Abstract
In thalassemia and other iron loading anemias, ineffective erythropoiesis and erythroid signaling molecules are thought to cause inappropriate suppression of a small peptide produced by hepatocytes named hepcidin. Previously, it was reported that the erythrokine GDF15 is expressed at very high levels in thalassemia and suppresses hepcidin expression. In this study, erythroblast expression of a second molecule named twisted gastrulation (TWSG1) was explored as a potential erythroid regulator of hepcidin. Transcriptome analyses suggest TWSG1 is produced during the earlier stages of erythropoiesis. Hepcidin suppression assays demonstrated inhibition by TWSG1 as measured by quantitative polymerase chain reaction (PCR) in dosed assays (1-1000 ng/mL TWSG1). In human cells, TWSG1 suppressed hepcidin indirectly by inhibiting the signaling effects and associated hepcidin up-regulation by bone morphogenic proteins 2 and 4 (BMP2/BMP4). In murine hepatocytes, hepcidin expression was inhibited by murine Twsg1 in the absence of additional BMP. In vivo studies of Twsg1 expression were performed in healthy and thalassemic mice. Twsg1 expression was significantly increased in the spleen, bone marrow, and liver of the thalassemic animals. These data demonstrate that twisted gastrulation protein interferes with BMP-mediated hepcidin expression and may act with GDF15 to dysregulate iron homeostasis in thalassemia syndromes.
Introduction
Systemic iron homeostasis in mammals is largely maintained by the effects of hepcidin,1 a small protein produced by hepatocytes. Hepcidin is regulated at the transcriptional and posttranscriptional levels by multiple extracellular signals related to iron homeostasis and inflammation. Erythropoiesis is also thought to regulate hepcidin expression through a variety of mechanisms including anemia-related hypoxia and erythropoietin production. β-Thalassemia syndromes are congenital anemias caused by mutations that reduce or abolish β-globin gene expression. Despite the common feature of decreased globin chain synthesis in all patients, there are prominent phenotypic variations in the disease that are not fully understood.2 In so-called “iron-loading” anemias like thalassemia, the diseased erythron dysregulates iron homeostasis by inhibiting hepcidin expression even in the presence of severe iron overload. Humans with thalassemia syndromes express very high levels of a cytokine named GDF15, and GDF15 present in thalassemia patients' sera inhibited hepatic hepcidin expression ex vivo.3 However, thalassemia sera also suppressed hepcidin expression to a lesser degree after immunoprecipitation of GDF15.3 It was therefore hypothesized that GDF15 may act with other molecules to suppress hepcidin.
In addition to clinical research in humans, murine models were developed for studies of thalassemia and hepcidin regulation. Mice with deletions of both the βminor and βmajor genes (th3 genotype) have a β-thalassemia intermedia phenotype in the heterozygous state. The homozygous deletion (th3/th3) results in death at the fetal stage of development.4-6 An adult mouse model of β-thalassemia major was established by transplantation of hematopoietic fetal liver cells harvested from th3/th3 embryos into irradiated syngeneic recipients.7 The transplanted th3/th3 mice manifested severe anemia due to low reticulocyte counts, profound splenomegaly, and extensive hepatic extramedullary hematopoiesis. In the th3/+ mice, erythropoiesis was characterized by a modest reduction in red blood cells (RBCs) and an increase in reticulocytes. The th3/th3 mice demonstrated almost complete absence of hemoglobinized erythroblasts, reticulocytes, and mature erythrocytes. In addition, the hallmark finding of erythroblast apoptosis at the polychromatophilic stage of differentiation in human β-thalassemia major was not reported in the murine model.8 Despite the differences in murine models and humans with thalassemia, associated iron overload is manifested in both species. However, the suppression of hepcidin expression was inconsistent.6,9 Because GDF15 is expressed among hemoglobinized erythroblasts at the final stages of human erythropoiesis, it is not predicted to be expressed at similarly high levels in the murine thalassemia model. Therefore, additional erythroid signals expressed at earlier stages during erythropoiesis may be contributing to hepcidin suppression in murine thalassemia. In this study, a cytokine named twisted gastrulation (human TWSG1, murine Twsg1) was explored as a second erythroid regulator of hepcidin in human and murine cells.
Methods
Clinical, animal, and cellular studies
Studies of primary human erythroblasts were approved by an Institutional Review Board at the National Institutes of Health (NIH), and donor informed consent was obtained in accordance with the Declaration of Helsinki. Primary human hepatocytes were obtained from Liver Tissue Procurement and Distribution System (Minneapolis, MN). Healthy donors were recruited for CD34+ mobilizations used for these studies and other efforts aimed toward understanding the erythroblast transcriptome. For ex vivo cultures, CD34+ cells were placed in medium containing 4 U/mL erythropoietin (EPO; Amgen). On culture days 7 and 14, the cells were sampled, counted, and examined for confirmation of erythroblast differentiation by microscopic examination and flow cytometry. For flow cytometry, immunostaining with antibodies directed against glycophorin A (GPA) and CD71 (Invitrogen) were performed using the BD FACSAria flow cytometer (BD Biosciences). Maturing erythroblasts were sorted into 5 gated populations on culture days 7 and 14 [CD71(low)/GPA(−), CD71(high)/GPA(−), CD71(high)/GPA(+), CD71(medium)/GPA(+), and CD71(low)/GPA(+)].
Hepcidin assays with recombinant proteins were performed under the conditions described by Tanno et al.3 Human TWSG1 (Peprotech), human bone morphogenic protein 2 (BMP2), and human BMP4 (R&D Systems) were used to determine the effects on hepcidin mRNA expression in human hepatoma cell line (HuH-7) and human primary hepatocytes. At 24 hours after stimulation, the cells were lysed for total RNA isolation, and cDNA was synthesized using FastLane Cell cDNA kit (QIAGEN) following the manufacturer's protocol. Primary mouse hepatocytes were isolated from 6-week-old C57Bl/6 mice by 2-step perfusion. The portal vein was cannulated using an 18-gauge angiocatheter, and the liver was flushed with modified Krebs-Henseleit Buffer (no calcium) for 2 minutes at 8 mL/minute. The perfusion was changed to collagenase buffer (0.2 mg/mL) for 5 minutes. The liver was removed, and the capsule punctured; cells were dissociated by shaking. Viable hepatocytes were isolated with Percoll purification by centrifugation and plated at 2.1 × 105 cells/cm2 on collagen-coated tissue culture plates. Cells were cultured in Williams E medium (Sigma-Aldrich) supplemented with 5% fetal bovine serum (FBS). Cells were treated with recombinant mouse Twsg1 (R&D Systems). At 24 hours after stimulation, cells were lysed for total RNA isolation, and cDNA was synthesized. Animal procedures were approved by the Institutional Animal Care and Use Committees (IACUCs) of University of California, Los Angeles (UCLA) and Pennsylvania State University.
For the determination of Twsg1 expression in spleen, liver, and bone marrow in the thalassemia mouse model, we harvested total RNA and protein from the organs of 5 female C57Bl/6 Hbbth3/th3, 13 female C57Bl/6 Hbbth3/+ and 7 female C57Bl/6 control mice (all at the age of 8 weeks). Before death, blood samples were obtained by retro-orbital puncture under ether anesthesia. Total RNA was isolated using TRIzol LS Reagent (Invitrogen), and cDNA was synthesized using Superscript III (Invitrogen) following the manufacturer's protocol.
Quantitative PCR analyses
Total RNA was used as template for all reverse transcriptase reactions. For TWSG1, Twsg1, and BMP2, 4, 6, and 9 mRNA analyses, copy number quantitation per nanogram cDNA was measured by comparing the cycle threshold (Ct) values with those generated from standard curves (20-20 000 000 copies). Standard curves were generated using plasmid DNA containing the appropriate template inserts. A 7900 Sequence Detector instrument (Applied Biosystems) was used for amplification. Gene-specific TaqMan probes (Applied Biosystems) were used for human TWSG1 (Hs00221028_m1), murine Twsg1 (Mm00452254_m1), human BMP2/4/6/9 (Hs00154192_m1, Hs00370078_m1, Hs00233470_m1, Hs00211913_m1), and murine Bmp2/4/6/9 (Mm01340178_m1,Mm00432087_m1, Mm01332882_m1, Mm03024080_m1). For hepcidin expression analyses, quantitative real-time polymerase chain reaction (PCR) was performed as previously described to determine the expression level relative to glyceraldehyde 3-phosphate dehydrogenase (GAPDH)3 or actin.10
Protein analyses
For Smad analyses, HuH-7 cells (cultured for 1 hour with 10 ng/mL BMP2 in presence or absence of 1000 ng/mL TWSG1) were lysed for whole cell extract by lysis buffer with phosphatase inhibitor (Active Motif). The protein extracts were electrophoresed (20 μg/lane) through NuPAGE gel (Invitrogen) and transferred to membranes. The membranes were probed with anti–human phospho-Smad1/5/8 antibody (Cell Signaling Technology), anti–human total Smad1 antibody (Santa Cruz Biotechnology), and anti–human β-actin antibody (Abcam) using secondary horseradish peroxidase (HRP)–conjugated antibodies (Amersham Pharmacia).
For murine Twsg1 protein detection, murine spleens were homogenized and extracted using T-PER tissue protein extraction reagent (Pierce). The protein extracts were centrifuged at 10 000g for 5 minutes. The supernatant (30 μg/lane) were used for immunoblot analysis with a rabbit polyclonal antibody against amino acids 65 to 78 of the mature Twsg1 protein (region conserved in the human and murine proteins).
Statistical analysis
Replicate data are expressed as means plus or minus SEM with significance calculated by Student t test.
Results
TWSG1 expression during erythropoiesis
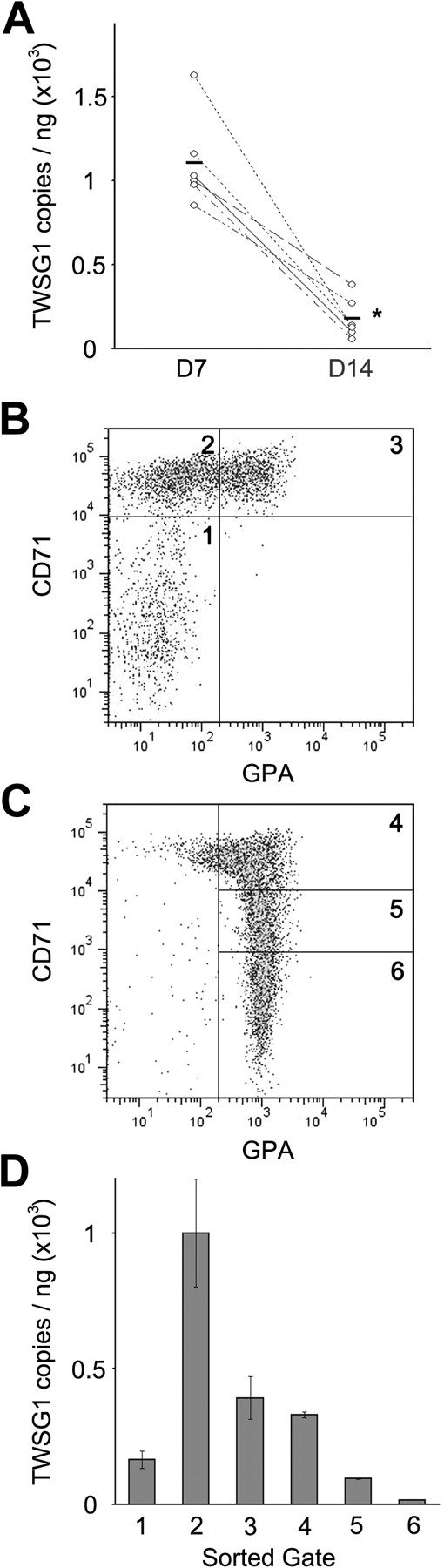
Ex vivo culture of human CD34+ cells was used to demonstrate TWSG1 expression in cultured human erythroblasts.3 The experimental approach in that study provided a robust transcriptional profile from cells using Affymetrix GeneChip Human Genome U133 Plus 2.0 Arrays. TWSG1 (probe set ID no. 225406_at) demonstrated a pattern of increased expression during the earlier stages of erythropoiesis (culture day 7) versus the mature, hemoglobinized precursors (culture day 14). Those array-based results were confirmed by quantitative PCR using cells from 6 individual donors as shown in Figure 1A. For a more complete description of TWSG1 expression during erythroid maturation, quantitative PCR analyses were performed on cells sorted on culture days 7 and 14, respectively. The degree of erythroblast maturation was defined by expression of erythroid markers (CD71 and GPA). As reported previously,11 the CD71(low)/GPA(−) population represents cells that did not increase expression of CD71 during the first week in erythropoietin-containing culture. The CD71(high)/GPA(−) phenotype denotes an immature erythroblast population that arises during the first week in culture in erythropoietin-containing medium, and the CD71(high)/GPA(+) phenotype consists mainly of proerythroblasts (Figure 1B). By culture day 14, the progressive loss of CD71 is detected with maturation of the cells into hemoglobinized precursor populations (Figure 1C). The corresponding TWSG1 mRNA expression levels were highest among the least mature erythroblasts [CD71(high)/GPA(−)], then reduced as the cells underwent erythroid maturation (Figure 1D).
TWSG1 expression in human erythropoiesis. (A) Microarray confirmation by quantitative PCR using erythroblasts from 6 separate donors (y-axis, copy number/ng total RNA). Each line associated values from the same donor on separate days. D7, culture day 7; D14, culture day 14. *P < .05. Bars represent the mean values. Flow cytometry analysis for CD71 and GPA surface expression on culture day 7 (B) and day 14 (C). Flow cytometric gates denoted as 1, 2, 3 (panel B) and 4, 5, 6 (panel C) were sorted for quantitative PCR quantitation of TWSG1. (D) TWSG1 expression level in the sorted populations according to the sorted gates shown in panels B and C.
TWSG1 expression in human erythropoiesis. (A) Microarray confirmation by quantitative PCR using erythroblasts from 6 separate donors (y-axis, copy number/ng total RNA). Each line associated values from the same donor on separate days. D7, culture day 7; D14, culture day 14. *P < .05. Bars represent the mean values. Flow cytometry analysis for CD71 and GPA surface expression on culture day 7 (B) and day 14 (C). Flow cytometric gates denoted as 1, 2, 3 (panel B) and 4, 5, 6 (panel C) were sorted for quantitative PCR quantitation of TWSG1. (D) TWSG1 expression level in the sorted populations according to the sorted gates shown in panels B and C.
TWSG1 inhibits up-regulation of hepcidin expression by BMP2/4
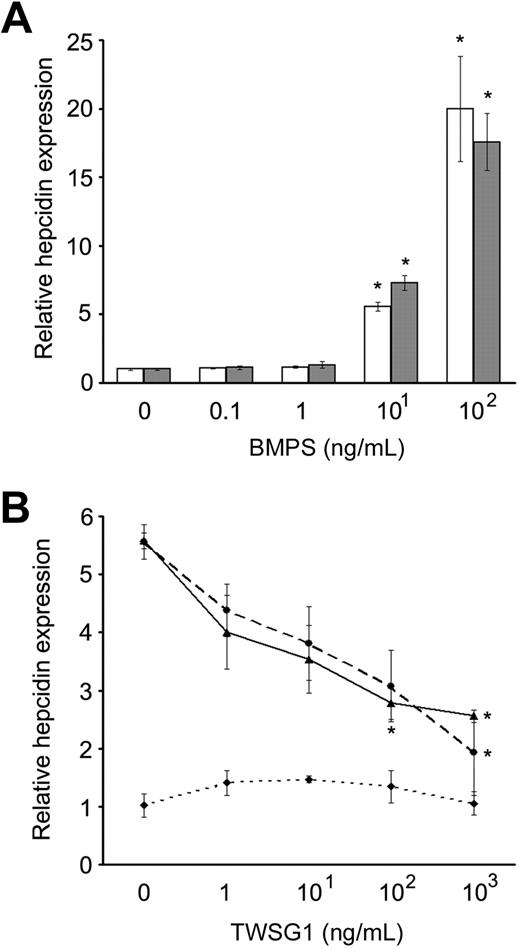
Several BMPs up-regulate hepcidin expression in liver cells.12 To determine whether TWSG1 affects BMP regulation of hepcidin expression, assays were performed using HuH-7 and human primary hepatocytes. In HuH-7 cells, dosing studies demonstrated significant up-regulation of hepcidin mRNA by BMP2 and BMP4 (Figure 2A). BMP2 (10 ng/mL) significantly elevated hepcidin expression in HuH-7 cells (5.7-fold increase, P = .021), and 10 ng/mL BMP4 also significantly elevated hepcidin expression in HuH-7 cells (7.3-fold increase, P = .005). Either BMP2 or BMP4 (100 ng/mL) elevated hepcidin in HuH-7 cells (20-fold increase; BMP2, P = .038; and BMP4, P = .015). In the absence of BMP supplements, TWSG1 did not significantly affect hepcidin expression in HuH-7 cells at doses up to 1000 ng/mL (Figure 2B). However, TWSG1 inhibited the increased expression of hepcidin in response to 10 ng/mL BMP2 or BMP4. In dosed-titrations, 100 ng/mL TWSG1 significantly reduced the BMP effect, and the reversal was nearly complete at 1000 ng/mL TWSG1.
TWSG1 inhibit up-regulation of hepcidin expression by BMP2/4 in human hepatoma cell line. (A) Relative hepcidin expression as a dosed response to BMP2 (□) and BMP4 (■) are shown. Final BMP concentrations are on the x-axis, and relative hepcidin expression on the y-axis (0 ng/mL assigned a value of 1 for comparison). (B) TWSG1 dose response of relative hepcidin expression in human HuH-7 cells in cultures supplemented with 10 ng/mL BMP2 (—), 10 ng/mL BMP4 (---), or in the absence of added BMP ( ). Bars represent mean values with asterisks signifying P < .05.
). Bars represent mean values with asterisks signifying P < .05.
TWSG1 inhibit up-regulation of hepcidin expression by BMP2/4 in human hepatoma cell line. (A) Relative hepcidin expression as a dosed response to BMP2 (□) and BMP4 (■) are shown. Final BMP concentrations are on the x-axis, and relative hepcidin expression on the y-axis (0 ng/mL assigned a value of 1 for comparison). (B) TWSG1 dose response of relative hepcidin expression in human HuH-7 cells in cultures supplemented with 10 ng/mL BMP2 (—), 10 ng/mL BMP4 (---), or in the absence of added BMP ( ). Bars represent mean values with asterisks signifying P < .05.
). Bars represent mean values with asterisks signifying P < .05.
For comparison with cell lines, BMP-mediated inhibition of hepcidin expression in human primary hepatocytes was also assayed. In human primary hepatocytes from 4 donors, the augmentation of hepcidin by BMP2 and BMP4 was more robust than in HuH-7 cells with several thousand-fold increases in hepcidin mRNA in each donor after 100 ng/mL supplementation of BMP2 or BMP4 (Figure 3A,C). Like HuH-7 cells, dosed TWSG1 (1-1000 ng/mL) did not directly affect hepcidin expression in primary hepatocytes (data not shown). Similarly, the BMP-mediated increases in hepcidin were significantly reduced in each of the donors (Figure 3B,D).
TWSG1 inhibits BMP2/4 up-regulation of hepcidin in primary human hepatocytes. Dose response of relative hepcidin expression levels to (A) BMP2 and (C) BMP4 are shown using primary hepatocytes from 4 separate donors (shaded bars). Final BMP concentrations are on the x-axis, and relative hepcidin expression on the y-axis (0 ng/mL assigned a value of 1 for comparison). In panels B and D, dosed response of relative hepcidin expression to TWSG1 in human primary hepatocytes cultured in 10 ng/mL BMP2 or BMP4, respectively. Lines represent results from 4 separate donors' cells from triplicate cultures. *P < .05.
TWSG1 inhibits BMP2/4 up-regulation of hepcidin in primary human hepatocytes. Dose response of relative hepcidin expression levels to (A) BMP2 and (C) BMP4 are shown using primary hepatocytes from 4 separate donors (shaded bars). Final BMP concentrations are on the x-axis, and relative hepcidin expression on the y-axis (0 ng/mL assigned a value of 1 for comparison). In panels B and D, dosed response of relative hepcidin expression to TWSG1 in human primary hepatocytes cultured in 10 ng/mL BMP2 or BMP4, respectively. Lines represent results from 4 separate donors' cells from triplicate cultures. *P < .05.
TWSG1 inhibits BMP-mediated phosphorylation of Smad proteins
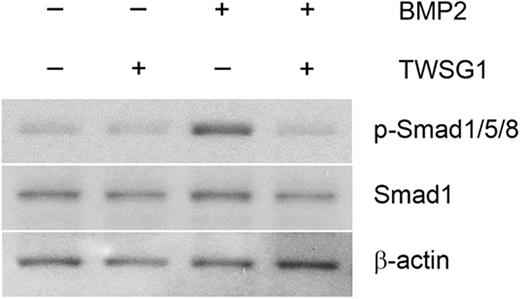
BMP2 and BMP4 regulate induction of hepcidin transcription in liver cells through Smad1/5/8 signaling.12,13 To determine whether TWSG1 affects this signaling pathway, Smad1/5/8 phosphorylation was performed using HuH-7 cell extracts. Low levels of phosphorylated Smad proteins were detected in controls and remained stable in the presence of TWSG1. However, 10 ng/mL BMP2 increased Smad1/5/8 phosphorylation compared with controls. While the BMP2 mediated increase in Smad1/5/8 phosphorylation was modest, both hepcidin expression and Smad phosphorylation returned to baseline levels with the addition of 1000 ng/mL TWSG1 (Figures 2B, 4).
TWSG1 inhibits BMP-mediated Smad phosphorylation. HuH-7 cells were cultured in the presence or absence of 1000 ng/mL TWSG1, 10 ng/mL BMP2, or both for 1 hour. Total cell lysates (20 μg/lane) were immunoblotted using a rabbit antiserum specific to human phosphorylated-Smad1/5/8 (p-Smad1/5/8). Anti–total Smad1 antibody and anti–β-actin antibody were used as internal controls.
TWSG1 inhibits BMP-mediated Smad phosphorylation. HuH-7 cells were cultured in the presence or absence of 1000 ng/mL TWSG1, 10 ng/mL BMP2, or both for 1 hour. Total cell lysates (20 μg/lane) were immunoblotted using a rabbit antiserum specific to human phosphorylated-Smad1/5/8 (p-Smad1/5/8). Anti–total Smad1 antibody and anti–β-actin antibody were used as internal controls.
Twsg1 inhibition of hepcidin in murine cells does not require BMP supplementation
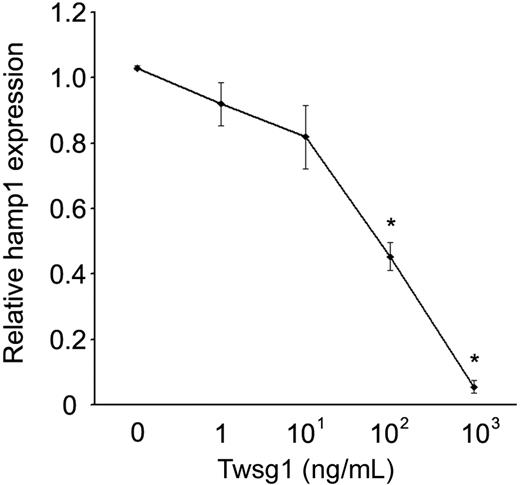
For comparison with studies in human cells, similar dosing studies were performed using primary murine hepatocytes. Unlike the human cells, mouse Twsg1 (1-1000 ng/mL) significantly suppressed hepcidin expression in the absence of BMP supplementation (Figure 5). The suppression showed a dose-response effect: 10 ng/mL Twsg1 resulted in an 18% reduction (not significant, NS), 100 ng/mL Twsg1 resulted in a 56% reduction (P < .001), and 1000 ng/mL Twsg1 resulted in a 94% reduction (P < .001).
Twsg1 dose response of hepcidin levels in primary murine hepatocytes. In primary hepatocytes from 4- to 6-week-old C57Bl/6 mice, dosed increased Twsg1 (x-axis) was added, and hepcidin (y-axis; hamp1) mRNA was quantified. *P < .05 compared with untreated cells.
Twsg1 dose response of hepcidin levels in primary murine hepatocytes. In primary hepatocytes from 4- to 6-week-old C57Bl/6 mice, dosed increased Twsg1 (x-axis) was added, and hepcidin (y-axis; hamp1) mRNA was quantified. *P < .05 compared with untreated cells.
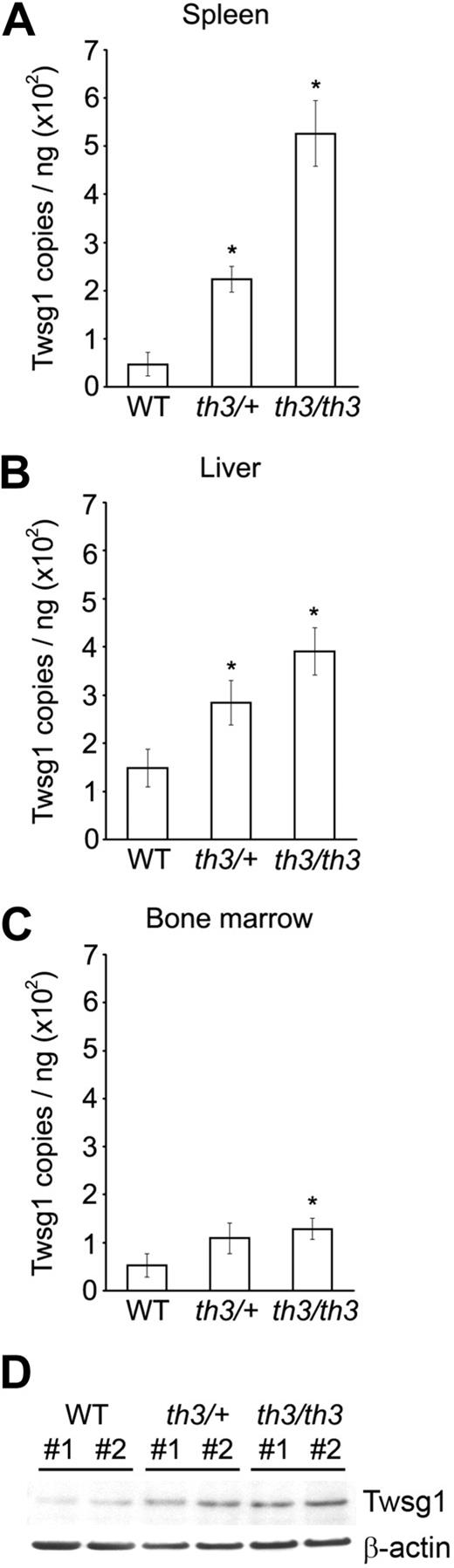
Increased Twsg1 expression in thalassemia mice
In addition to studies of cell lines and primary hepatocytes from healthy animals, Twsg1 expression was studied in thalassemia mice. Quantitative PCR using the β-thalassemia murine model (Hbbth3/+ β-thalassemia intermedia mouse model, n = 13; Hbbth3/th3 β-thalassemia major mouse model, n = 5) revealed a significant increase in Twsg1 mRNA levels among the spleen, bone marrow, and liver (Figure 6A-C). The most significant increase was detected in the spleen (Hbbth3/+, 2.2E02 ± 2.7E01 copies/ng total RNA, P = .001; Hbbth3/th3, 5.3E02 ± 6.8E01 copies/ng total RNA, P = .001) compared with wild-type mice (4.7E01 ± 2.4E01 copies/ng total RNA, n = 7; Figure 6A). Twsg1 expression levels in the murine liver were also significantly higher in both thalassemia types (Hbbth3/+, 2.8E02 ± 4.6E01 copies/ng total RNA, P = .038; Hbbth3/th3, 3.9E02 ± 4.9E01 copies/ng total RNA, P = .004) compared with wild-type mice (1.5E02 ± 4.0E01 copies/ng total RNA; Figure 6B). Bone marrow expression of Twsg1 was also elevated, but reached statistical significance only in the Hbbth3/th3 mice (Hbbth3/+, 1.1E02 ± 3.2E01 copies/ng total RNA, P = .18; Hbbth3/th3, 1.3E02 ± 2.2E01 copies/ng total RNA, P = .038) compared with wild-type mice (5.3E01 ± 2.5E01 copies/ng total RNA; Figure 6C). Twsg1 protein expression was confirmed by immunoblot analysis using spleen extract from Hbbth3/+ and Hbbth3/th3 mice (Figure 6D). As shown, expression of Twsg1 protein was elevated in both thalassemia types compared with wild-type mice.
Twsg1 mRNA expression in murine thalassemia. Murine Twsg1 mRNA in (A) spleen, (B) liver, and (C) bone marrow from wild-type mice (WT, n = 7), Hbbth3/+ β-thalassemia intermedia mice (th3/+, n = 13), and Hbbth3/th3 β-thalassemia major mice (th3/th3, n = 5) was determined by quantitative PCR using total RNA isolated from the tissues. Bars represent mean values; *P < .05 compared with wild-type. (D) Murine Twsg1 protein (Twsg1) expression in spleen from wild-type mice (WT), Hbbth3/+ β-thalassemia intermedia mice (th3/+), and Hbbth3/th3 β-thalassemia major mice (th3/th3) was detected by immunoblotting using a rabbit anti–serum-specific Twsg1. Anti–β-actin antibody were used as internal control. Splenic tissues from 2 mice were studied for comparison (#1, #2).
Twsg1 mRNA expression in murine thalassemia. Murine Twsg1 mRNA in (A) spleen, (B) liver, and (C) bone marrow from wild-type mice (WT, n = 7), Hbbth3/+ β-thalassemia intermedia mice (th3/+, n = 13), and Hbbth3/th3 β-thalassemia major mice (th3/th3, n = 5) was determined by quantitative PCR using total RNA isolated from the tissues. Bars represent mean values; *P < .05 compared with wild-type. (D) Murine Twsg1 protein (Twsg1) expression in spleen from wild-type mice (WT), Hbbth3/+ β-thalassemia intermedia mice (th3/+), and Hbbth3/th3 β-thalassemia major mice (th3/th3) was detected by immunoblotting using a rabbit anti–serum-specific Twsg1. Anti–β-actin antibody were used as internal control. Splenic tissues from 2 mice were studied for comparison (#1, #2).
Discussion
In this study, TWSG1 was explored as a candidate hepcidin regulatory protein that is expressed during erythropoiesis. TWSG1 and GDF15 were originally discovered in this context through transcriptome analyses.3 In contrast to GDF15, the highest-level expression of TWSG1 was detected at early stages of erythroblast differentiation before hemoglobinization of the cells. TWSG1 was named according to the characteristically twisted phenotype during gastrulation of Drosophila melanogaster.14 Later, it was determined that Twsg1 functions as an agonist/antagonist of BMPs.15-17 The BMP-regulatory function of Twsg1 may be modified by additional proteins in the extracellular milieu including chordin and crossveinless-2 proteins.18 In murine loss-of-function studies, Twsg1 demonstrated an abnormal craniofacial development.19 In the murine thymus, Twsg1 enhances T-cell development by blocking BMP2 and BMP4 protein signaling.20 Very little is known about the expression and function of twisted gastrulation protein in humans.
Hepcidin plays a central role in the regulation of iron absorption and availability in mammals.21 To date, iron, hypoxia, interleukins, GDF15, BMPs, hemojuvelin, and TMPRSS6 all contribute to physiologic and pathologic regulation of hepcidin expression in hepatocytes.22 The data suggest another layer of complexity exists with regard to hepcidin expression and subsequent regulation of iron homeostasis. TWSG1 suppressed hepcidin in human hepatocytes through a BMP-dependent mechanism that did not cause increased cellular toxicity or affect the expression of albumin (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Phosphorylation studies further suggest TWSG1 acts by inhibiting the BMP-dependent activation of Smad-mediated signal transduction. In comparison to human cells, hepcidin expression in murine hepatocytes was also suppressed by Twsg1 even in the absence of supplemental BMP proteins. Increased hepatic expression of several BMPs in murine liver may explain this result (supplemental Figure 2), but the relative contribution of each molecule toward the overall regulation of hepcidin is difficult to quantitate. As such, the data suggest the regulation of hepcidin expression is complex and strongly suggest that multiple regulatory mechanisms have evolved to maintain hepcidin expression under a variety of nutritional and inflammatory states.
The phenotypic differences between the human and murine thalassemia are not well understood and may be due to interspecies differences at several cellular levels including: secreted molecules, signal transduction pathways, and transcriptional regulation of the hepcidin gene. Overproduction of GDF15 to levels that are sufficient to suppress hepcidin in human hepatocytes was previously demonstrated in thalassemia patients.3 GDF15 expression at higher levels in humans with thalassemia is thought to be due to erythroblast precursor cell apoptosis at the later stages of erythropoiesis.8 However, GDF15 dysregulation in mice was not associated with an iron phenotype, and the murine thalassemia model studied here is associated with less apoptosis and erythroblast maturation arrest at very earlier stages of maturation.3,6 In addition, extramedullary hepatic and splenic hematopoiesis is robust in murine thalassemia.7 The greatly expanded population of immature erythroblasts combined with extramedullary erythropoiesis may thus cause an increase in Twsg1 expression detected in this study (Figure 6). The increased BMP expression in the murine liver reported here and in association with iron loading23 may further enhance Twsg1 effects in mice. Additional genetic manipulations and methods are being developed to strictly control expression of Twsg1 and other hepcidin-regulating molecules in thalassemia mice to determine whether Twsg1 is the major regulator of hepcidin suppression in murine thalassemia.
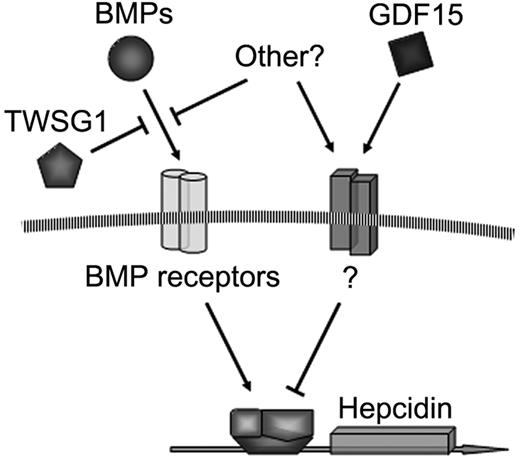
As shown in Figure 7, it is proposed that TWSG1 and GDF15 act together to inappropriately inhibit hepcidin expression in thalassemia. Due to the expansion of erythropoiesis, TWSG1 expressed during the early stages of erythropoiesis acts indirectly by inhibiting BMP-mediated expression of hepcidin. Such a mechanism is supported by the recent discovery that iron-fed mice increase hepcidin expression secondary to increased BMP expression.23 In thalassemia, overexpression of TWSG1 would inhibit the host's ability to sense and respond to iron loading. In addition to iron regulation, the ability of TWSG1 to dysregulate BMP and Smad signaling may be further compelling in the context of crippling bony disease in thalassemia. In humans, apoptosis of more mature erythroblasts may further suppress hepcidin via pathologic expression of GDF15. The increased expression of TWSG1 may also impact erythropoiesis in thalassemia in another way. Analysis of stress erythropoiesis in mice demonstrated that BMP4 is a key signal that promotes the expansion of a specialized population of stress erythroid progenitors.24 Stress erythroblasts have a greater capacity to generate new erythrocytes than bone marrow steady state erythroid progenitors. In addition to effects upon hepatocytes, overexpression of Twsg1 in Hbbth3 mice could inhibit the BMP4-dependent expansion stress erythroblasts, which in turn would exacerbate the anemia. Thus, the pathologic combination of TWSG1 and GDF15 expression in thalassemia may contribute to the clinical triad of iron overload, bony disease, and ineffective erythropoiesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (Bethesda, MD).
National Institutes of Health
Authorship
Contribution: T.T. performed experiments, assembled data, and wrote the manuscript; P.P. generated the Hbbth3/+ and Hbbth3/th3 mice and harvested tissues for analysis; O.S. performed erythroblast cultures and cell harvesting; S.N. performed bioinformatics analysis; C.B. performed flow cytometric analysis; A.B. performed erythroblast culture and cell harvesting; Y.T.L. collected and assembled array data; J.B.G. performed experiments, analyzed data, and edited the manuscript; O.H. performed transplantations on Hbbth3/th3 mice; T.G. analyzed data and edited the manuscript; R.F.P. designed and supervised the Hbbth3/th3 mouse experiments and contributed to manuscript writing; and J.L.M. contributed to experimental conception and design and manuscript writing and assisted and supervised the research team.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jeffery L. Miller, Molecular Medicine Branch, NIH, Bldg 10, Rm 9N311, 10 Center Dr, Bethesda, MD 20892; e-mail: jm7f@nih.gov.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal