Abstract
Human T-cell leukemia virus type 1 (HTLV-1) and type 2 (HTLV-2) retroviruses infect T lymphocytes. The minus strand of the HTLV-1 genome encodes HBZ, a protein that could play a role in the development of leukemia in infected patients. Herein, we demonstrate that the complementary strand of the HTLV-2 genome also encodes a protein that we named APH-2 for “antisense protein of HTLV-2.” APH-2 mRNA is spliced, polyadenylated, and initiates in the 3′-long terminal repeat at different positions. This transcript was detected in all HTLV-2–infected cell lines and short-term culture of lymphocytes obtained from HTLV-2 African patients tested and in 4 of 15 HTLV-2–infected blood donors. The APH-2 protein is 183 amino acids long, is localized in the cell nucleus, and is detected in vivo. Despite the lack of a consensus basic leucine zipper domain, APH-2 interacts with cyclic adenosine monophosphate-response element binding protein (CREB) and represses Tax2-mediated transcription in Tax2-expressing cells and in cells transfected with an HTLV-2 molecular clone. Altogether, our results demonstrate the existence of an antisense strand–encoded protein in HTLV-2, which could represent an important player in the development of disorders, such as lymphocytosis, which is frequently observed in HTLV-2 patients.
Introduction
Human T-cell leukemia virus type 1 (HTLV-1) and type 2 (HTLV-2) retroviruses belong to the primate T lymphotropic virus family.1 They share several features, including a similar genomic organization, similar cellular receptor complexes, the presence of a pX region that encodes a viral Tax transactivator protein, and several accessory proteins.2 Whereas HTLV-1 is the causal agent of both adult T-cell leukemia/lymphoma (ATLL), a fatal disease that occurs in 1% to 5% of infected persons, and HTLV-associated myelopathy/tropical spastic paraparesis (HAM/TSP), a progressive myelopathy, HTLV-2 infection has been associated with HAM/TSP but not ATLL.3 Several distinctions have been reported between Tax1 and Tax2.4-11 These differences might account in part for the distinctive outcomes observed in HTLV-1 versus HTLV-2–infected patients. However, the low expression of tax1 mRNA in ATLL patients represents an observation that is difficult to reconcile with the suggested primary role played by the viral protein in the maintenance of a transformed stage.
HBZ (HTLV-1 bZIP factor) is a protein that was shown to be encoded by the complementary strand of the HTLV-1 genome. The discovery of HBZ provided a promising explanation for the mechanism of ATLL development.12 HBZ transcripts initiate in the 3′ long terminal repeat (LTR) and are either unspliced or spliced, thereby producing 2 isoforms, HBZ (or usHBZ) and HBZ-SP1 (also named HBZ-SI or sHBZ).13-15 The HBZ-SP1 transcript is present in primary cells from healthy carriers TSP/HAM and correlates with disease severity16 but also in ATLL patients as well as in infected cell lines, and its level is 4-fold higher than that of unspliced HBZ mRNA in cell lines.17 Furthermore, HBZ-SP1 expression is correlated with the HTLV-1 proviral load.17 Even if HBZ mRNA levels are lower (10-fold) than tax/rex mRNA levels, they are much more abundant (10- to 1000-fold) than p12, p27/p12, p30, and p13 accessory gene transcripts.18 The HBZ protein is 209 amino acids long,12 whereas HBZ-SP1 encodes for a 206-amino-acid isoform.13,14 Both proteins contain a basic leucine zipper (bZIP) domain at their COOH-terminus and only differ in their N-terminal sequence. HBZ possesses 3 different nuclear localization signals, 2 of which are necessary for its localization to the nucleus.19 In this cellular organelle, HBZ forms nuclear bodies that are different from promyelocytic leukemia bodies or splicing factor compartments,19 whereas HBZ-SP1 is found in the nucleolus.14,20
In vitro, both isoforms down-regulate Tax1-mediated viral transcription, although a recent report has suggested that HBZ-SP1 is a stronger repressor of Tax1-mediated transcription.21 Tax1 inhibition is linked to the ability of both isoforms to interact either with the CREB and CREB-2 transcription factors and/or to sequester the CBP/p300 transcription coactivators.22,23 HBZ is also capable of forming dimers with c-Jun, JunD, or JunB, which consequently modify their transcriptional activity.20,24-26 Experiments using either HBZ-SP1 transgenic mice or rabbits infected with wild-type or HBZ-deficient viruses led several investigators to suggest that HBZ was responsible for (1) the proliferation of CD4+ lymphocytes cells and (2) the viral persistence in vivo, but not for cellular transformation.15,27,28
Recent data obtained from a cohort of HTLV-2–infected carriers demonstrated that the total lymphocyte count is increased by 7% to 10% in these persons compared with noninfected blood donors.29 We therefore hypothesized that HTLV-2 complementary strand might encode a protein that would behave like HBZ and could participate in the cell proliferation in vivo and act negatively on viral expression.
Methods
Cell lines
HTLV-2–transformed C19, MO, and BJAB/Gu,30 HTLV-1 transformed C91/PL, uninfected Jurkat, and CEM were grown in RPMI medium; 293T and HeLa cell lines were grown in Dulbecco modified Eagle medium in the presence of 10% fetal bovine serum and antibiotics and were maintained at 37°C in 5% CO2. Primary T cells were cultured in RPMI containing 10% fetal bovine serum in the presence of antibiotics and interleukin-7.
HTLV-2 carrier samples
Peripheral blood mononuclear cells (PBMCs; 5 million/sample) were obtained from 15 different HTLV-2–infected carriers who are enrolled in the HOST (patient information, supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article).31 The proviral load has previously been determined and ranged from 101 to 105 per 106 PBMCs. Research use of biologic specimens from the subjects in the HTLV Outcomes Study (HOST) was approved by the Committee on Human Research of the University of California San Francisco, and all subjects gave written informed consent in accordance with the Declaration of Helsinki. Short-term cultures of PBMCs were also obtained from 2 HTLV-2–infected Pygmies (59011, Pyl116).
Plasmids
pH6neo32,33 was kindly provided by Dr P. Green (Departments of Veterinary Biosciences, Molecular Virology, Immunology, and Medical Genetics, The Ohio State University, Columbus, OH). pHTLV-2ΔEcoRI construct was generated by cloning the 4269 pb EcoRI fragment (nucleotide 4683-8952) in pBlueScript KS. Antisense protein of HTLV-2 (APH-2) cDNA was cloned into the pEGFP-C2 (Clontech) and pcDNA3.1-Myc-His (Invitrogen) vectors using the EcoR1 and EcoR1/HindIII restriction sites, respectively.
Transfections
293T cells were transfected using the Polyfect reagent (QIAGEN), whereas HeLa cells were transfected with the Effectene reagent (QIAGEN).5,34 Jurkat cells were transfected by electroporation with the Gene Pulser System (Bio-Rad). Primary T cells were transfected by electroporation with the Nucleofector reagent (Amaxa).
RT-PCR, 5′ and 3′ RACE analyses
Total RNA was extracted using the TRIzol reagent (Invitrogen) and treated twice with the DNAseI RNAse-free DNA set (QIAGEN).35 PolyA+ RNA was purified using the Oligotex mRNA Mini Kit (QIAGEN). Reverse-transcribed polymerase chain reaction (RT-PCR) analyses were conducted using the RT primer 24-2 (5′-ctatacactccaactgctgatgcc-3′; Figure 1). Total RNA (5 μg for transfected cells, 10 μg for infected cells) was mixed with 0.5 μM of the specific RT primer. The RNA:RT primer mix was heated at 70°C for 5 minutes, cooled on ice for 5 minutes, and incubated for 2 hours at 42°C in the presence of 1× avian myeloblastosis virus reaction buffer (1 mM dNTPs, 10 U SUPERase·In RNAse inhibitor [Ambion] and 15 U avian myeloblastosis virus reverse transcriptase [USB]). cDNAs were amplified in the presence of 1.25 U Taq DNA polymerase, 1× ThermoPol buffer, 20 μM dNTPs, 1.5 μM of each primer using a T gradient thermocycler (Biometra). Primers added to the PCRs were the reverse primer 21-2 (5′-CTCTCCCGGCGCTTTTGATCC-3′) and forward primers 19-1 (5′-CGTATGTTGGCCCCGGGAC-3′) or 22-1 (5′-CTCGACCTGAGAGGAGACTTAC-3′), 21-1 (5′-GGGGTAATCGTCAGAGCCTTG-3′) (Figure 1A). PCR conditions were as follows: denaturation at 94°C for 5 minutes followed by 35 cycles (94°C 1 minute, 60°C 1 minute, 72°C 1 minute) and a final extension at 72°C for 5 minutes. For amplification of the total encoding APH-2 cDNA, PCR amplification was conducted using the 22-1 forward and 22-6 reverse (5′-TGTATCCTCCGCCAAATCCAGG-3′) primers (positioned at nt 293-314 and 2381-2402 of the antisense strand, respectively). The amplified product was cloned and used either for generating the APH-2 expression vectors or as a probe for Northern blot analyses.
Both the 5′ and 3′ extremities of the antisense transcript were amplified from isolated polyA+ RNA using the SMART RACE cDNA Amplification kit (Clontech) and the FirstChoice RLM-RACE kit (Ambion), respectively. For the 5′ end, the cDNA was synthesized with a modified oligo dT and ligated to a supplied anchor at its 5′ end. Two subsequent PCR rounds were conducted with the 5′ RACE outer and inner primers along with the 24-2 and 21-2 transcript-specific reverse primers (Figure 1A). For the 3′end, cDNA synthesis was performed in the presence of the supplied 3′RACE adapter complementary to the polyA tail. PCR was conducted in the presence of the 3′RACE outer and inner primers and the forward 21-3 primer (5′-GAGAAGTTGGTGGAGGCTGAG-3′) derived from the sequence downstream of the HTLV-2 antisense APH-2 open reading frame (ORF; nt 3224-3244 of the antisense strand). PCR products were cloned and sequenced.
RT-PCR analyses in infected cell lines
A total of 5 μg total RNA (TRIzol) was used for the RT-PCR experiment with the SuperScript Reverse Transcriptase II and RNAse Out ribonuclease inhibitor (Invitrogen) kits. During the RT step, the APH-2–specific 24-2 primer was used. The cDNA was amplified with a seminested PCR using the 19-1/21-2 primer set for the first PCR followed by the 22-1/21-2 primer set for the second PCR. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers were described elsewhere.36
Northern blot analyses
A total of 4 μg polyA+ RNA was denaturated in formaldehyde/formamide for 10 minutes at 70°C, chilled on ice, and separated on a 1.5% agarose gel containing 20 mM MOPS (3-(N-morpholino)propanesulfonic acid) and 17% formaldehyde. The gel was transferred onto a Hybond-XL nylon membrane (GE Healthcare) and hybridized with a 32P-labeled APH-2 encoding cDNA. The membrane was revealed (PhosphorImager BioRad Molecular Image FX).
Generation of APH-2 antibodies
Two APH-2 peptides were synthesized (Eurogentec), coupled to KLH, and injected to New Zealand white rabbits. After several booster injections, the serum (SPY557) was tested for the presence of anti–APH-2 antibodies. As a negative control, a preimmune serum was also tested.
Western blot analyses
Twenty-four hours after transfection, cells were lysed as described.5 Samples were loaded into 12% Bis-Tris gel (Invitrogen), subjected to electrophoresis, transferred onto a PVDF membrane, and incubated with the appropriate primary antibody: anti-APH-2 (SPY557), anti-GFP (Clontech), anti-Myc 1:1000 (Upstate), anti-p24 (ZeptoMetrix Corporation), anti-GAPDH (Santa Cruz Biotechnology), or anti–β-tubulin (Santa Cruz Biotechnology). Membranes were developed using the SuperSignal West Pico Chemiluminescent substrate Kit (Pierce Chemical). Membranes were reprobed with the anti–β-tubulin or anti-GAPDH antibodies. Band intensities were calculated using the Alpha EaseFC Flurochem 8900 software (Alpha Innotech Corporation).
Coimmunoprecipitation
Cells were lysed in radioimmunoprecipitation assay buffer. Cross-linking was performed between 2 μg of a rabbit polyclonal anti-CREB antibody (SC-186; Santa Cruz Biotechnology) and the Bio-Adembeads Protein G magnetic beads (Ademtech). Beads were incubated overnight with 400 μg protein, and the immunoprecipitate was eluted in a glycine 50 mM-Tween 0.65%, pH 2.7 solution. The eluate was subjected to Western blot analysis using anti–APH-2 antibodies (see “Western blot analyses”).
GST pull-down
APH-2 cDNA was subcloned into the pGEX-4T vector. Glutathione S-transferase (GST) pull-down experiments were performed with glutathione-agarose beads equilibrated in 0.5× Superdex buffer. GST fusion proteins were incubated with glutathione-agarose beads and washed. Purified p300 was then added to the beads and incubated overnight.22 Bound proteins were eluted and resolved on a 10% sodium dodecyl sulfate–polyacrylamide gel. p300 was detected with a rabbit anti-p300 antibody (N-15; Santa Cruz Biotechnology).
Immunofluorescence analyses
HeLa, Jurkat, and primary T cells were analyzed as described.5 Cells were incubated with a rabbit polyclonal anti-nucleolin antibody (Affinity Bioreagents) or with a mouse monoclonal anti-SC35 antibody (Sigma-Aldrich). Samples were then incubated with a fluorescein isothiocyanate–conjugated mouse anti-Myc antibody (Sigma-Aldrich), a secondary Dylight 549–conjugated anti–rabbit IgG (Pierce Chemical) antibody, or a secondary Fluorolink Cy3-labeled anti–mouse IgG (Amersham) antibody. Images were acquired with an Upright microscope Zeiss Axioplan 2 imaging, with the Zeiss Axiovision 4.4 software.
Luciferase assays
The 293T cells were transfected with pHTLV-2-LTR-luc or pHTLV-1-LTR-luc plasmids together with pSG5M-Tax2 or pSG5M-Tax15 and APH-2-Myc or HBZ-SP1-Myc13 as described.4,5 Reporter activities were assayed using the Dual-Luciferase reporter assay system (Promega) as described.5 For the experiments performed with the HTLV-2 pH6neo proviral DNA clone, luciferase activities were measured using the MLX microplate luminometer (Dynex Technologies). The β-galactosidase activity was measured using the Galacto-Light kit (Applied Biosystems). For detection of the p19 antigen, cell supernatants were used for p19 enzyme-linked immunosorbent assay (HTLV-I/II p19 Antigen ELISA; ZeptoMetrix Corporation).
Results
Existence of a putative ORF in the HTLV-2− strand
We first performed an analysis on HTLV-2 full-length proviral DNA sequences available in GenBank (ie, Gab [Y13051], K96 [AF326584], AF412314, SP-WV [AF139382], G2 [AF074965], G12 [L11456], RP329 [AF326583], and Efe2 [Y14365]). This revealed the presence of a putative ORF in all of the analyzed strains except for MO, which had been used for generating the HTLV-2 infectious molecular clone pH6neo.33 We therefore sequenced this proviral DNA and discovered that the GenBank MO sequence (accession no. M10060) contained a series of errors. After correction, it appeared that MO also possessed a similar antisense ORF. As HTLV-1 HBZ, this ORF is located between the Tax and Env ORFs in the HTLV-2 genome (Figure 1A). The putative protein encoded from this ORF was therefore named APH-2 for “antisense protein of HTLV-2.”
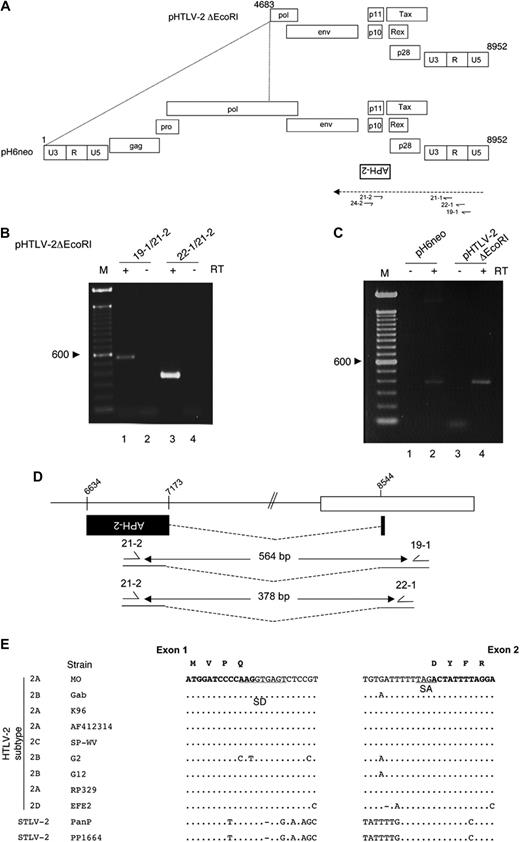
HTLV-2 antisense transcripts are detected and are spliced. (A) The HTLV-2 molecular clone pH6neo provirus is depicted with all known genes and is adapted from Feuer and Green.2 The positioning of the putative APH-2 ORF from the antisense strand is indicated below the proviral DNA. A 5′ deleted version of this vector termed pHTLV-2 ΔEcoRI is also presented and encompasses nt 4683 to 8952. The primers used for the RT-PCR experiments are also represented with their positioning. The dashed line represents the hypothetical antisense transcript. (B-C) The 293T cells were transfected either with pHTLV-2 ΔEcoRI and/or (C) with pH6neo. RNA was extracted and analyzed by RT-PCR for the presence of antisense transcripts using the primer combinations 19-1/21-2 and 22-1/21-2. (C) The 22-1/21-2 primer set that allows the amplification of the spliced transcript was used. When indicated, RT was omitted during the RT step. Control indicates no cDNA; M, 100-bp marker. (D) The position of exon 1 and exon 2 of the APH-2 transcript (■) is presented. The size of the RT-PCR products is also indicated. (E) The splice donor and acceptor sequences of the APH-2 transcript are conserved. DNA sequences containing the APH-2 splice donor and splice acceptor sites were compared between various HTLV-2 and STLV-2 isolates retrieved from GenBank. The modified sequence of the MO proviral DNA was used as a reference. Bold letters represent exonic sequence; underlined, SD and SA sequences. The HTLV-2 subtype is indicated on the left of the panel. Predicted amino acid sequence of the spliced APH-2 transcript at the splice junction are also presented above the exonic nucleotide sequence.
HTLV-2 antisense transcripts are detected and are spliced. (A) The HTLV-2 molecular clone pH6neo provirus is depicted with all known genes and is adapted from Feuer and Green.2 The positioning of the putative APH-2 ORF from the antisense strand is indicated below the proviral DNA. A 5′ deleted version of this vector termed pHTLV-2 ΔEcoRI is also presented and encompasses nt 4683 to 8952. The primers used for the RT-PCR experiments are also represented with their positioning. The dashed line represents the hypothetical antisense transcript. (B-C) The 293T cells were transfected either with pHTLV-2 ΔEcoRI and/or (C) with pH6neo. RNA was extracted and analyzed by RT-PCR for the presence of antisense transcripts using the primer combinations 19-1/21-2 and 22-1/21-2. (C) The 22-1/21-2 primer set that allows the amplification of the spliced transcript was used. When indicated, RT was omitted during the RT step. Control indicates no cDNA; M, 100-bp marker. (D) The position of exon 1 and exon 2 of the APH-2 transcript (■) is presented. The size of the RT-PCR products is also indicated. (E) The splice donor and acceptor sequences of the APH-2 transcript are conserved. DNA sequences containing the APH-2 splice donor and splice acceptor sites were compared between various HTLV-2 and STLV-2 isolates retrieved from GenBank. The modified sequence of the MO proviral DNA was used as a reference. Bold letters represent exonic sequence; underlined, SD and SA sequences. The HTLV-2 subtype is indicated on the left of the panel. Predicted amino acid sequence of the spliced APH-2 transcript at the splice junction are also presented above the exonic nucleotide sequence.
Detection of a spliced APH-2 transcript in cells transfected with HTLV-2 proviral DNA
We then reasoned that an HTLV-2 antisense transcript encoding APH-2 would initiate from the 3′-LTR and be spliced as it is the case for HTLV-1 HBZ-SP1. Antisense primers spanning different positions in the 3′-LTR were designed and RT-PCR experiments conducted with RNA extracts obtained from 293T cells transfected either with the pH6neo vector or with pHTLV-2 ΔEcoRI, which is deleted from nt 1 to 4683 (Figure 1A). This latter construct can therefore not initiate sense transcription and is assumed to lead to increased antisense transcription. RT-PCR was performed using different sets of primers. These RT-PCR analyses clearly demonstrated that HTLV-2 antisense transcription occurs in HTLV-2–transfected 293T cells (Figure 1B lanes 1 and 3). The size of the specific amplicons (Figure 1B,D) was shorter than expected from an unspliced transcript (564 bp instead of 1935 bp and 378 bp instead of 1749 bp, respectively). These results are indicative of a potentially spliced APH-2 RNA. RT-PCR analyses were performed with primers 22-1/21-2 on RNA extracts obtained from cells transfected with the pH6neo clone and confirmed the presence of a spliced APH-2 mRNA (Figure 1C lane 2). Although this technique is not quantitative, as previously reported for HTLV-1 HBZ, the RT-PCR signal was stronger in RNA extracts obtained from pHTLV-2 ΔEcoRI-transfected cells (Figure 1C lane 2 vs lane 4). No signals were detected when RT was omitted from the reaction mix (Figure 1B lanes 2 and 4; Figure 1C lanes 1 and 3).
PCR products obtained in Figure 1B (lanes 1 and 3) were sequenced and were confirmed to correspond to antisense transcripts that initiated in the 3′-LTR and were spliced to the 5′ end of the deduced APH-2 ORF (Figure 1D). Splice donor (SD) and splice acceptor (SA) sites were located at position 8544 and 7173 of the antisense strand of pH6neo proviral DNA, respectively. The full-length ORF region of APH-2 would therefore span 1923 bp of the HTLV-2–negative strand with an intronic region of 1371 bp. Analyzing the full-length genome sequences of various HTLV-2 and STLV-2 (the simian homolog of HTLV-2) strains present in the GenBank database revealed that the SD and SA consensus sequences (underlined in Figure 1E) were always conserved, with the exception of the G2 strain, which showed a slight variation in the SD consensus sequence. The ATG initiation codon positioned in exon 1 (Figure 1E) was also conserved. The resulting spliced transcript should lead to a protein with 4 amino acids derived from exon 1 added to the remaining APH-2 amino acid sequence (Figure 1E).
APH-2 mRNA is present in HTLV-2–infected cells and in HTLV-2 carriers in vivo
We next tested whether the APH-2 spliced transcript was present in 3 HTLV-2–infected cell lines (MO, C19, and Gu), 2 short-term cultures of lymphocytes obtained from HTLV-2 African carriers (590101 and Pyl116), or in noncultured PBMCs obtained from 15 different HTLV-2 blood donors. A seminested RT-PCR experiment was performed using primers located in exons 1 and 2, leading to an expected signal of 378 bp. APH-2 mRNA was indeed detected in all HTLV-2–infected cell lines (Figure 2A lanes 2 and 3; Figure 2B lanes 3 and 7), in the 2 samples from HTLV-2 African carriers (Figure 2B lanes 5 and 9), and in 4 of 15 HTLV-2 carriers (Figure 2C lanes 9, 13, 16, and 17). No signal was amplified from HTLV-1–infected cell lines (Figure 2A lane 6) or from uninfected Jurkat and CEM cells (Figure 2A lanes 4 and 5). The RT-PCR products were cloned and sequenced and were demonstrated to be genuine APH-2 cDNAs originating from spliced transcripts (supplemental Figure 2). Of note, in lane 10, the PCR products obtained in the absence of RT corresponded to residual DNA. Because they belong to HTLV-2 subtype B, and as expected, APH-2 sequences from HTLV-2 Pygmies or from the Gu cell lines were different from those of the MO and C19 cell lines or from US former blood donors, which belong to HTLV-2 subtype A. APH-2 sequences from blood donors were almost all identical among themselves and with the Mo sequence. This is expected because the HTLV-2 sequence variation is extremely low in this region of the provirus.
The spliced APH-2 mRNA is detected in HTLV-2–infected cell lines and in PBMCs from HTLV-2–infected patients. (A) Total RNA was extracted from C19 and MO (HTLV-2–infected), Jurkat, and CEM (HTLV noninfected) and C91/PL (HTLV-1–infected) cell lines. RT was performed using the 24-2 primer and was followed by a seminested PCR using the 19-1/21-2 and 22-1/21-2 primers. Lanes 1 and 2 represent C19; lane 3, MO; lane 4, Jurkat; lane 5, CEM; lane 6, C91/PL; lane 7, H2O. (B) Same experimental conditions as in panel A. Lanes 1 and 2 represent RNA extract from Jurkat cells; lanes 3 and 4, from C19 cells; lanes 5 and 6, from 590101; lanes 7 and 8, from GU; lanes 9 and 10, from Pyl116; lane 11, H2O. (C) Same experimental conditions as in panel A. Lanes 2 and 3 represent 400 ng and 40 ng, respectively, of RNA extracted from MO cells used for RT-PCR analysis; lanes 4 and 12, H2O; lanes 10 and 11, RNA extract from C19 cells; lane 22, RNA extract from Jurkat cells; lanes 5 to 9 and 13 to 21, RNAs extracted from PBMC of 15 HTLV-2–infected persons. (Bottom of both panels) The same samples were analyzed for the presence of GAPDH mRNA by RT-PCR. In panel A (lane 1), panel B (lanes 2, 4, 6, 8, and 10), and panel C (lanes 1 and 10), RT was omitted during the RT step. M indicates 100-bp marker. The 2 parts of the image shown in panel C (top panel) are from the same gel.
The spliced APH-2 mRNA is detected in HTLV-2–infected cell lines and in PBMCs from HTLV-2–infected patients. (A) Total RNA was extracted from C19 and MO (HTLV-2–infected), Jurkat, and CEM (HTLV noninfected) and C91/PL (HTLV-1–infected) cell lines. RT was performed using the 24-2 primer and was followed by a seminested PCR using the 19-1/21-2 and 22-1/21-2 primers. Lanes 1 and 2 represent C19; lane 3, MO; lane 4, Jurkat; lane 5, CEM; lane 6, C91/PL; lane 7, H2O. (B) Same experimental conditions as in panel A. Lanes 1 and 2 represent RNA extract from Jurkat cells; lanes 3 and 4, from C19 cells; lanes 5 and 6, from 590101; lanes 7 and 8, from GU; lanes 9 and 10, from Pyl116; lane 11, H2O. (C) Same experimental conditions as in panel A. Lanes 2 and 3 represent 400 ng and 40 ng, respectively, of RNA extracted from MO cells used for RT-PCR analysis; lanes 4 and 12, H2O; lanes 10 and 11, RNA extract from C19 cells; lane 22, RNA extract from Jurkat cells; lanes 5 to 9 and 13 to 21, RNAs extracted from PBMC of 15 HTLV-2–infected persons. (Bottom of both panels) The same samples were analyzed for the presence of GAPDH mRNA by RT-PCR. In panel A (lane 1), panel B (lanes 2, 4, 6, 8, and 10), and panel C (lanes 1 and 10), RT was omitted during the RT step. M indicates 100-bp marker. The 2 parts of the image shown in panel C (top panel) are from the same gel.
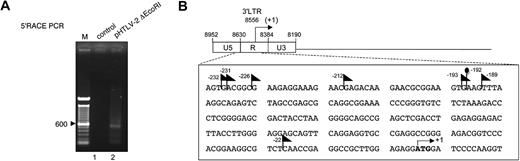
HTLV-2 antisense transcription initiates at different positions in the 3′-LTR
We determined the transcription initiation sites of the APH-2 transcript after transfection of 293T with pHTLV-2 ΔEcoRI or in HTLV-2–infected MO cells. RNA was analyzed using the SMART RACE kit and a reverse primer positioned in exon 2, close to the splice junction. Cloning and sequencing of the amplified products generated by 5′ RACE (Figure 3A lane 2) led to the identification of several major initiation sites positioned in the 3′-LTR (exclusively in the R and U5 regions) and spanning a total of 210 nt in transfected cells (Figure 3B black triangle). The position of these initiation sites is in agreement with the size of the signal observed in Figure 3A. In Mo, only one site of initiation was reproducibly detected (Figure 3B black oval).
HTLV-2 antisense transcription initiates in the 3′-LTR. (A) The 5′RACE PCR analysis was conducted using RNA samples from 293T cells transfected with pHTLV-2 ΔEcoRI (lane 2). The cDNA was synthesized with a modified oligo dT and ligated to a supplied anchor at their 5′ end. Two subsequent PCR rounds were conducted with the 5′RACE outer and inner primers along with the transcript-specific 24-2 and 21-2 reverse primers As a negative control (lane 1), final PCR amplification was performed with water (ie, no cDNA). The resulting amplified products were run on an agarose gel. M indicates 100-bp marker. (B) The position of the identified transcription initiation sites of APH-2 mRNA (◣ represents 293T;  , Mo) is depicted in the 3′-LTR. Nucleotide positioning is relative to the APH-2 ATG initiation codon numbered as +1. The numbering in the proviral DNA corresponds to the sense strand.
, Mo) is depicted in the 3′-LTR. Nucleotide positioning is relative to the APH-2 ATG initiation codon numbered as +1. The numbering in the proviral DNA corresponds to the sense strand.
HTLV-2 antisense transcription initiates in the 3′-LTR. (A) The 5′RACE PCR analysis was conducted using RNA samples from 293T cells transfected with pHTLV-2 ΔEcoRI (lane 2). The cDNA was synthesized with a modified oligo dT and ligated to a supplied anchor at their 5′ end. Two subsequent PCR rounds were conducted with the 5′RACE outer and inner primers along with the transcript-specific 24-2 and 21-2 reverse primers As a negative control (lane 1), final PCR amplification was performed with water (ie, no cDNA). The resulting amplified products were run on an agarose gel. M indicates 100-bp marker. (B) The position of the identified transcription initiation sites of APH-2 mRNA (◣ represents 293T;  , Mo) is depicted in the 3′-LTR. Nucleotide positioning is relative to the APH-2 ATG initiation codon numbered as +1. The numbering in the proviral DNA corresponds to the sense strand.
, Mo) is depicted in the 3′-LTR. Nucleotide positioning is relative to the APH-2 ATG initiation codon numbered as +1. The numbering in the proviral DNA corresponds to the sense strand.
Position of the polyA site
The APH-2 mRNA 3′ end was next mapped using 3′RLM-RACE. Using a primer positioned downstream of the APH-2 stop codon, we amplified a 590-bp fragment (Figure 4A). Sequencing of this fragment demonstrated that the polyA tail was positioned at a distance of 1449 nt from the APH-2 stop codon. As in the HTLV-1 HBZ transcript,13 the cleavage site needed for the addition of the polyA tail was located in a TA dinucleotide positioned 17 nucleotides downstream of the polyA signal and a few nucleotides from a GT-rich segment, which represents another typical consensus sequence for polyA addition (Figure 4B). These consensus sequences were highly conserved among other various HTLV-2 strains (Figure 4B).
Identification of the polyA addition site of the APH-2 transcript. (A) RNA samples obtained from 293T cells transfected with the pHTLV-2 ΔEcoRI plasmid (lane 2) were analyzed by 3′ RLM-RACE using the 21-3 primer. Lane 1 represents PCR amplification in the absence of any cDNA. M indicates 100-bp marker. (B) Position of the polyA addition site next to a consensus polyA signal and a GT-rich consensus sequence. The sequences of the APH-2 mRNA and of the 3′ polyA tail are shown. ■ represents the coding portion of the APH-2 spliced transcript. HTLV-2 sequences retrieved from GenBank were also compared using MO as a reference sequence. Comparisons were focused on the AATAAA polyA signal (position 5185 on the sense strand), the cleavage site deduced from our 3′RACE results and the GT-rich sequence. (C) PolyA+ RNA was extracted from pHTLV-2 ΔEcoRI-transfected 293T cells (lane 1) or from nontransfected cells (lane 2) and was run on a 1.5% agarose gel. After migration and transfer, the membrane was hybridized with a probe corresponding to the coding segment of the APH-2 cDNA and signals were revealed with a PhosphorImager. An RNA marker was migrated in parallel and corresponding molecular weights are indicated on the left side of the panel.
Identification of the polyA addition site of the APH-2 transcript. (A) RNA samples obtained from 293T cells transfected with the pHTLV-2 ΔEcoRI plasmid (lane 2) were analyzed by 3′ RLM-RACE using the 21-3 primer. Lane 1 represents PCR amplification in the absence of any cDNA. M indicates 100-bp marker. (B) Position of the polyA addition site next to a consensus polyA signal and a GT-rich consensus sequence. The sequences of the APH-2 mRNA and of the 3′ polyA tail are shown. ■ represents the coding portion of the APH-2 spliced transcript. HTLV-2 sequences retrieved from GenBank were also compared using MO as a reference sequence. Comparisons were focused on the AATAAA polyA signal (position 5185 on the sense strand), the cleavage site deduced from our 3′RACE results and the GT-rich sequence. (C) PolyA+ RNA was extracted from pHTLV-2 ΔEcoRI-transfected 293T cells (lane 1) or from nontransfected cells (lane 2) and was run on a 1.5% agarose gel. After migration and transfer, the membrane was hybridized with a probe corresponding to the coding segment of the APH-2 cDNA and signals were revealed with a PhosphorImager. An RNA marker was migrated in parallel and corresponding molecular weights are indicated on the left side of the panel.
We then performed a Northern blot analysis using the coding region of the APH-2 cDNA as a probe. Interestingly, a signal lower than 2.5 kb was specifically detected in pHTLV-2 ΔEcoRI-transfected cells (Figure 4C lane 1 vs lane 2). The size of the transcript corresponds to the 2.3 kb predicted APH-2 mRNA when taking into consideration both 5′ and 3′ RACE data. Altogether, these results demonstrate that APH-2 mRNA initiates at different positions in the 3′-LTR, is spliced, and is polyadenylated.
APH-2 and HBZ-SP1 do not have a similar localization in transfected cells
Analysis of the resulting APH-2 protein sequence (Figure 5A) demonstrated that, unlike HBZ, it does not contain a classic bZIP domain12 because 7 instead of 6 amino acids were present between the sixth and the seventh leucine of this nonconventional bZip domain. However, APH-2 harbors an LXXLL motif at its COOH-terminus (amino acids 179-183) as well as a central IXXLL (amino acids 64-68) motif. Similar domains were recently shown to be involved in the binding of HTLV-1 HBZ to the KIX domain of CBP/p300 and to interfere with the ability of HTLV-1 Tax to bind to these transcription coactivators.22
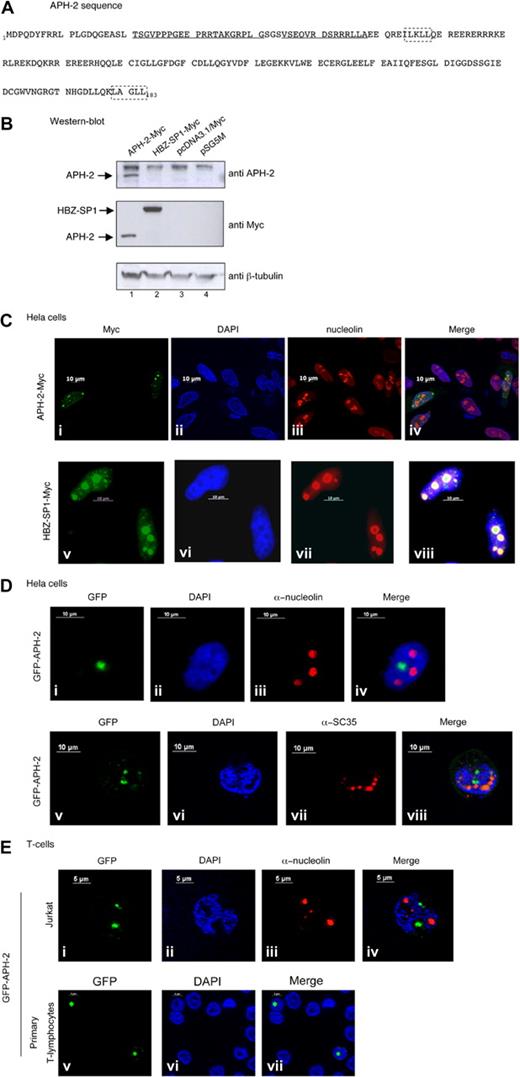
APH-2 localizes to the nucleus. (A) Amino acid sequence of the APH-2 protein. The amino acid sequence of APH-2 was deduced from the MO strain. Underlining represents the peptides used for the immunization of the rabbits; dashed boxes, 2 potential LXXLL motifs. (B) Cell lysates (50 μg) from 293T cells transfected with the APH-2-Myc or HBZ-SP1-Myc expression vector or 2 control empty vectors (parental pcDNA3.1/Myc and pSG5M vectors) were subjected to electrophoresis on a 12% Bis-Tris gel and analyzed by Western blot with anti-Myc, anti–APH-2, or anti–β-tubulin antibodies. pcDNA3.1/Myc parental vector was used as a negative control. (C-E) Intracellular localization of APH-2 in (C-D) HeLa and (E) Jurkat or primary T lymphocytes transfected with HBZ or APH-2 expression vectors. (Ci-iv) APH-2-Myc. (Cv-viii) HBZ-SP1-Myc. (D-E) GFP-APH-2. Twenty-four hours after transfection, cells were fixed and stained as described in “Immunofluorescence analyses.” Cells were finally mounted in 4,6-diamidino-2-phenylindole-containing mounting medium. Images are representative of the entire population of transfected cells.
APH-2 localizes to the nucleus. (A) Amino acid sequence of the APH-2 protein. The amino acid sequence of APH-2 was deduced from the MO strain. Underlining represents the peptides used for the immunization of the rabbits; dashed boxes, 2 potential LXXLL motifs. (B) Cell lysates (50 μg) from 293T cells transfected with the APH-2-Myc or HBZ-SP1-Myc expression vector or 2 control empty vectors (parental pcDNA3.1/Myc and pSG5M vectors) were subjected to electrophoresis on a 12% Bis-Tris gel and analyzed by Western blot with anti-Myc, anti–APH-2, or anti–β-tubulin antibodies. pcDNA3.1/Myc parental vector was used as a negative control. (C-E) Intracellular localization of APH-2 in (C-D) HeLa and (E) Jurkat or primary T lymphocytes transfected with HBZ or APH-2 expression vectors. (Ci-iv) APH-2-Myc. (Cv-viii) HBZ-SP1-Myc. (D-E) GFP-APH-2. Twenty-four hours after transfection, cells were fixed and stained as described in “Immunofluorescence analyses.” Cells were finally mounted in 4,6-diamidino-2-phenylindole-containing mounting medium. Images are representative of the entire population of transfected cells.
APH-2 cDNA was cloned into the pcDNA3.1-Myc expression vector, allowing the production of a Myc-tagged APH-2 protein (referred to APH-2-Myc throughout the text) and the expression vector was transfected into 293T cells. An HBZ-SP1-Myc expression vector was also used as a control. Both Myc and APH-2 antibodies led to the detection of an APH-2 protein having an approximate 25-kDa size (Figure 5B lane 1). HBZ-SP1-Myc was detected with the Myc antibody, but not with the anti-APH-2 antibody (lane 2). The polyclonal APH-2 antibody is therefore specific to APH-2.
APH-2-Myc (Figure 5Ci-iv) and HBZ-SP1-Myc expression vectors (Figure 5Cv-viii) were transfected in HeLa cells. A strong nuclear signal was observed in cells transfected with the APH-2-Myc construct. The APH-2-Myc protein was not nucleolar as shown by comparison with the antinucleolin staining (Figure 5Ciii). Of note, in some cells, part of the APH-2 signal was detected outside the nucleus. As previously published, HBZ-SP1 was nucleolar (Figure 5Cv-viii). Localization results were then obtained with the GFP-APH-2 construct (Figure 5Di-iv). Splicing factor compartments (SFCs) correspond to perichromatin fibrils and interchromatin granule cluster and are involved in the storage of splicing factors. To determine whether APH-2 colocalizes with SFCs, cells were transfected with the GFP–APH-2 construct and stained with anti-SC35 antibodies (Figure 5Dv-viii). This allowed us to show that APH-2, like HBZ, is not associated with SFCs. This was confirmed by a vertical view using the Zeiss Axiovision 4.4 software (supplemental Figure 3).
Because HTLV viruses preferentially infect T cells in vivo, APH-2 localization was also determined in Jurkat T cells (Figure 5Ei-iv) as well as in primary T lymphocytes (Figure 5Ev-vii). As in HeLa cells, GFP-APH-2 localized in the nucleus and did not colocalize with the nucleolus. Altogether, these results demonstrate that APH-2 protein is a nuclear but not nucleolar protein.
APH-2 interacts with CREB but not with p300
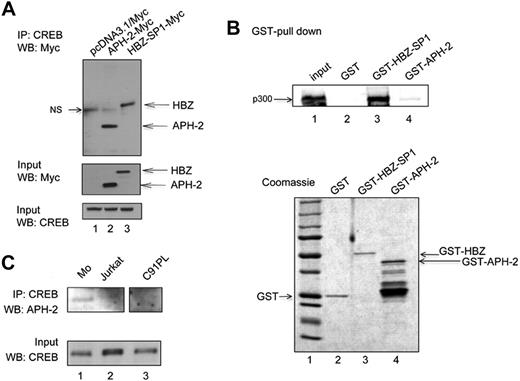
HBZ-SP1 down-regulates HTLV-1 transcription by interacting with CREB-2 and CREB through its bZIP domain.23 HBZ-SP1 also interacts with CBP/p300 coactivators through LXXLL-like motifs and prevents their recruitment to the viral promoter.22 Therefore, at least 2 domains of HBZ-SP1 play a role in the down-regulation of viral expression. We wanted to determine whether, despite the lack of a canonical bZIP domain, APH-2 interacts with CREB. A series of APH-2/CREB coimmunoprecipitation experiments was performed (Figure 6A). This allowed us to determine that, despite the lack of a canonical bZIP domain, APH-2 binds CREB (Figure 6A lane 2). As a control, HBZ-SP1 did also bind CREB (Figure 6A lane 3), whereas no detection of CREB was observed in immunoprecipitates of extracts from pCDNA3.1-Myc–transfected cells (Figure 6A lane 1). As controls, APH-2, HBZ, and CREB input levels were assessed (Figure 6A bottom panel).
APH-2 binds to CREB but not to p300 and is expressed in vivo. (A) Cell lysates (400 μg) obtained from 293T cells transfected with pcDNA3.1/Myc backbone vector (lane 1), APH-2-Myc (lane 2), or HBZ-SP1-Myc (lane 3) were subjected to immunoprecipitation with a rabbit polyclonal anti-CREB antibody followed by Western blot with a mouse monoclonal anti-Myc antibody. Input proteins (50 μg) are shown in the middle (anti-Myc) and bottom (anti-CREB) panels. (B) Purified p300 (0.2 pmol) was incubated with 5 pmol GST, GST-HBZ, or GST-APH-2. Bound proteins were detected by Western blot analysis with an anti-p300 antibody. Input protein (100%) is shown in lane 1. (Bottom panel) Purified GST, GST-HBZ, and GST-APH-2 (5 pmol of each) are shown by Coomassie blue staining after sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Lane 1 represents molecular weight marker. (C) Cell lysates (400 μg) obtained from MO (lane 1), Jurkat (lane 2), and C91PL (lane 3) were subjected to immunoprecipitation with a rabbit polyclonal anti-CREB antibody followed by Western blot with a rabbit polyclonal anti-APH-2 antibody. Input proteins (50 μg) are shown in the bottom (anti-CREB) panels. (Top panel) These lanes correspond to different part of the same gel.
APH-2 binds to CREB but not to p300 and is expressed in vivo. (A) Cell lysates (400 μg) obtained from 293T cells transfected with pcDNA3.1/Myc backbone vector (lane 1), APH-2-Myc (lane 2), or HBZ-SP1-Myc (lane 3) were subjected to immunoprecipitation with a rabbit polyclonal anti-CREB antibody followed by Western blot with a mouse monoclonal anti-Myc antibody. Input proteins (50 μg) are shown in the middle (anti-Myc) and bottom (anti-CREB) panels. (B) Purified p300 (0.2 pmol) was incubated with 5 pmol GST, GST-HBZ, or GST-APH-2. Bound proteins were detected by Western blot analysis with an anti-p300 antibody. Input protein (100%) is shown in lane 1. (Bottom panel) Purified GST, GST-HBZ, and GST-APH-2 (5 pmol of each) are shown by Coomassie blue staining after sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Lane 1 represents molecular weight marker. (C) Cell lysates (400 μg) obtained from MO (lane 1), Jurkat (lane 2), and C91PL (lane 3) were subjected to immunoprecipitation with a rabbit polyclonal anti-CREB antibody followed by Western blot with a rabbit polyclonal anti-APH-2 antibody. Input proteins (50 μg) are shown in the bottom (anti-CREB) panels. (Top panel) These lanes correspond to different part of the same gel.
Because APH-2 contains an LXXLL and an LXXLL-like (ILKLL) motif, we also wondered whether it could also bind to p300. We performed GST pull-down assays using recombinant purified proteins (Figure 6B). As shown in Figure 6B, p300 was found to bind specifically to HBZ, but only weakly to APH-2 (Figure 6B lanes 3 and 4). As a control, the GST protein alone did not interact with p300 (lane 2). Altogether, these results revealed that APH-2 binds to CREB but not to p300.
APH-2 is expressed in vivo
To detect the endogenous APH-2 protein, we then performed a CREB/APH-2 coimmunoprecipitation using HTLV-2 Mo protein extracts. As expected, a signal corresponding to endogenous APH-2 was detected in Mo extracts (Figure 6C lane 1), but not in Jurkat or C91PL cell extracts (Figure 6C lanes 2 and 3). As controls, CREB input levels were assessed (Figure 6C bottom panel).
APH-2 down-regulates Tax2-mediated HTLV-2 LTR activation
Given the fact that APH-2 interacts with CREB, we hypothesized that, similarly to HBZ, APH-2 could also act as a viral repressor. The 293T cells were transfected with an HTLV-2 LTR reporter construct, an HTLV-2 Tax expression vector, and increasing amounts of the APH-2 expression vector (Figure 7A left panel). We could reproducibly observe that APH-2 exerted a negative effect on Tax2-mediated transcription and that this effect was dose-dependent, whereas APH-2 had no effect in the absence of Tax2. As a control, a similar experiment was conducted with HBZ-SP1 (Figure 7A right panel). The levels of expression of Tax1, Tax2, HBZ-SP1, and APH-2 were determined (Figure 7B).
APH-2 represses Tax2-dependent activation of transcription from the HTLV-2 LTR. (A) The 293T cells were transiently transfected with 125 ng pHTLV-2-LTR-luc (left) or pHTLV-1 LTR-luc (right), 500 ng pSG5M-Tax2 (left), or pSG5M-Tax1 (right) together with increasing amounts (125, 250, 500, and 1000 ng in both cases) of APH-2-Myc (left) or of HBZ-SP1-Myc (right). The graphs represent an average of 3 (left) or 2 (right) independent experiments. (B) A total of 50 μg protein extracts from lysates obtained from transfected 293T cells were subjected to electrophoresis and probed for Myc, Tax2, Tax1, and β-tubulin. (C) The 293T cells were transiently transfected with 250 ng pH6neo, 62.5 ng pHTLV-2-LTR-luc, 50 ng pRcActin-LacZ together with increasing amounts (0, 62.5, 125, 250, and 500 ng) of the APH-2-Myc vector. Cells were lysed 48 hours after transfection, and luciferase activities were measured using the MLX microplate luminometer (Dynex Technologies). The β-galactosidase activity was measured using the Galacto-Light kit (Applied Biosystems) according to the manufacturer's suggestions. Luciferase activities are presented as normalized relative light units (RLU/β-gal) and correspond to the calculated mean ±SD of 3 transfected samples normalized by the measured β-galactosidase activity. (D) The 293T cells were transiently transfected with 2.5 μg pH6neo, increasing amounts (0, 500, 1000, 2000, and 4000 ng) of the APH-2-Myc vector. Cell lysates were subjected to electrophoresis and probed for p24 or GAPDH. (E) For detection of p19 antigen in supernatants, 293T cells were transiently transfected with 250 ng pH6neo with increasing amounts (0, 62.5, 125, 250, and 500 ng) of APH-2-Myc vector. Cell supernatants were used for p19 enzyme-linked immunosorbent assay according to the manufacturer's instructions (HTLV-I/II p19 Antigen ELISA; ZeptoMetrix Corporation). Experiments were done in triplicate, and values are representative of the calculated mean ± SD.
APH-2 represses Tax2-dependent activation of transcription from the HTLV-2 LTR. (A) The 293T cells were transiently transfected with 125 ng pHTLV-2-LTR-luc (left) or pHTLV-1 LTR-luc (right), 500 ng pSG5M-Tax2 (left), or pSG5M-Tax1 (right) together with increasing amounts (125, 250, 500, and 1000 ng in both cases) of APH-2-Myc (left) or of HBZ-SP1-Myc (right). The graphs represent an average of 3 (left) or 2 (right) independent experiments. (B) A total of 50 μg protein extracts from lysates obtained from transfected 293T cells were subjected to electrophoresis and probed for Myc, Tax2, Tax1, and β-tubulin. (C) The 293T cells were transiently transfected with 250 ng pH6neo, 62.5 ng pHTLV-2-LTR-luc, 50 ng pRcActin-LacZ together with increasing amounts (0, 62.5, 125, 250, and 500 ng) of the APH-2-Myc vector. Cells were lysed 48 hours after transfection, and luciferase activities were measured using the MLX microplate luminometer (Dynex Technologies). The β-galactosidase activity was measured using the Galacto-Light kit (Applied Biosystems) according to the manufacturer's suggestions. Luciferase activities are presented as normalized relative light units (RLU/β-gal) and correspond to the calculated mean ±SD of 3 transfected samples normalized by the measured β-galactosidase activity. (D) The 293T cells were transiently transfected with 2.5 μg pH6neo, increasing amounts (0, 500, 1000, 2000, and 4000 ng) of the APH-2-Myc vector. Cell lysates were subjected to electrophoresis and probed for p24 or GAPDH. (E) For detection of p19 antigen in supernatants, 293T cells were transiently transfected with 250 ng pH6neo with increasing amounts (0, 62.5, 125, 250, and 500 ng) of APH-2-Myc vector. Cell supernatants were used for p19 enzyme-linked immunosorbent assay according to the manufacturer's instructions (HTLV-I/II p19 Antigen ELISA; ZeptoMetrix Corporation). Experiments were done in triplicate, and values are representative of the calculated mean ± SD.
We next wanted to evaluate the effect of APH-2 on viral transcription from a complete HTLV-2 proviral DNA. We hypothesize that, in the proviral context, APH-2 should be able to down-regulate Tax2-mediated LTR transactivation and indeed should result in a more potent inhibition of the luciferase expression driven by the HTLV-2 LTR than in experiments in which Tax2 expression relied on a heterologous (CMV) promoter. Results showed that APH-2 expression reduced the LTR-driven luciferase activity in a dose-dependent fashion (Figure 7C) and that the observed inhibition (up to 85.3%) was more important than the one obtained when Tax2 was overexpressed. The viral production was also monitored by quantifying p24 levels (Figure 7D) and the amount of p19 protein in the supernatants of transfected cells (Figure 7E). These data demonstrate that APH-2 potently impaired viral production and that this effect probably resulted from its ability to negatively regulate viral sense transcription.
Discussion
The existence of antisense transcription has been controversial.37,38 The demonstration that retroviral antisense transcription exists has recently been obtained for 2 human retroviruses (ie, HTLV-1 and HIV-1).12,39 In HTLV-1, the HBZ protein, which is encoded from such an antisense transcript, is the subject of numerous studies. HBZ binds to several transcription factors or transcription coactivators and alters their functions.20,22-24,26,40,41 Interestingly, HBZ transgenic mice in which the transgene is expressed under the CD4 promoter/enhancer do not develop severe pathologies but demonstrate an increased percentage of CD4+ T lymphocytes in the splenocytes and display a higher proliferation of this cellular population.15
Given the fact that HTLV-2–infected persons have a persistent lymphocytosis,29 we wondered whether HTLV-2 could also encode a protein that would be functionally related to HBZ. The existence of an “HBZ-like” protein in HTLV-2 has been ruled out by several authors,12,18 whereas others suggested that such a protein might exist in HTLV-2.42 These discrepancies are the result of 2 facts: (1) the HTLV-2 prototype strain (MO),33 which has been used for searching an antisense ORF, contains several sequence errors that prematurely interrupt the putative ORF; and (2) there is an antisense ORF in other HTLV-2 strains, but its sequence does not contain a classic bZIP sequence.
In this report, we first showed that a spliced mRNA corresponding to APH-2 is expressed in vitro in transfected cells and in all tested HTLV-2–infected cell lines (regardless of the viral subtype). This transcript is structured similarly to the HBZ transcript; that is, it is spliced, initiates in the 3′-LTR at multiple sites, and is polyadenylated. The length of the intron and of the 5′ and 3′ untranslated regions is also similar to that of the HBZ transcript. These similarities imply that antisense transcription in the HTLV (and STLV) family has been conserved among the different viruses. Indeed, 2 reports suggested that HTLV-3 and HTLV-4 proviruses also contain an antisense ORF.35,43 The positioning of both APH-2 and HBZ ORFs, which is similar might be favored to avoid undesirable antisense effect on Tax2 sense transcripts. Our results also suggest that the unspliced APH-2 transcript if it exists, is present in very low amount because it could not be detected during our RT-PCR analyses. This is in contrast to HTLV-1 HBZ, which produces a substantial amount of unspliced transcript encoding for a different HBZ isoform.14,17,44 Further studies are therefore required to search for the presence of the putative unspliced APH-2 transcript.
Two recent reports clearly demonstrated that HTLV-1 HBZ-SP1 mRNA is expressed in vivo in almost all HTLV-1–infected persons and that its level is correlated with the proviral load.16,17 Our current results indicate that APH-2 is expressed in vivo in a significant proportion of persons and in all cells derived from HTLV-2–infected patients, whether these are long- or short-term cultures. A study based on a much larger number of HTLV-2 persons using quantitative PCR and quantitative RT-PCR techniques is required to address this issue. This study will allow us to examine whether the HTLV-2 proviral load and the levels of APH-2 and Tax2 transcripts are correlated. Whether APH-2 is expressed at the protein level in infected patients and elicits a humoral or a cellular response is not known and will also be investigated.
In 1995,45 Green et al made a deletion in the HTLV-2 pH6neo, which mostly corresponds to the APH-2 ORF (729pH6neo ΔUT). They did not observe a difference between cells stably transfected with the wild-type versus the deleted version of the HTLV-2 proviral DNA and concluded that this region was dispensable in vitro. A similar study was performed later with HTLV-1.27 In that case, too, the authors concluded that biologic properties of HBZ mutant viruses were indistinguishable from wild-type HTLV-1 in vitro. However, mutating HBZ sequence results in a decreased proviral load as well as antibody response in vivo. Strikingly, inoculating rabbits with HTLV-2 729pH6neo ΔUT-infected cells also led to a decreased proviral load, suggesting that APH-2 plays a role in vivo too.46
Our results have also led us to demonstrate clear differences between HBZ-SP1 and APH-2 at the level of nuclear localization. Indeed, HBZ-SP1 is specifically retained in nucleolar structures, unlike APH-2. Differences at the levels of protein-protein or nucleic acid-protein interactions might contribute to these divergent nuclear localization and could be dependent both on differences in amino acid sequence and/or in posttranslational modifications. In addition, a nucleolar localization signal might be specifically present in HBZ-SP1. Future studies will be required to address these issues.
Despite its lack of a bZIP consensus sequence, APH-2 abrogates Tax-dependent increase in transcription levels initiated from the HTLV-2 5′-LTR probably through its binding to the cellular CREB protein. As stated in “Results,” a recent report clearly established that the 2 LXXLL-like motifs that are located within the N-terminal sequence of HBZ-SP1 allow the protein to bind CBP/p300 coactivators and to squelch them.22 Interestingly, although APH-2 presents such motifs in its amino acid sequence, we did not observe an interaction between APH-2 and p300 in GST pull-down assays. Whether this is the result of the position of these motifs in the APH-2 sequence, ie, central and COOH-terminal, whereas those of HBZ are located at the N-terminal end, remains to be evaluated. It would also be interesting to define whether APH-2 mRNA also plays a role, as suggested for HBZ-SP1.15,21
In conclusion, we have demonstrated that the HTLV-2 minus strand is also transcribed and that the encoded APH-2 protein shares some functional similarities with HTLV-1 HBZ even if, unlike HBZ, APH-2 does not contain a classic bZIP domain. Interestingly, the latter has been found to bind to several Jun transcription factor family members that are involved in the control of different target genes. These genes are important in regulating a series of biologic processes, including proliferation, differentiation, apoptosis, and transformation. Nevertheless, it is still possible that, despite the lack of this sequence, APH-2 is accountable for the cell proliferation, which leads to the persistent lymphocytosis observed among HTLV-2 carriers. If this is the case, the lack of hematologic disease could be solely the result of the intrinsic properties of Tax2.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Mrs Emmanuelle Perret (www.pfid.org) from the Plateforme d'Imagerie Dynamique, Imagopole, Institut Pasteur for her help with the imaging; Dr M. A. Thoulouze for the generous gift of primary human T cells; and A. Vargas for her help with the RACE-PCR experiments.
R.M. was supported by Inserm and is now supported by Ecole Normale Supérieure de Lyon. E.D. and C.J. were supported by the Ministère de la Recherche. N.L.K. was supported by the Croucher Foundation. This work was supported by Agence Nationale de Recherches sur le Sida et les hépatites virales (J.-M.M.) and the Cancer Research Society (B.B.). I.C. was supported by Agence Nationale de Recherches sur le Sida. M.H. was supported by an institutional Hydro-Quebec scholarship. S.L. holds a Canadian Institutes of Health Research PhD scholarship. E.L.M. was supported by US National Heart, Lung, and Blood Institute (grant R01-HL-62235 for the HOST cohort study and grant K24-HL-75036). B.B. holds a Research Chair of Canada (Tier 2).
National Institutes of Health
Authorship
Contribution: B.B., J.-M.M., I.L., and R.M. designed the research; M.H., E.D., I.C., C.J., I.L., N.L.K., and S.L. performed research; A.G. and E.L.M. contributed vital new reagents and edited the manuscript; and B.B., J.-M.M., and R.M. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Renaud Mahieux, Unité Inserm U758, Ecole Normale Supérieure de Lyon, 46, allée d'Italie, 69364 Lyon cedex 07, France; e-mail: renaud.mahieux@ens-lyon.fr; or Benoît Barbeau, Département des Sciences Biologiques, Université du Québec à Montréal, 2080 Saint-Urbain Montréal, QC, Canada H2X 3X8; e-mail: barbeau.benoit@uqam.ca.
References
Author notes
*M.H. and E.D. contributed equally to this study.
†B.B. and R.M. contributed equally to this study.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal