Abstract
Being central players in thrombosis and hemostasis, platelets react in manifold and complex ways to extracellular stimuli. Cell-matrix and cell-cell interactions are mandatory for initial adhesion as well as for final development of stable plugs. Primary interfaces for interactions are plasma membrane proteins, of which many have been identified over the past decades in individual studies. However, due to their enucleate structure, platelets are not accessible to large-scale genomic screens and thus a comprehensive inventory of membrane proteins is still missing. For this reason, we here present an advanced proteomic setup for the detailed analysis of enriched platelet plasma membranes and the so far most complete collection of platelet membrane proteins. In summary, 1282 proteins were identified, of which more than half are termed to be of membrane origin. This study provides a brief overview of gene ontology subcellular and functional classification, as well as interaction network analysis. In addition, the mass spectrometric data were used to assemble a first tentative relative quantification of large-scale data on the protein level. We therefore estimate the presented data to be of major interest to the platelet research field and to support rational design of functional studies.
Introduction
Platelets are essential mediators of hemostasis and are well known for their major role in thrombotic events. In the context of increasing numbers of cardiovascular diseases, platelets are of premier interest for scientific research. Owing to their unique enucleate ultrastructure (still including organelles such as Golgi, endoplasmic reticulum, or mitochondria), common molecular biology–based methods can hardly be applied. Their megakaryocyte-derived maturation process renders platelets inconvenient targets for genome-based research approaches. Although mouse knockout models were generated for a range of functional studies, they are limited mostly to known protein components of the platelet system, estimated to be of functional importance. Moreover, protein synthesis in platelets is limited, and a direct correlation between mRNA profiling and protein presence is problematic at best. Based on these limitations, modern proteome analysis might be a key asset for the analysis of the platelet proteome and its so far unknown components. The derived knowledge of these newly identified proteins represents a rich source for the rational design of functional studies.
Despite their major functional importance a complete inventory of the platelet proteome is far from being accessible. Due to the development of biomolecular mass spectrometry, a series of medium- to large-scale studies was conducted upon the platelet proteome. Early studies aiming for a complete overview of platelet proteins most often used 2-dimensional polyacrylamide gel electrophoresis (2D-PAGE) as primary separation technique, frequently in conjunction with peptide mass fingerprint identification of proteins. However, as demonstrated by Moebius et al1 2D-PAGE–based studies of complete platelets2,3 were reasonably unsuccessful in elucidating membrane components compared with targeted membrane purifications in combination with suitable separation techniques,1 which achieved identification of nearly 300 proteins, of which approximately half were of membrane origin. Of these, G6B was later confirmed by Senis et al to be a novel immunoreceptor tyrosine-based inhibitory motif protein4 as part of an additional study on the platelet membrane proteome. Still, only 46 plasma membrane components could be observed within a total of136 membrane proteins in this study, thereby indicating a potential gap of knowledge regarding protein membrane constituents. Plasma membrane proteins are among the key targets for platelet drug and functional research, since they are the primary interface to extracellular stimuli and therefore also for pharmaceutical treatment of platelet-related disorders. We therefore sought to increase the identification rate of platelet membrane proteins with a major focus on the plasma membrane.
For plasma membrane enrichment, several methods have been established in the past, with density gradients being the most common. In addition, lectins have been used to additionally purify plasma membrane glycoproteins.1 However, due to our experience with efficient enrichment of rat brain plasma membranes by 2-phase aqueous partitioning systems,5,6 we adopted this technique for platelet membranes. Two-phase partitioning is based on sorting of vesicles upon their physicochemical surface properties (such as hydrophilicity/hydrophobicity and net surface charge, possibly due to their phospholipid composition)7 within a defined 2-polymer system. A common system is the polyethyleneglycol (PEG)/dextran system, where plasma membranes show the highest affinity for the more hydrophobic upper PEG phase in comparison with other membrane vesicles.8 We already successfully used this technique for analysis of plasma membrane N-glycosylation sites on human platelets.9,10 In contrast, the current study intends to provide a general overview of membrane proteins within these preparations to allow for a more global assessment of the membrane protein composition.
Here, 3 major strategies were pursued for protein identification: (1) For a global analysis, separation of proteins by 1-dimensional sodium dodecyl sulfate–polyacrylamide gel electrophoresis (1D-SDS-PAGE) followed by nano–liquid chromatography (LC)–tandem mass spectrometry detection of peptides, basically as described previously6 was conducted. (2) To address issues commonly encountered with PAGE separation of membrane proteins, highly complex peptide mixtures from membrane proteins were separated by strong cation exchange and reversed-phase chromatography within a MudPIT setup (Multidimensional Protein Identification Technology) prior to mass spectrometric detection.11 (3) To reduce the high complexity of peptide mixtures prior detection, Combined Fractional Diagonal Chromatography (COFRADIC) was used to isolate N-terminal, methionine- or cysteine-containing peptides means chemical derivatization and standardized liquid chromatography.12,13 Thus, in combination with aqueous 2-phase partitioning, this 3-pronged approach enabled a comprehensive analysis of the platelet membrane proteome that clearly exceeds our previous knowledge about the plasma membrane inventory.
Methods
Materials
Unless stated otherwise, chemicals were purchased from Merck KGaA in analytical quality or better. Water (18 MΩ) was obtained from an Elga Labwater system.
Platelet purification
Human platelets were prepared based on procedures described previously9,14,15 from fresh apheresis concentrates (leukocyte depleted, ∼ 2 × 105 leukocytes, ∼ 6 × 105 erythrocytes, and 2-4 × 1011 platelets/250 mL; Department of Transfusion Medicine, University Würzburg) by additional differential centrifugation steps to further diminish potential cellular contaminations; a detailed procedure is given in the supplemental methods, available on the Blood website; see the Supplemental Materials link at the top of the online article. Use of platelets was approved by the ethics committee of the University of Würzburg and donors were informed upon use of concentrates for research purposes in accordance with the Declaration of Helsinki.
Plasma membrane enrichment
Platelet plasma membranes were enriched as previously described by 2-phase partitioning.9,16 Briefly, 2-phase systems consisting of 6.3% polyethyleneglycol 3350 and dextran T500 (Roth) each in 15 mM Tris, pH 7.8, were equilibrated overnight at 6°C. For separation of 100 mg (wet weight) platelets, a 20-mL system (1:1 vol/vol of the equilibrated PEG and dextran phases) was used. Lysis was accomplished by repetitive ultrasonic bursts (6 × 15 seconds) and subsequent phase separation was achieved by centrifugation at 500g for 10 minutes at 6°C. The top PEG phase was extracted twice with equal volumes of fresh dextran phase. The final upper PEG phase was diluted 1:1 with water, and membranes were pelleted by ultracentrifugation at 100 000g for 1 hour at 4°C in a TLA 100.4 rotor (Beckman Coulter). Removal of cytoplasmic and peripheral membrane proteins was achieved by 2-fold carbonate extraction in 100 mM sodium carbonate, pH 11.5.17 Pellets were stored at −80°C until further use.
1D-PAGE and digests
Platelet membrane pellets from 2-phase partitioning systems were reconstituted in 1 × LDS-sample buffer (Invitrogen). After incubation at 75°C for 15 minutes, samples were applied to 4% to 12% Bis-Tris gels using a MOPS buffer system (NuPAGE-Novex; Invitrogen). Protein separation was followed by colloidal Coomassie staining.15 Subsequently, gel lanes were cut in 1-mm slices and bands were treated as described previously.5 After reduction and alkylation, proteins were tryptically digested in-gel and peptides were extracted by 0.1% trifluoroacetic acid.
COFRADIC
A detailed description of COFRADIC experimental procedures is given in the supplemental methods.
Mass spectrometry
Nano-LC-tandem mass spectrometry of 1D-PAGE and COFRADIC fractions.
Tryptic peptides from in-gel digestion of 1D-PAGE bands were separated by nano-LC and detected by a Qtrap4000 mass spectrometer (Applied Biosystems) as described previously.9 In turn, mass spectrometric detection of COFRADIC fractions on a QStar Elite Q-TOF (Applied Biosystems) was preceded by nano-LC separation, essentially as described previously.18
Multidimensional protein identification technology.
MudPIT analyses were conducted following a variation of a protocol by Wolters et al.11 A detailed methods section is given in the supplemental methods.
Data evaluation
Shotgun approaches.
Raw data were transformed into Mascot19 generic format using either plug-ins for Analyst 1.4.2 (mascot.dll, MatrixScience) for Qtrap data or ltq dta.exe for LTQ-XL data. The resulting peak lists were searched against a concatenated forward/reversed human subset of the Swiss-Prot database20 (http://www.expasy.org, 20 834 sequences21 ) using either Mascot (MatrixScience) or Omssa22 as search engines. For Omssa and Mascot, the following search parameters were used: Trypsin was chosen as protease with one miscleavage site allowed, carbamidomethylation (C) was set as fixed, and oxidation (M) was set as variable modification. Precursor and tandem mass spectrometry (MS/MS)–ion tolerances were limited to 0.05 Da (Qtrap) and 0.01 Da (LTQ), respectively. Subsequent to searches, the result-files of Omssa and Mascot were combined in Masssieve (http://www.proteomecommons.org/dev/masssieve).23 As filter criteria, P value cutoffs were set to .05 for Mascot and .01 for Omssa. Furthermore, hits were limited to proteins that feature at least 2 different significantly identified peptides. Thereby, a false-positive rate of less than 1% was achieved.
COFRADIC.
COFRADIC data were searched against a human subset of the Swiss-Prot database (see “Shotgun approaches”) using Mascot (MatrixScience). Mascot generic files were generated using the plug-in to Analyst QS 2.0 (mascot.dll; MatrixScience). Although the different approaches necessitated individual settings, the following general parameters were used: Both precursor and MS/MS tolerance were set to 0.2 Da, and one missed cleavage was allowed. Depending upon the type of peptide that was sorted for the Mascot, modification parameters were set accordingly as seen in Table 1. All resulting peptide identifications were manually validated regarding presence of ion series, dominant fragmentation patterns (eg, adjacent to proline), and overall signal intensity.
Mascot search parameters for COFRADIC approaches
| . | Fixed modification . | Variable modification . |
|---|---|---|
| N-terminal COFRADIC (protease: Arg-C/P) | Oxidation to sulphoxide (M), trideutero-acetylation (K), carbamidomethylation (C) | Pyroglutamate (N-term Q), pyrocarbamidomethyl cysteine (N-term alkylated C), acetylation (N-term), trideutero-acetlyation (N-term) |
| Met-/Cys-COFRADIC (protease: trypsin) | Oxidation to sulphoxide (M), carbamidomethylation (C) | Pyroglutamate (N-term Q), pyrocarbamidomethyl cysteine (N-term alkylated C), acetylation (N-term) |
| . | Fixed modification . | Variable modification . |
|---|---|---|
| N-terminal COFRADIC (protease: Arg-C/P) | Oxidation to sulphoxide (M), trideutero-acetylation (K), carbamidomethylation (C) | Pyroglutamate (N-term Q), pyrocarbamidomethyl cysteine (N-term alkylated C), acetylation (N-term), trideutero-acetlyation (N-term) |
| Met-/Cys-COFRADIC (protease: trypsin) | Oxidation to sulphoxide (M), carbamidomethylation (C) | Pyroglutamate (N-term Q), pyrocarbamidomethyl cysteine (N-term alkylated C), acetylation (N-term) |
Spectral counting.
A version of the exponentially modified protein abundance index (emPAI)24 approach was used to gain quantitative information of protein abundance by spectral counting. Therefore, the number of possible tryptic peptides (Nobservable) was calculated for each protein with resulting peptides in the range of m/z 760-4000 Da. emPAI indices were calculated as ln(10̂[Nobserved/{5 × Nobservable}]). The number of observed peptide hits was extracted from the Masssieve result lists for each protein. Quantification was performed for discrete protein hits from shotgun data only. Differentiable hits share peptides with other proteins, whereas the COFRADIC approach concentrates on subsets of peptides, rendering both data sets unsuitable for the spectral counting approach.
Results
General overview
By the combined approaches of MudPIT, 1D-PAGE coupled to nano-LC-tandem mass spectrometry, as well as COFRADIC, a total of 1282 proteins were identified within the platelet membrane preparations derived from aqueous 2-phase partitioning. Thereof, 498 proteins were identified with peptide-centric COFRADIC on single-peptide basis, whereas a total of 1202 proteins were accessible by the combined results of the 2 shotgun-based approaches. A major intersection of 418 proteins was determined between shotgun and COFRADIC data sets.
Shotgun data
Mass spectra of the 2 shotgun approaches were searched by 2 independent search engines, Omssa25 and Mascot.26 Results from 3 MudPIT and 3 independent 1D-PAGE experiments were directly combined after searches. They showed a large correlation of the engines on the peptide level with 7841 peptides being identified by Omssa and 6895 peptides identified by Mascot, with a major intersection of 5918 peptides. The parallel use of Mascot and Omssa therefore extended the results by approximately 14% (based on Mascot hits) and crosswise confirmed the majority of hits for each search engine. After data evaluation of result files in Masssieve, only protein hits with at least 2 valid peptide identifications were accepted. This limitation alongside a false-positive discovery rate ranging below 1% enabled presentation of a highly reliable and accurate data set. Therefore, we also refrained from introducing proteins with only one identified peptide, which would have resulted in more than 2000 protein identifications. To further remove ambiguous data, a differentiation between discrete protein hits (934) and differentiable protein hits (268) was made. These proteins are either identified by discrete peptides that are shared with no other protein sequence, or they are partially identified by shared peptides. However, additional peptides always guarantee the correct annotation of the protein (isoform). Indeed, about three quarters of identifications were based on 3 or more peptides, and the average sequence coverage was 21.8%. Other proteins, which could not be identified unambiguously (superset, subsumable, equivalent; total of 167 protein features), were discarded for the current analysis. For further use, all data including the latter are presented in a supplemental table along with identified peptide sequences.
Combined fractional diagonal chromatography
Using a peptide-centric strategy, 3 approaches aiming for N-terminal, and cysteine- and methionine-containing peptides were used, identifying a total of 498 proteins. Methionine- and cysteine-based COFRADIC strategies were able to identify 274 and 219 proteins, respectively, whereas N-terminal COFRADIC enabled 160 protein identifications (Figure 1).
Combined fractional diagonal chromatography results. (A) Distribution of the 498 proteins identified by COFRADIC with respect to the 3 different approaches. (B) Exemplary scheme of N-terminal COFRADIC. Upon blockage of free amines (i) and tryptic digest (ii), fractions are collected during a first-dimension high-performance liquid chromatography run (iii). Due to subsequent blockage by TNBS, internal peptides are shifted during a second high-performance liquid chromatography run (iv), whereas true N-terminal peptides are eluted in the same timeframe as before (iii). (C) COFRADIC results are mostly covered as well by identifications based on shotgun approaches. (D) List of selected N-termini elucidated by N-terminal COFRADIC from platelet membrane proteins. N-termini often differ from literature (Swiss-Prot database) or are newly assigned.
Combined fractional diagonal chromatography results. (A) Distribution of the 498 proteins identified by COFRADIC with respect to the 3 different approaches. (B) Exemplary scheme of N-terminal COFRADIC. Upon blockage of free amines (i) and tryptic digest (ii), fractions are collected during a first-dimension high-performance liquid chromatography run (iii). Due to subsequent blockage by TNBS, internal peptides are shifted during a second high-performance liquid chromatography run (iv), whereas true N-terminal peptides are eluted in the same timeframe as before (iii). (C) COFRADIC results are mostly covered as well by identifications based on shotgun approaches. (D) List of selected N-termini elucidated by N-terminal COFRADIC from platelet membrane proteins. N-termini often differ from literature (Swiss-Prot database) or are newly assigned.
The 3 approaches exhibited highly complementary results shown by the 360 proteins, which were covered by only one of the approaches each. This stresses the necessity for parallel application of all 3 analysis variants. Although the identification of the proteins is based on individual mass spectra, all peptide hits were manually validated to account for correct annotation and identification by completeness of ion series. In addition, all spectra are listed in the pride database for public access (http://www.ebi.ac.uk/pride/init.do; experiment accession no. 8127 [methionine], no. 8128 [N-terminal], and no. 8129 [cysteine]).27 COFRADIC enabled protein identification ranging from high abundant proteins, such as integrin alpha-IIb and other integrins (integrin alpha-5 or alpha-6), down to low abundant proteins, such as the G-protein–coupled receptor PAR1. In combination with the 2-phase aqueous partitioning system, COFRADIC enabled the identification of 234 proteins (47%) with at least one predicted transmembrane domain (prediction by TMHMM 2.028 ). For comparison, we mapped the IPI-based accessions of another COFRADIC-based study on whole platelets lysates29 back to Swiss-Prot accession,30 yielding only a share of 17% proteins with at least one TMD (66 of 385). Clearly, aqueous 2-phase partitioning-based membrane purification caused a nearly 3-fold increase in membrane protein identification rate.
Although only 80 proteins were exclusively identified by COFRADIC, the approach also yields additional protein-related information. In case of N-term COFRADIC, the in vivo existing N-termini are determined, of which a few examples are shown in Figure 1 (for a complete list, compare supplemental table). In comparison with the Swiss-Prot database, those N-termini often concur with literature as shown for GpIX, JAM1, or GpIB-beta. In the case of less known proteins, however, the N-termini are so far unknown and can now be added to the database information or be used for functional experiments. However, it should be noted that N-terminal processing might also occur during the apheresis used for production of the platelet concentrates.
Enrichment of membrane proteins
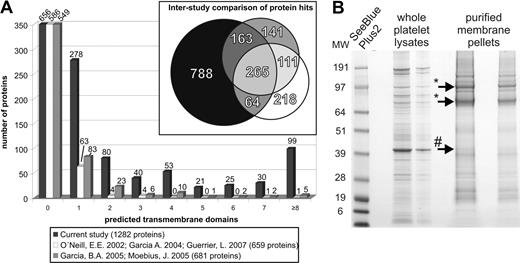
Being targeted at membrane proteins and more specifically those located in the plasma membrane, an analysis of potential TMDs was conducted using TMHMM 2.0 for the complete data set and in addition also for several previous studies found in the literature. The results are summarized in the bar diagram of Figure 2A. Clearly, the current study provides a tremendous increase of protein identifications featuring at least one TMD. Although studies using whole platelet lysates (O'Neill/Garcia/Guerrier) detected 63 proteins in this case, the current study features a more than 4-fold increase. In addition, in comparison with more focused studies on plasma membranes or microparticles, the current analysis encompasses a far superior number of multimembrane-pass proteins, for example, 30 proteins with 7 transmembrane domains including several G-protein–coupled receptors such as PAR1, PAR4, or the potential G-protein–coupled receptor 92. Nearly a hundred proteins were predicted with equal or more than 8 TMDs including adenylate cyclases 3, 5, and 6, proteins of the solute carrier families 12, 22, 23, and 40, as well as many less studied proteins such as the 2-pore calcium channel protein 1. In conclusion, TMHMM 2.0 analyses confirmed the improved discovery rate of membrane proteins by determining 626 membrane-spanning proteins.
TMHMM-based interstudy comparison and membrane protein enrichment. (A) Prediction of membrane spanning proteins was done by TMHMM 2.0; comparison with several published studies on the platelet proteome clearly shows an increased number of identified membrane proteins by the current approach. (B) CBB-G250–stained 1D-SDS-PAGE of whole platelet lysates and enriched plasma membranes by aqueous 2-phase partitioning (* indicates gpIIb/IIIa; #, actin; proteins were identified by mass spectrometric sequencing).
TMHMM-based interstudy comparison and membrane protein enrichment. (A) Prediction of membrane spanning proteins was done by TMHMM 2.0; comparison with several published studies on the platelet proteome clearly shows an increased number of identified membrane proteins by the current approach. (B) CBB-G250–stained 1D-SDS-PAGE of whole platelet lysates and enriched plasma membranes by aqueous 2-phase partitioning (* indicates gpIIb/IIIa; #, actin; proteins were identified by mass spectrometric sequencing).
To enable a more direct impression of the purification success, Figure 2B depicts the Coomassie G250–stained SDS-PAGE of whole platelet lysates in comparison with the purified membranes derived from aqueous 2-phase partitioning. Although soluble proteins such as actin (hash key marker) are largely depleted, plasma membrane proteins such as gpIIb/IIIa (asterisks) are heavily enriched. This observation is congruent with Moebius et al,1 showing a similar distribution of actin and gpIIb/IIIa, although with a higher share of actin remaining. This, however, is attributed to the additional, second carbonate extraction step17 in the current study, obviously removing larger parts of the membrane cytoskeleton.
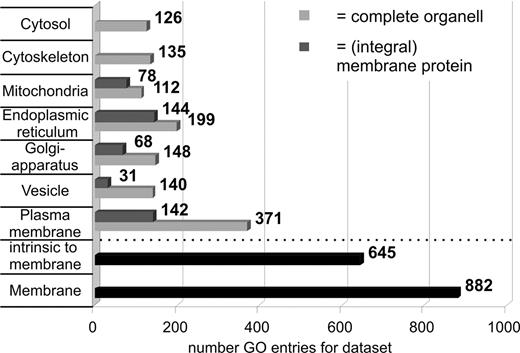
The subcellular distribution of proteins was estimated by their GO annotations.31 Therefore, Ontologizer 2.032 was used to sort proteins as shown by Figure 3. Evidently, a high number of plasma membrane components (371) are present within the 1282 protein data set, including 142 integral plasma membrane components. Furthermore, a range of proteins from other membrane-bound compartments, such as endoplasmic reticulum (ER) (199), Golgi apparatus (148), and vesicles (140), has been identified. A comparison of accessions between summed ER, Golgi, and vesicle (388) and plasma membrane proteins (371) yields a major intersection of 116 proteins. Obviously those proteins have a certain distribution or are shuttling between the mentioned organelles. However, a major share of the proteins has no determined subcellular localization at all, judged by their GO terms, and represents protein species not yet characterized.
Gene ontology (GO)–based subcellular annotation of identified proteins. Clearly, high numbers of membrane-associated proteins (882) were identified with a major share of 371 belonging directly to the plasma membrane compartment.
Gene ontology (GO)–based subcellular annotation of identified proteins. Clearly, high numbers of membrane-associated proteins (882) were identified with a major share of 371 belonging directly to the plasma membrane compartment.
Functional characterization
A complete functional characterization of the current protein survey seems improbable, especially due to the diverse functions of identified proteins. To provide some estimate of functional subclasses, we used GO annotations to highlight protein classes of potential interest.
Being the primary interface for extracellular stimuli, the platelet plasma membrane is supposed to contain a large selection of proteins involved in signal transduction. Indeed, 290 proteins were returned by this entry via their GO terms, including 156 proteins with signal transduction activity and encompassing 104 entries with receptor activity. Despite this high number, it has to be noted that many other activities may not be reflected by GO terms, yet. Although it is far beyond the scope of this study to discuss all membrane proteins in total, some examples of membrane-residing protein classes will be briefly shown: G-protein–coupled receptors (GPCRs), proteins of cellular adhesion, and membrane ordering proteins. Furthermore, a complete list of proteins sorted by GO terms into (functional) classes is presented within the supplemental table or may rapidly be assembled using Ontologizer (Table 2).
Algorithms and tools suitable for analysis of platelet proteome data
| Name . | Description . | Source . |
|---|---|---|
| UniProt | Curated database with linked information sources, retrieval of fasta sequences, etc | http://www.uniprot.org/33 |
| Protein identifier cross reference service (PICR) | Cross-reference algorithm for accession exchange between different formats (eg, NCBI vs Swiss-Prot or gene name vs Swiss-Prot accession) | http://www.ebi.ac.uk/Tools/picr/34 |
| Expasy | Proteomics server for various prediction and sequence analysis programs (eg, prediction of phosphorylation and glycosylation sites, TMDs, import sequences, domains) | http://www.expasy.org/tools/ |
| TMHMM 2.0 | Prediction of transmembrane domains | http://www.cbs.dtu.dk/services/TMHMM/ |
| Simple Modular Architechture Research Tool (SMART) | Prediction and annotation of protein domains, internal links to PFAM | http://smart.embl-heidelberg.de/smart/batch.pl35 |
| ProtFun 2.2 | Ab initio prediction of protein function, enzymatic properties and possible gene ontology, ab initio predictions of protein function from sequence. | http://www.cbs.dtu.dk/services/ProtFun/36 |
| STRING | STRING is a database of known and predicted protein interactions. The interactions include direct (physical) and indirect (functional) associations | http://string.embl.de/37 |
| Unified Human Interactome (UniHI) | Interaction network prediction | http://theoderich.fb3.mdc-berlin.de:8080/unihi/home38 |
| GoMiner | Evaluation of gene ontology classification for large scale data | http://discover.nci.nih.gov/gominer/39 |
| Ontologizer | Evaluation of gene ontology classification for large scale data, comparison between datasets | http://compbio.charite.de/index.php/ontologizer2.html |
| Bioinformatic Harvester III | Harvester crawls and cross-links the following bioinformatic sites: 4DXp, AceView, BLAST, Biocompare, CDART, CDD, ensEMBL, Entrez, FishMap, Galaxy, UCSC GenomeBrowser, gfp-cDNA, Google-Scholar, gopubmed, Harvester42, H-Inv, HomoloGene, iHOP, IPI, MapView, MGI, MINT, Mitocheck, OMIM, PolyMeta, PSORT II, RGD, SMART, SOSUI SOURCE, STRING, TAIR, Unigene, UniprotKB, Wikipedia, WikiProtein | http://harvester.fzk.de/harvester/40 |
| Reactome (including SkyPainter) | Curated knowledgebase of biologic pathways | http://www.reactome.org41 |
| KEGG PATHWAY database | Collection of manually drawn pathway maps representing our knowledge on the molecular interaction and reaction networks (also accessible via STRING) | http://www.genome.jp/kegg/pathway.html42 |
| iHOP | Hyperlinked information about proteins | http://www.ihop-net.org/UniPub/iHOP/43 |
| Name . | Description . | Source . |
|---|---|---|
| UniProt | Curated database with linked information sources, retrieval of fasta sequences, etc | http://www.uniprot.org/33 |
| Protein identifier cross reference service (PICR) | Cross-reference algorithm for accession exchange between different formats (eg, NCBI vs Swiss-Prot or gene name vs Swiss-Prot accession) | http://www.ebi.ac.uk/Tools/picr/34 |
| Expasy | Proteomics server for various prediction and sequence analysis programs (eg, prediction of phosphorylation and glycosylation sites, TMDs, import sequences, domains) | http://www.expasy.org/tools/ |
| TMHMM 2.0 | Prediction of transmembrane domains | http://www.cbs.dtu.dk/services/TMHMM/ |
| Simple Modular Architechture Research Tool (SMART) | Prediction and annotation of protein domains, internal links to PFAM | http://smart.embl-heidelberg.de/smart/batch.pl35 |
| ProtFun 2.2 | Ab initio prediction of protein function, enzymatic properties and possible gene ontology, ab initio predictions of protein function from sequence. | http://www.cbs.dtu.dk/services/ProtFun/36 |
| STRING | STRING is a database of known and predicted protein interactions. The interactions include direct (physical) and indirect (functional) associations | http://string.embl.de/37 |
| Unified Human Interactome (UniHI) | Interaction network prediction | http://theoderich.fb3.mdc-berlin.de:8080/unihi/home38 |
| GoMiner | Evaluation of gene ontology classification for large scale data | http://discover.nci.nih.gov/gominer/39 |
| Ontologizer | Evaluation of gene ontology classification for large scale data, comparison between datasets | http://compbio.charite.de/index.php/ontologizer2.html |
| Bioinformatic Harvester III | Harvester crawls and cross-links the following bioinformatic sites: 4DXp, AceView, BLAST, Biocompare, CDART, CDD, ensEMBL, Entrez, FishMap, Galaxy, UCSC GenomeBrowser, gfp-cDNA, Google-Scholar, gopubmed, Harvester42, H-Inv, HomoloGene, iHOP, IPI, MapView, MGI, MINT, Mitocheck, OMIM, PolyMeta, PSORT II, RGD, SMART, SOSUI SOURCE, STRING, TAIR, Unigene, UniprotKB, Wikipedia, WikiProtein | http://harvester.fzk.de/harvester/40 |
| Reactome (including SkyPainter) | Curated knowledgebase of biologic pathways | http://www.reactome.org41 |
| KEGG PATHWAY database | Collection of manually drawn pathway maps representing our knowledge on the molecular interaction and reaction networks (also accessible via STRING) | http://www.genome.jp/kegg/pathway.html42 |
| iHOP | Hyperlinked information about proteins | http://www.ihop-net.org/UniPub/iHOP/43 |
In a recent study, Amisten et al44 probed for GPCR-derived mRNA in platelet transcripts, identifying 28 GPCRs and quantifying 12 receptors on the transcript level. In our current data set, 13 GPCRs are either GO annotated (AVPR1A, CCR4, CD97, CXCR4, LPAR5/GPR92, P2RY1, P2RY12, PTAFR, PTGDR, PTGIR, XPR1) or otherwise known GPCRs (PAR1, PAR4). Only 6 of them (AVPR1A, GPR92, P2RY1, P2RY12, PAR1, and PTGIR) were determined by both studies. Clearly, proteomics is able to complement mRNA profiling by proving the presence (eg, of GPR92) for the first time on the protein level for platelet samples.
Apart from signal transduction, membrane proteins also have additional functions, such as cellular adhesion. By GO terms, 86 proteins implicated in cell adhesion have been found and also 47 proteins present or involved in cell junctions. Among them are several proteins known to locate to either cell-cell junctions17 or anchoring junctions16 such as ITGA2B, ITGAV, TLN1, VASP, or VCL. Furthermore, ILK (integrin-linked kinase) was identified, which interacts with PINCH-1 and PINCH-2 at extracellular matrix adhesion sites.45 Cellular adhesion has a profound role in the stability of platelet plugs/aggregates and the current list of proteins potentially includes those important for cellular adhesion, although not currently annotated by GO. As examples, junctional adhesion molecules A and C (JAM1, JAM3), endothelial cell-selective adhesion molecule, intercellular adhesion molecule 2, and platelet endothelial cell adhesion molecule can be seen.
Having no obvious adhesion properties, tetraspanins feature another interesting subgroup of proteins that are supposed to have membrane ordering function in various cell types.46,47 A recent study by Protty et al48 identified 19 tetraspanins in megakaryocytes by mRNA profiling. Upon raising of antibodies, it was possible to detect tetraspanin 9 (TSN9) as novel platelet tetraspanin being colocalized with GpVI in membrane microdomains. In contrast, the current study features 13 tetraspanin identifications (CD9/37/63/81/82/151, TSN2/4/9/14/15/18/33) of which TSN2 and TSN15 were exclusively identified, although the remaining 11 proteins were covered as well by Protty et al.48 However, in addition it was possible to prove existence of CD37, CD81, and Tspan4 on the protein level by mass spectrometric sequencing without the need for initially raising antibodies.
Platelet function is dependent on defined distributions of ions within the cell. Most notably are Ca2+ ions, which are quickly released into the cytoplasm upon activation leading, for example, to cytoskeletal rearrangements. We identified 91 proteins with transmembrane ion transporter activity, of which 23 account for ion channels. furthermore, 24 are ion transporters with ATPase activity such as plasma membrane calcium transporting ATPase1 (PMCA1), which removes Ca2+ ions from the cytosol. Among the 23 ion channels, some CLCN isoforms (chloride channel protein, CLCN3, CLCN4, CLCN6, CLCN7) as well as CLIC1 (chloride intracellular channel protein 1), ABCC4 (multidrug resistance-associated protein 4), and TTYH3 (protein tweety homolog 3) are potentially responsible for chloride transport, with TTYH3 being a potentially Ca2+-activated large conductance calcium channel with proposed implications in cellular signaling.49 Besides 3 known potassium channels (KCNA2, KCNA3, and KCNK6), a total of 8 Ca2+ ion channel proteins were retrieved by GO annotations. Inositol 1,4,5-trisphosphate receptor types 1 and 2 are responsible for Ca2+ release from the ER upon stimulation by inositol 1,4,5-trisphosphate. Meanwhile, platelet ORAI-1 (calcium release-activated calcium channel protein 1) is a store-operated calcium channel mediating Ca2+ influx to the cytosol following depletion of intracellular Ca2+ stores and channel activation by the Ca2+ sensor, STIM1 (stromal interaction molecule 1).50,51 Besides the well-known ATP-activated P2X purinoceptor 1 (P2RX1), 3 transient receptor potential (TRP) ion channels were detected (TRPC6, TRPM4, and TRPV2). TRPM4 is a potential calcium-activated nonselective cation channel and appears to provide a mechanism allowing for cell depolarization in a Ca2+-dependent manner.52 TRPC6 is supposed to be involved in receptor-activated, diacyl-glycerol–mediated cation entry53 in platelets, whereas little is known about TRPV2. Interestingly, we could not detect TRPC1 on human platelets by proteomic means, although its presence was shown previously for mice.54 A further, although yet uncharacterized potential member of calcium channel proteins was identified with TPCN1 (2 pore calcium channel protein 1). TPCN1 has 12 predicted TMDs (by TMHMM 2.0) and 2 cation channel domains as predicted by SMART (aa's 143 to 319 and aa's 478 to 686).55,56 It might function as a voltage-gated Ca2+ channel across the plasma membrane (by similarity; Uniprot annotation). Supplementing the information regarding calcium channels, we identified 101 proteins that are reported to bind calcium by GO. These include integrins as well as, for example, the ER calcium-sensor STIM1.50,51
Relative quantification of platelet proteins
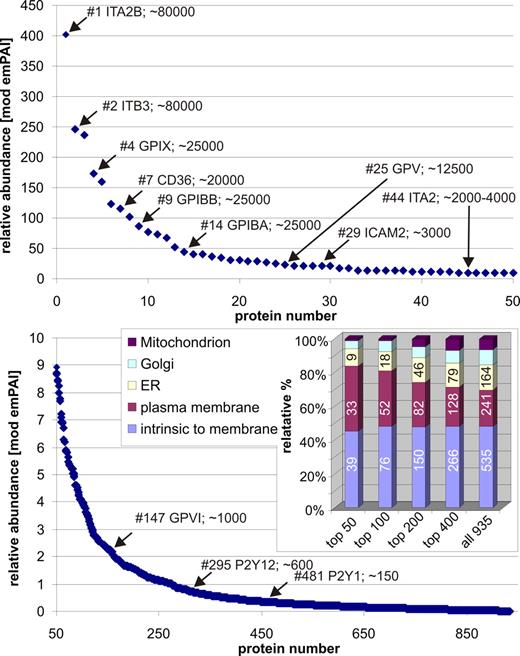
Inherently, mass spectrometry is a qualitative technique used for identification of compounds by their fragmentation patterns. However, under certain conditions, quantitative information may be deduced from mass spectrometric data sets. During recent years, a number of techniques for relative and absolute quantitation of proteins by mass spectrometry have been introduced. Although some approaches rely on chemical labeling, other label-free techniques have gained interest over recent years. One of them, called exponentially modified Protein Abundance Index (emPAI), relies on spectral counting.24 Mass spectrometric detection of peptides is partially dependent on the concentration of the respective compound. Assuming, a high abundant peptide is detected more often during a mass spectrometric analysis, this fact enables an approximate relative quantification of identified proteins based on the number of peptide hits identified for each protein. We used a modified version of the emPAI index to generate an approximate estimation of platelet protein abundance within the current study set in relation to published data. For 935 discrete protein hits derived from the shotgun data, the ratio of observable to observed peptide hits was calculated and results are depicted in Figure 4 (a complete list of emPAI ratios is presented in the supplemental table). Indeed, the highest emPAI indices were calculated for integrin IIb/IIIa, which is present at approximately 80 000 copies/platelet.57 In addition, the components of the gpIb-IX-V complex (∼ 25 000 copies/platelet gpIb/gpIX and ∼ 12 500 copies/platelet gpV58,59 ) are present among entries with high emPAI indices (Figure 4 top panel) as well. At slightly lower emPAI values, ICAM-2 (3000 copies/platelet60 ) and integrin alpha-2 (2000-4000 copies/platelet61 ) fit into the scheme as well. In terms of low abundant proteins, the data correlate as well with the copy numbers of gpVI, P2Y12, and P2Y1 with 1000, approximately 600, and approximately 150 copies/platelet,57 respectively. Therefore, the current data set can be potentially used to differentiate between high, medium, and low abundant protein species. Although the absolute copy number of an individual protein might not always fits with its position in emPAI ranking (eg, CD36 with 20 000 copies/platelet61,62 ; prior gpIBB and gpIBA with ∼ 25 000 copies each59 ), a general accordance was found. However, a major problem remains: the different reported copy numbers for proteins in the literature.
Relative quantification of proteins by exponentially modified protein abundance indices (emPAI). Relative quantification for enriched plasma membrane samples was done by spectral counting within the shotgun-based approaches for 935 proteins (supplemental table). emPAI values were compared with literature estimations of protein abundance and showed overall good correlation. The inset shows gene ontology–based subcellular classifications for groups from high to low emPAI-ranking proteins. Reaching low emPAI values, low abundant plasma membrane components are mixed with contaminants by other compartments.
Relative quantification of proteins by exponentially modified protein abundance indices (emPAI). Relative quantification for enriched plasma membrane samples was done by spectral counting within the shotgun-based approaches for 935 proteins (supplemental table). emPAI values were compared with literature estimations of protein abundance and showed overall good correlation. The inset shows gene ontology–based subcellular classifications for groups from high to low emPAI-ranking proteins. Reaching low emPAI values, low abundant plasma membrane components are mixed with contaminants by other compartments.
Furthermore, spectral counting may enable quality control of membrane preparations. As can be seen in the inset of Figure 4 (bottom panel), we combined the quantitative information of emPAI values with GO annotation for this purpose. Within the top 100 abundant proteins of the 2-phase membrane separation, 76 proteins were intrinsic to a membrane and 52 proteins could be confirmed to be plasma membrane constituents. The percentage of proteins derived from ER, Golgi apparatus, and mitochondria was minor in comparison (18, 10, 2, respectively). In addition, GO allows for multiple localizations, so a protein might be present in Golgi apparatus and plasma membrane as well. Tracing the shares of ER, Golgi, and mitochondrial contaminations from top 50 to all935 discrete protein hits, the share of contaminations constantly rises in comparison with the plasma membrane share. This argues for the majority of impurities to be rather low abundant. In turn, the relative percentage of proteins intrinsic to membranes remains fairly constant throughout the data sets. Furthermore, the purification of membranes can also be monitored looking for actin abundance, which is present in high concentrations of up to 2 000 000 copies/platelet. The calculated emPAI value of 36.9 for actin within the current preparation indicates in turn an abundance of 20 000 to 25 000 copies. This is in good agreement with the fact that actin may be tightly associated with the plasma membrane63 in addition to its abundant soluble or cytoskeletal localization. Other proteins such as VASP (∼ 80 000 copies/platelet64 ; emPAI 0.86) or coagulation factor V (emPAI 0.01)/multimerin 1 (emPAI 1.1) might indicate low abundance of soluble cytoplasmic (VASP) and vesicular proteins. Furthermore, the absence of several marker proteins for white blood cells such as CD4, CD8, CD15, CD16, CD19, as well as the low emPAI value (1.17) for the high abundant CD45 leukocyte marker and very low levels of leukosialin (which is present at 150 000 copies on human lymphocytes,65 but low abundant on platelets66 ; emPAI 0.46) might argue for a low-level contamination by white blood cells—which might possibly be due to platelet-leukocyte aggregates. Moreover, serum albumin was detected with a minor emPAI value (1.05) arguing for low abundance of serum protein in the membrane preparations—in contrast to approximately 60% serum albumin content in the circulating plasma.
Evaluation of protein interactions
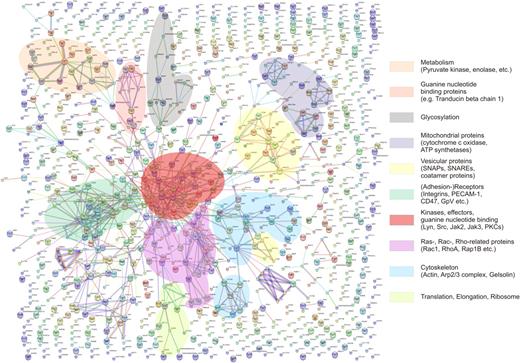
Knowledge of the platelet proteome has been constantly increasing over the years. In parallel to the progress of proteome works, bioinformatics tools have been developed by other groups for the large-scale analysis of proteomic data sets. In this context, we used the STRING algorithm67,68 (http://string.embl.de) to assess the scope of already known protein-protein interactions for the complete list of 1282 identified proteins. For data evaluation, the complete protein list was submitted as batch data, all evidence levels were accepted, and only hits with high confidence (0.9) were used with a network depth of 1 and an edge scaling factor of 80%. The results are presented in Figure 5 (a scalable image is given in supplemental table). From the network analysis, a clustering of components was clearly observable. The central groups of the data set were composed of adhesion-mediating receptors and kinases or effectors. This result confirms the presence of a large number of known plasma membrane components (several integrins, gpV, gpIX, PECAM-1, etc). Since these receptors initiate a range of downstream cascades, the occurrence of downstream effectors was to be expected as well. Therefore, protein kinases (PKA, PKC, Jak1-3, Src, Syk, etc) were identified alongside Ras-, Rac-, and Rho-related proteins, which can be easily deduced from the supplied lists in the supplemental table. Since membrane proteins are connected to the cytoskeleton, for example, via actin-binding proteins, a range of actin- and cytoskeleton-associated proteins was found as a clustered group as well. They include components of the Arp2/3 complex as well as vinculin or VASP, which is a known interactor of actin.69 Furthermore, coatomer and vesicle-associated proteins such as SNAPs and SNAREs were identified as interacting units within the data set. As can be deduced from Figure 5, also proteins relating to metabolism, glycosylation processes, as well as proteins of mitochondrial origin could be found to be tightly associated by network analysis.
Protein interaction network of 1282 identified proteins. Interaction analysis was performed by STRING (scalable picture can be found in the supplemental table). Several clusters of direct interaction partners are marked and correspond, for example, to adhesion receptors, effector proteins, or vesicular proteins (sorting based on function-derived, eg, by GO terms and Swiss-Prot entry information). However, the majority of proteins are without direct interaction partners, thereby arguing for a more extensive near-membrane proteome.
Protein interaction network of 1282 identified proteins. Interaction analysis was performed by STRING (scalable picture can be found in the supplemental table). Several clusters of direct interaction partners are marked and correspond, for example, to adhesion receptors, effector proteins, or vesicular proteins (sorting based on function-derived, eg, by GO terms and Swiss-Prot entry information). However, the majority of proteins are without direct interaction partners, thereby arguing for a more extensive near-membrane proteome.
In general, a first survey of the membrane proteome by STRING already revealed a total of 858 protein-protein interactions present in databases. However, evident from Figure 5, a large number of proteins were not associated with any other membrane or soluble protein. By introducing 250 nodes (see supplemental table for image), representing additional interaction partners, the number of interactions could be raised to 2669. These 250 nodes reflect known interaction partners, which, however, were not part of the current data set.
Discussion
Platelet function in thrombosis and hemostasis is enabled by an intricate interplay of various proteins, small messengers, and ions on various levels. Current research on platelet function is driven mostly by studies on individual proteins, many of which are also studied in knockout models or using antibodies. The results of the current proteome study on membrane proteins can supplement these studies on various levels, especially regarding important plasma membrane interface proteins.
The assembled data set of 1282 proteins, of which 788 proteins were not covered by several proteomic approaches before (Figure 2A), offers a premier repository for rational design of upcoming studies. Comparison of currently studied components with the presented accessions can return formerly unknown members of functional protein classes in platelets, as shown briefly in the “Functional characterization” section for the tetraspanins. To deduce suitable targets for functional research and extend the scientific value of the presented platelet protein collection, accession lists from the supplemental material can conveniently be evaluated by a multitude of bioinformatic algorithms, of which some are proposed in Table 2. Examples thereof have already been given by TMD prediction and STRING interaction networks. Moreover, STRING data may serve as origin for potential downstream analysis of signaling pathways by rational search for known binding partners, which were so far uncovered in platelets but known, for example, in other cell types. In addition, shotgun-derived semiquantitative information offers a first estimation of protein abundance and thereby influences the choice of future research targets. The immunoglobulin receptor G6B might serve as an example. Although the receptor was initially described on platelets by proteomic approaches in 2005,1,29 its function remained elusive until the recent works of Senis et al.4 The current data set ranks G6B with an emPAI score of 122 among the high abundant receptors on the platelet surface with possibly profound impact on platelet function. However, no direct conclusions from apparent abundance to functional importance should be drawn without further validation. The mass spectrometric detection of a proteins/peptides within the given workflow is dependent, for example, on ionization properties of peptides, individual protein behavior during membrane purification, or even varying abundance due to biologic differences in platelet populations.
Furthermore, the presented repository on identified peptides is a valid source for sequences to be used in antibody generation. Obviously, proteins may be modified by a multitude of posttranslational modifications (eg, glycosylation). However, the given peptides in the current study have been unambiguously identified in their indicated form (see supplemental table) and can therefore avoid choice of unsuitably modified sequences for antibody generation. Lastly, COFRADIC results were shown to contain valuable information on N-terminal protein sequences, supplementing existing database information.
In total, we estimate the current study to exert a profound influence on the platelet research field and stimulate research on new target proteins for thrombosis and hemostasis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The financial support by the Ministerium für Innovation, Wissenschaft, Forschung und Technologie des Landes Nordrhein-Westfalen and by the Bundesministerium für Bildung und Forschung is gratefully acknowledged. Parts of the work were furthermore supported by the SonderforschungsbereichGrant 688 (A.S.).
Authorship
Contribution: U.L., S.W., K.L., and R.P.Z. performed experiments and collected/analyzed data; U.L., A.S., D.A.W., and U.W. designed the research; and U.L., S.W., K.L., and R.P.Z. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Albert Sickmann, Institute for Analytical Sciences, Bunsen-Kirchhoff-Str 11, 44139 Dortmund, Germany; e-mail: albert.sickmann@isas.de.