV-Cat or RSV hGATA-1. The cells were divided at harvest, and half were used for the dual luciferase reporter assay and half for Western blotting with HRP-conjugated anti-p53 (top) and with anti-GAPDH antibody (bottom).
GATA-1 can inhibit transactivation by p53 in 6C2 cells. (A) All samples were transfected with 150 ng of CMV Renilla luciferase and 0.5 μg of p53 Luc reporter plasmid; and 0.25 μg of CMV p53 was added to all but the first sample. Increasing amounts of RSV-Cat, empty RSV expression vector, or RSV hGATA-1 were added (1, 2, and 3 μg of DNA). The dual luciferase reporter assay was performed on aliquots of the samples, and firefly luciferase activity normalized to Renilla luciferase is plotted. The error bars indicate the standard deviations and n = 3. Means of the 2 highest DNA concentrations of RSV-Cat and empty vector were compared with means for equal weights of hGATA-1 by Student t test; *P < .05 and **P < .005. (B) RSV-Cat and hGATA-1 were compared in 6 transfections. Results were normalized to Renilla luciferase activity. The firefly luciferase value for 1 μg of RSV-Cat was then set to 100%, and all other values were normalized to this. The error bars indicate the standard deviation and n = 6. We compared means for equal weights of the 2 highest DNA concentrations of the RSV-Cat and hGATA-1 plasmids to test for statistical significance by Student t test; **P < .005. (C) A comparable transfection was performed adding CMV Renilla luciferase, p53 Luc reporter plasmid, and CMV p53 to all samples and cotransfecting increasing amounts of RSV-Cat or RSV hGATA-1. The cells were divided at harvest, and half were used for the dual luciferase reporter assay and half for Western blotting with HRP-conjugated anti-p53 (top) and with anti-GAPDH antibody (bottom).
Whereas inhibition in the transactivation assay is reciprocal between the p53 and GATA-1 proteins, we hypothesize that p53 inhibition of GATA-1 does not serve an important biologic function in red cell development. GATA-1 levels far exceed p53 levels at all stages of red cell development, and p53 null mice have normal erythroid cells,45,46 although possible compensation by p53 homologs p63 and p73 has not been ruled out. Consequently, only the inhibition of p53 by GATA-1 has been studied further.
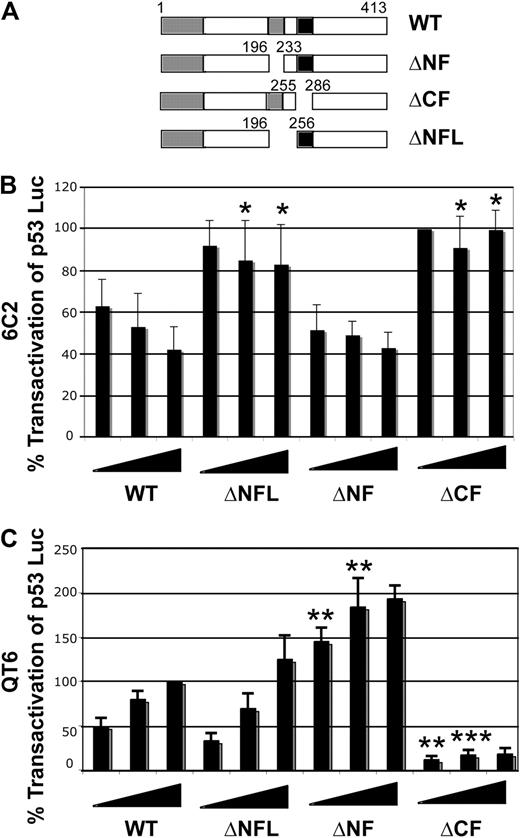
To determine which regions of GATA-1 are required for the inhibition of p53, a series of deletion mutants of mouse GATA-1 in pEF-BOS (Figure 6A) were tested in the transactivation assay in 6C2 cells. Wild-type GATA-1 and a construct missing the amino-terminal zinc-finger domain (ΔNF) inhibit p53 activity to similar extents (Figure 6B). In contrast, constructs missing the carboxyl-terminal zinc finger (ΔCF) or the amino-terminal finger and the linker region (ΔNFL) do not (Figure 6B), indicating that the carboxyl-terminal zinc finger and the linker between the 2 zinc fingers are sufficient for the inhibition of p53 activity, but the amino-terminal zinc-finger domain is not required. The ΔNFL mutant is capable of activation of FR7Luc, a GATA-1–responsive reporter gene, in QT6 fibroblasts (and in 6C2 cells; data not shown), whereas the ΔCF mutant is not (Figure 6C). This is consistent with the fact that the ΔCF mutant lacks the major DBD of GATA-1. The activation levels seen with the ΔNFL mutant are similar to wild-type GATA-1, indicating that it accumulates stably in cells. Thus, the ΔNFL mutant can transactivate a GATA-responsive reporter (Figure 6C) and bind to DNA (data not shown), and yet does not inhibit transactivation by p53.
The linker between the zinc fingers and the CF of GATA-1 are required for p53 inhibition. (A) Schematic of deletion mutants of mouse GATA-1. (B) Transfections into 6C2 cells were performed, as described in the Figure 5 legend. A total of 150 ng of CMV Renilla luciferase, 0.5 μg of p53 Luc reporter plasmid, and 0.25 μg of CMV p53 was present in all samples, whereas various amounts of mouse wild-type GATA-1 expression vector or mutants were added as indicated (1, 2, or 3 μg). After normalization to Renilla luciferase activity, the firefly luciferase value for 1 μg of ΔCF was set to 100%, and the values for all other samples were correspondingly adjusted. The error bars indicate the standard deviation, and the experiments were performed between 4 and 7 times. We compared means for equal weights of the 2 highest DNA concentrations of ΔNFL, ΔNF, and ΔCF to wild type by Student t test, and *P < .05. (C) CMV Renilla luciferase and 1 μg of FR7Luc, a GATA-1–responsive reporter plasmid, were tranfected into QT6 fibrobasts with increasing amounts of mouse GATA-1 expression plasmid, as indicated (0.5, 1, or 2 μg). Normalization and error bars are as in (B), with values for 2 μg of wild-type GATA-1 set to 100%. The means for equal weights of the 2 lowest concentrations of wild-type were compared with those of all other constructs by Student t test; **P < .005 and ***P < .0005.
The linker between the zinc fingers and the CF of GATA-1 are required for p53 inhibition. (A) Schematic of deletion mutants of mouse GATA-1. (B) Transfections into 6C2 cells were performed, as described in the Figure 5 legend. A total of 150 ng of CMV Renilla luciferase, 0.5 μg of p53 Luc reporter plasmid, and 0.25 μg of CMV p53 was present in all samples, whereas various amounts of mouse wild-type GATA-1 expression vector or mutants were added as indicated (1, 2, or 3 μg). After normalization to Renilla luciferase activity, the firefly luciferase value for 1 μg of ΔCF was set to 100%, and the values for all other samples were correspondingly adjusted. The error bars indicate the standard deviation, and the experiments were performed between 4 and 7 times. We compared means for equal weights of the 2 highest DNA concentrations of ΔNFL, ΔNF, and ΔCF to wild type by Student t test, and *P < .05. (C) CMV Renilla luciferase and 1 μg of FR7Luc, a GATA-1–responsive reporter plasmid, were tranfected into QT6 fibrobasts with increasing amounts of mouse GATA-1 expression plasmid, as indicated (0.5, 1, or 2 μg). Normalization and error bars are as in (B), with values for 2 μg of wild-type GATA-1 set to 100%. The means for equal weights of the 2 lowest concentrations of wild-type were compared with those of all other constructs by Student t test; **P < .005 and ***P < .0005.
The amino-terminal zinc finger of GATA-1 is not required for p53 inhibition, but is critical to other GATA-1 functions, so we restored it to the construct in 2 forms. Amino acids 196-232 were restored to the ΔNFL mutant to generate the ΔNL mutant, and this protein does not inhibit p53 (supplemental Figure 2A,B). A construct restoring 196-234 to the ΔNFL mutant to generate the ΔNL2 mutant (supplemental Figure 2A) was also tested, and this mutant inhibits p53, but less efficiently than ΔNFL (data not shown), suggesting that amino acids 233 and 234 may be involved, but are not sufficient for interaction with p53. Both the ΔNL and ΔNL2 mutants can transactivate a GATA-responsive reporter (supplemental Figure 2C) and bind to the GATA cofactor, FOG-1 (data not shown).
Plasmids with the promoters of p53-responsive genes driving luciferase reporter genes were obtained and assayed in 6C2 cells. Consistent with our results above, both the BAX and p21 promoters are partially inhibited by wild-type GATA-1, but not by the ΔNFL mutant (data not shown). The observation that the ΔNF mutant inhibits p53, whereas the ΔNFL mutant does not, demonstrates a strict requirement for the linker between the GATA-1 zinc-finger domains for p53 inhibition, and these results correlate perfectly with results of our GST pulldown assay (above). The linker region and the carboxyl-terminal zinc finger are required both for p53TAD binding and inhibition of p53 activity.
GATA-1 L plus CF and p53TAD interact strongly and specifically in vitro
To compare the binding affinity of the 2 subdomains within p53TAD for the zinc-finger domains of GATA-1, we determined dissociation constants (KD) by ITC (supplemental Figure 3). By ITC, p53TAD (p531-73) bound GATA-1 ZFD (GATA-1199-317) with apparent KD values of 77 (± 16) nM and GATA-1 L plus CF (GATA-1228-317) with an apparent KD of 67 (± 7) nM (supplemental Figure 3). In contrast, we were not able to detect any interaction between either p53TAD1 (p531-40) or p53TAD2 (p5340-73) with either GATA-1 L plus CF (supplemental Figure 3) or GATA-1 ZBD by ITC. The ITC results are consistent with our GST pulldown and transactivation assays. Taken together, the results indicate that the minimal region of GATA-1 required for the interaction with p53TAD and for the inhibition of p53 activity is the linker region plus the carboxyl-terminal zinc-finger domain.
The inhibition of p53 activity by GATA-1 requires the interaction of p53TAD with GATA-1
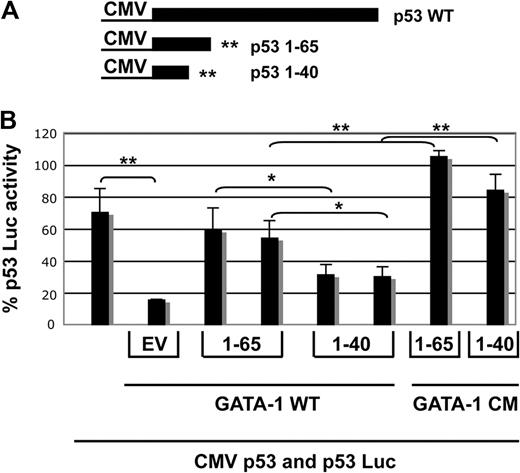
A competition assay was developed to examine the mechanism of inhibition of p53 by GATA-1. Two truncation mutants of p53 producing p531-65 or p531-40 were constructed in CMVp53. These truncation mutants that no longer activate p53-responsive reporters (data not shown) were expressed along with wild-type p53 and GATA-1 in 6C2 cells. In the presence of wild-type GATA-1, p531-65 increased the expression of p53 Luc to 80% of the levels seen with p53 alone, whereas p531-40 did not (Figure 7B bars 1-6). A GATA-1 mutant (GATA-1 CM; “Methods”) that does not inhibit p53 (data not shown) was used as a control, and neither p53 fragment affected the activation of p53Luc by p53 in the presence of this mutant GATA-1 (Figure 7B bars 1, 7, and 8). These p53 fragments have no effect on the activity of p53 in the absence of GATA-1, and GATA-1 does not affect the activity of p53 Luc in the absence of p53 (data not shown). Taken together, these results indicate that p531-65 competes with wild-type p53 for GATA-1, and this reduces the inhibitory effect of GATA-1; p531-40, which does not bind to GATA-1, did not compete. Thus, the interaction of p531-65 with GATA-1 is essential for the inhibition of p53.
p531-65 can compete with wild-type p53 for binding to GATA-1 and reduce the inhibition of p53. (A) A schematic of constructs made by inserting translation termination signals in the CMV p53 expression vector to generate peptides 1-65 and 1-40 is shown. (B) A transfection assay in which CMV Renilla luciferase, p53 Luc reporter plasmid, and CMV p53 were present in all samples and expression plasmids for mGATA-1 were added to samples 2-6 along with 6 μg of empty vector (bar 2), 5 or 6 μg of CMV p53 1-65 (bars 3 and 4), or CMVp53 1-40 (bars 5 and 6), as indicated. For the last 2 samples, GATA-1CM, a mutant that does not interfere with transactivation by p53, was used instead of wild-type GATA-1, and 6 μg of CMV 1-65 (bar 7) or 6 μg of CMV 1-40 (bar 8) was added. Normalization is as described in the Figure 6 legend with the average of 5 μg of GATA-1 CM 65 and 40 samples set to 100%. Results are the average of 3 experiments, and error bars indicate the standard deviation. Means of equal weights of DNA were compared by Student t test, as indicated; *P < .05 and ** P < .005.
p531-65 can compete with wild-type p53 for binding to GATA-1 and reduce the inhibition of p53. (A) A schematic of constructs made by inserting translation termination signals in the CMV p53 expression vector to generate peptides 1-65 and 1-40 is shown. (B) A transfection assay in which CMV Renilla luciferase, p53 Luc reporter plasmid, and CMV p53 were present in all samples and expression plasmids for mGATA-1 were added to samples 2-6 along with 6 μg of empty vector (bar 2), 5 or 6 μg of CMV p53 1-65 (bars 3 and 4), or CMVp53 1-40 (bars 5 and 6), as indicated. For the last 2 samples, GATA-1CM, a mutant that does not interfere with transactivation by p53, was used instead of wild-type GATA-1, and 6 μg of CMV 1-65 (bar 7) or 6 μg of CMV 1-40 (bar 8) was added. Normalization is as described in the Figure 6 legend with the average of 5 μg of GATA-1 CM 65 and 40 samples set to 100%. Results are the average of 3 experiments, and error bars indicate the standard deviation. Means of equal weights of DNA were compared by Student t test, as indicated; *P < .05 and ** P < .005.
Discussion
The observation that GATA-1 interacts strongly with p53 in vitro caused us to consider that GATA-1 directly influences p53 and that there may be interdependency between these 2 regulatory networks in erythroid cells. We have shown that GATA-1 is a potent inhibitor of p53 transactivation in erythroid precursor cells and that this inhibition requires a region of the GATA-1 DBD critical to a high-affinity interaction with the TAD of p53. This inhibition is competed by p53 fragments that bind to GATA-1, but not by ones that do not. Mutants of GATA-1 that do not inhibit p53 transactivation can still bind to DNA and to FOG-1, and transactivate GATA-responsive reporter genes like native GATA-1.
The region of the GATA-1 DBD required for p53 inhibition correlates with the region required for binding to the p53TAD. Residues 228-317 of GATA-1 are sufficient for maximal binding to p53, and significantly lower binding is seen when residues 228-251 from the linker of GATA-1 are deleted (Figure 2D). In the transactivation assay, the GATA-1 ΔNF mutant (removal of residues 196-233) still inhibits p53. This mutant removes the cysteine residues of the amino-terminal zinc finger, but retains most of the linker region, including several residues that are part of the helix associated with the amino-terminal zinc finger. In contrast, the GATA-1 ΔNL and ΔNL2 mutants, which contain the amino-terminal zinc-binding domain, but lack either residues 234-254 or residues 236-254 from the linker, do not inhibit p53 activity efficiently. Residues within the linker between the 2 zinc fingers of GATA-1 are critical for both the in vitro binding and the inhibition of p53 transactivation.
All GATA-1 DNA-binding mutants tested to date are defective in the inhibition of p53 transactivation. This is consistent with the requirement for the complete carboxyl-terminal zinc finger of GATA-1 (the major DBD) for p53 binding, and does not necessarily indicate that DNA binding by GATA-1 is required for the inhibition of p53. Our NMR studies indicate that the p53TAD does not disturb the GATA-1/DNA complex in vitro, suggesting that a ternary complex of the 3 molecules does not occur (Figure 4). This is consistent with the fact that the affinity of the GATA-1 DNA interaction is 10-fold higher than that of GATA-1 and p53TAD, and there is overlap between the regions of GATA-1 required for its interaction with DNA and with p53. We do not know if a ternary complex forms at p53 DNA-binding sites, but this is plausible because the p53 DBD is not required for the interaction with GATA-1. It is important to remember that GATA-1 is present at very high concentration in erythroid cells and could either sequester p53 from its targets, or bind to p53 at its targets and inactivate it. Further experiments are required to determine whether GATA-1 and p53 are able to form a ternary complex on a p53 DNA-binding site.
Whereas the inhibition of p53 by GATA-1 has been seen in 2 types of erythroid cells, we have not observed it in nonerythroid cells, and thus, cell-type–specific modifications of either GATA-1 or p53 may be important. Alternately, the cell-type specificity may be the result of a significantly different balance between the concentrations of the 2 proteins in other cell types. Both proteins can be toxic at high concentrations, and it is difficult to express GATA-1 at the normally high levels that occur in red blood cells in nonhematopoietic cells. High levels of GATA-1 may be required to prevent cell death induced by ectopically expressed p53, and this may be unattainable except in an erythroid cell. In this case, the inhibition of p53 by GATA-1 would be an erythroid cell-specific event.
GATA-1 has been linked to leukemias in mice and humans. GATA 1.05 knockdown mice, producing 5% of normal levels of GATA-1, develop erythroleukemia at 3 to 5 months of age and B-cell leukemia at 1 year; 1.05 fetal erythroid cells are unable to differentiate due to lack of GATA-1, but escape the apoptosis seen in GATA-1 null cells and are hyperproliferative.47 After birth, secondary mutations in this unchecked cell population lead to overt transformation and disease; the inhibition of p53 by GATA-1 observed in this study could facilitate this cell expansion.
In humans, GATA-1 mutations are associated with transient myeloproliferative disorder (TMD) and with acute megakaryoblastic leukemia (AMKL) in Downs Syndrome (DS): both diseases occur frequently in DS patients.22 TMD and AMKL patients produce GATA-1s, a shorter form of GATA-1 lacking the N-terminal transactivation domain that is translated from an internal ATG: GATA-1s is a critical factor in these diseases. DS TMD is usually a transient disease, but 20% to 30% of cases develop into DS AMKL derived from the original clone of diseased cells with identical mutations in GATA-1.22 In several experimental systems, substitution of GATA-1s for the native protein has resulted in transient hyperproliferation of fetal megakaryocyte progenitors, but leukemia was not observed consistent with the requirement for trisomy 21.22,47 It is thought that during DS fetal development, mutations producing GATA1s occur at high incidence, leading to expansion of megakaryocyte progenitors to generate TMD. These cells undergo secondary mutation leading to transformation and AMKL.22 The p53 tumor suppressor pathway is evidently unable to prevent the accumulation of these cells, but p53 mutations are observed in approximately 21% of cases, and are thus not prerequisite for DS AMKL.48 GATA-1s has the complete DBD and should interact with p53 as native GATA-1. It would be interesting to investigate whether the interaction between GATA-1 and p53 described in this study prevents the p53 response, allowing the expansion of this cell type. The interaction could also be relevant in preventing the activation of the p53 pathway in other leukemias that express hematopoietic GATA factors. Targeting the interaction in diseased cells may be therapeutic.
Inhibition in the transactivation assay is reciprocal between the p53 and GATA-1 proteins, and our competition assay (Figure 7) shows that p53TAD is involved. We propose that only the inhibition of p53 by GATA-1 is functionally important, and may be needed to prevent the induction of the p53 pathway and to avoid apoptosis and/or senescence during red cell development. To accomplish this, GATA-1 regulates p53 and several proteins usually controlled by p53 to cause cell cycle withdrawal to facilitate enucleation and the formation of functional erythrocytes.
There is precedent for the participation of GATA-1 in this type of regulation, although the mechanism may not be identical. The association of PU.1 with GATA-1 through the DBDs of both proteins and the PU.1 TAD also results in a reciprocal inhibition of activity, which specifies the choice of either the myeloerythroid or myelolymphoid lineages.24,42,49,50 Each transcription factor drives cells toward a specific hematopoietic potential, while simultaneously inactivating the master regulator that specifies alternate lineages. The similarity of the TADs of p53 and PU.1 suggests that both the transactivation and the inhibitory functions of this domain have been conserved between the 2 proteins.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Masayuki Yamamoto for mouse GATA-1 expression clones, and Dr Sam Lee for reporter plasmids. We thank Stoney Simons for a glucocorticoid receptor expression vector and reporter gene. We thank Gerardo Ferbeyre for helpful discussions and the p53 expression vectors. We thank Connie Noguchi for advice and discussions. We also thank Gary Felsenfeld for advice and support.
This work was supported by Canadian Institutes of Health Research Grant MOP-135604 (Ottawa, ON; to J.G.O.) and by National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (Bethesda, MD). The microcalorimeter for the ITC experiments was purchased with funds from National Sciences and Engineering Research Council (Ottawa, ON).
National Institutes of Health
Authorship
Contribution: C.D.T. designed research, performed research, contributed vital new reagents, analyzed data, and wrote the paper. C.M. designed research, performed research, contributed vital new reagents, analyzed data, and wrote the paper. P.A. performed research and contributed vital new reagents. P.D.L. designed research and performed research. J.G.O. designed research, performed research, contributed vital new reagents, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: All the authors declare no competing financial interests.
Correspondence: Cecelia D. Trainor, Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bldg 5, Rm 208, Bethesda, MD 20892; e-mail: ceceliat@mail.nih.gov; and James G. Omichinski, Département de Biochimie, Université de Montréal C.P. 6128 Succursale Centre-Ville, Montréal, QC H3C 3J7 Canada; e-mail: jg.omichinski@umontreal.ca.
References
Author notes
*C.D.T. and C.M. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal