Abstract
Plerixafor (AMD3100) and granulocyte colony-stimulating factor (G-CSF) mobilize peripheral blood stem cells by different mechanisms. A rhesus macaque model was used to compare plerixafor and G-CSF–mobilized CD34+ cells. Three peripheral blood stem cell concentrates were collected from 3 macaques treated with G-CSF, plerixafor, or plerixafor plus G-CSF. CD34+ cells were isolated by immunoselection and were analyzed by global gene and microRNA (miR) expression microarrays. Unsupervised hierarchical clustering of the gene expression data separated the CD34+ cells into 3 groups based on mobilization regimen. Plerixafor-mobilized cells were enriched for B cells, T cells, and mast cell genes, and G-CSF–mobilized cells were enriched for neutrophils and mononuclear phagocyte genes. Genes up-regulated in plerixafor plus G-CSF–mobilized CD34+ cells included many that were not up-regulated by either agent alone. Two hematopoietic progenitor cell miR, miR-10 and miR-126, and a dendritic cell miR, miR-155, were up-regulated in G-CSF–mobilized CD34+ cells. A pre-B-cell acute lymphocytic leukemia miR, miR-143-3p, and a T-cell miR, miR-143-5p, were up-regulated in plerixafor plus G-CSF–mobilized cells. The composition of CD34+ cells is dependent on the mobilization protocol. Plerixafor-mobilized CD34+ cells include more B-, T-, and mast cell precursors, whereas G-CSF–mobilized cells have more neutrophil and mononuclear phagocyte precursors.
Introduction
Granulocyte colony-stimulating factor (G-CSF) has been the standard hematopoietic growth factor for increasing the levels for circulating hematopoietic stem cells (HSCs) in both healthy subjects donating cells for allogeneic transplantation and patients donating cells for autologous transplantation. G-CSF–mobilized HSCs collected by apheresis have been used for transplantation, immune therapy, and the treatment of cardiac ischemia. Another HSC-mobilizing agent, plerixafor (AMD3100), has been used in large animal models,1,2 with G-CSF to mobilize stem cells for autologous transplants3 and is currently being evaluated as a single agent to mobilize HSCs for allogeneic donor transplants.4
The mechanisms by which plerixafor and G-CSF alter HSC trafficking and mobilize HSCs are different, suggesting that HSCs with different intrinsic properties may be mobilized by these agents. Plerixafor, as a CXC chemokine receptor 4 (CXCR4) antagonist, mobilizes HSCs within 6 hours5-7 by disrupting the engagement of stem cell surface CXCR48,9 with its ligand SDF-1 (CXCL12), which is expressed on marrow osteoblasts.10,11 In contrast, G-CSF mobilizes stem cells indirectly by down-regulating the expression of CXCL12 on marrow osteoblasts and by releasing neutrophil and monocyte proteolytic enzymes, including neutrophil elastase, cathepsin G, and matrix metalloproteinase-9, which in turn degrade important HSC-trafficking and adhesion molecules c-kit, VCAM-1, and CXCR4.12 Animal models suggest that plerixafor mobilizes a CD34+ cell population different from that mobilized by G-CSF,2,13,14 due to differences in mechanisms of mobilization.
To explore differences between plerixafor and G-CSF mobilization, we used a rhesus macaque HSC-mobilization model to compare CD34+ cells mobilized by G-CSF and plerixafor. We also compared CD34+ cells mobilized with G-CSF and plerixafor in combination. The combination of G-CSF plus plerixafor was tested because it is being used clinically, and when these agents are used together, more CD34+ cells are mobilized than when either agent is used alone.3,15 The CD34+ cells were compared using global gene and microRNA expression analysis.
Methods
Study design
Young rhesus macaques (Macaca mulatta) were studied before and after the completion of 3 different mobilization protocols. The protocols involved the administration of 5 days of G-CSF, one dose of plerixafor, and 5 days of G-CSF plus one dose of plerixafor. Peripheral blood stem cell (PBSC) concentrates were collected by apheresis after the mobilization protocol was complete. Three rhesus macaques were given all 3 mobilization protocols, and PBSCs were collected from each of these 3 animals. Each course of PBSC mobilization was separated by at least 6 weeks. Two monkeys were given plerixafor first, one was given G-CSF first, and all 3 were given plerixafor plus G-CSF last (Table 1). The CD34+ cells were isolated from the PBSCs by using CD34 monoclonal antibodies and magnetic particles. The CD34+ cells were analyzed using cDNA gene and miRNA expression microarrays.
Composition of PBSC products collected from 3 rhesus macaque and the yield of CD34+ cells isolated from each product
| Mobilizing agents . | Animal ID . | Collection date . | PBSC products . | Isolated CD34+ cells . | ||
|---|---|---|---|---|---|---|
| Mononuclear cells/mL, ×109 . | Granulocytes/mL, ×109 . | Total no. . | Purity (%) . | |||
| G-CSF | ||||||
| RQ4766 | 3/28/2008 | 49 | 163 | 1.5 × 107 | 86.5 | |
| 2RC104 | 1/22/2008 | 90 | 81 | 2.5 × 107 | 98.8 | |
| RQ4963 | 1/23/2008 | 30 | 184 | 2.0 × 107 | 96.4 | |
| Mean ± SD | 56 ± 31 | 143 ± 54 | 2.0 ± 0.5 × 107 | 93.9 ± 6.5 | ||
| Plerixafor | ||||||
| RQ4766 | 11/14/2007 | 103 | 30 | 1.5 × 107 | 82.0 | |
| 2RC104 | 3/11/2008 | 99 | 28 | 2.6 × 107 | 95.1 | |
| RQ4963 | 10/31/2007 | 107 | 61 | 2.0 × 107 | 82.0 | |
| Mean ± SD | 103 ± 4 | 40 ± 18 | 2.0 ± 0.6 × 107 | 86.4 ± 7.6 | ||
| Plerixafor + G-CSF | ||||||
| RQ4766 | 7/14/2008 | 63 | 215 | 8.9 × 107 | 78.8 | |
| 2RC104 | 7/12/2008 | 56 | 215 | 12.5 × 107 | 99.5 | |
| RQ4963 | 7/16/2008 | 55 | 266 | 6.4 × 107 | 85.5 | |
| Mean ± SD | 58 ± 4 | 233 ± 29 | 9.3 ± 3.1 × 107 | 88.0 ± 10.6 | ||
| Mobilizing agents . | Animal ID . | Collection date . | PBSC products . | Isolated CD34+ cells . | ||
|---|---|---|---|---|---|---|
| Mononuclear cells/mL, ×109 . | Granulocytes/mL, ×109 . | Total no. . | Purity (%) . | |||
| G-CSF | ||||||
| RQ4766 | 3/28/2008 | 49 | 163 | 1.5 × 107 | 86.5 | |
| 2RC104 | 1/22/2008 | 90 | 81 | 2.5 × 107 | 98.8 | |
| RQ4963 | 1/23/2008 | 30 | 184 | 2.0 × 107 | 96.4 | |
| Mean ± SD | 56 ± 31 | 143 ± 54 | 2.0 ± 0.5 × 107 | 93.9 ± 6.5 | ||
| Plerixafor | ||||||
| RQ4766 | 11/14/2007 | 103 | 30 | 1.5 × 107 | 82.0 | |
| 2RC104 | 3/11/2008 | 99 | 28 | 2.6 × 107 | 95.1 | |
| RQ4963 | 10/31/2007 | 107 | 61 | 2.0 × 107 | 82.0 | |
| Mean ± SD | 103 ± 4 | 40 ± 18 | 2.0 ± 0.6 × 107 | 86.4 ± 7.6 | ||
| Plerixafor + G-CSF | ||||||
| RQ4766 | 7/14/2008 | 63 | 215 | 8.9 × 107 | 78.8 | |
| 2RC104 | 7/12/2008 | 56 | 215 | 12.5 × 107 | 99.5 | |
| RQ4963 | 7/16/2008 | 55 | 266 | 6.4 × 107 | 85.5 | |
| Mean ± SD | 58 ± 4 | 233 ± 29 | 9.3 ± 3.1 × 107 | 88.0 ± 10.6 | ||
Mobilization and collection of PBSC concentrates
CD34+ cells were mobilized, and PBSC concentrates were collected from rhesus macaques that were housed and handled in accordance with the guidelines set by the Committee on Care and Use of Laboratory Animals of the Institute of Laboratory Animal Resources, National Research Council (DHHS publication no. NIH 85-23). The protocol was approved by the Animal Care and Use Committee of the National Heart, Lung, and Blood Institute, National Institutes of Health.
G-CSF mobilization involved the administration of 10 μg/kg G-CSF (Filgrastim; Amgen) subcutaneously daily for 5 days. When plerixafor (Genzyme) was used to mobilize peripheral blood mononuclear cells (PBMCs), a single dose of 1 mg/kg plerixafor was given subcutaneously, and when both G-CSF and plerixafor were given, 10 μg/kg G-CSF was given for 5 days and 1 mg/kg plerixafor was given on the fifth day. PBSC concentrates were collected by apheresis approximately 2 hours after the last dose of G-CSF or plerixafor. The PBSCs were collected using a CS3000 plus blood cell separator (Baxter Healthcare Corp, Fenwal Division).16 The concentrates were processed to remove contaminating red blood cells and granulocytes by using Ficoll gradient separation. CD34+ cells were isolated by immunomagnetic separation as previously described.16 Purification was determined following immunoselection using phycoerythrin-conjugated murine anti-rhesus/human CD34 antibody (clone 563; BD Biosciences), which recognizes a different epitope from that used in the immunoselection procedure. The processed CD34+ cells were frozen and stored at −80°C until analysis.
Analysis of gene expression with a 17 500 gene cDNA microarray
Total RNA was extracted from the rhesus samples and amplified into antisense RNA (aRNA). In addition, total RNA from human PBMCs pooled from 6 healthy subjects was extracted and amplified into aRNA to serve as a constant reference. Test and reference RNA were labeled with Cy5 (red) and Cy3 (green), respectively, and cohybridized to a custom-made 17.5K cDNA (UniGene cluster) microarray. The microarrays were printed in the Immunogenetics Section, Department of Transfusion Medicine (DTM), Clinical Center (CC), National Institutes of Health with a configuration of 32 × 24 × 23 and contained 17 500 elements. The clones used for printing the arrays included a combination of the Research Genetics RG_HsKG_031901 8k clone set and 9000 clones selected from the RG_Hs_seq_ver_070700 40k clone set. The 17 500 spots included 12 072 uniquely named genes, 875 duplicated genes, and approximately 4000 expression sequence tags. A complete list of genes included in the Hs-CCDTM-17.5k-1px printing is available at http://nciarray.nci.nih.gov/gal_files/index.shtml.
Analysis of miRNA expression with a microarray
Two μg total RNA from each sample were labeled with Hy5 and the reference sample (total RNA from Epstein-Barr virus-transformed lymphoblastoid cell lines) with Hy3 by using a miRCURY LNA Array Power Labeling Kit (Exiqon) following the recommended protocol by the manufacturer's instruction. A commercially available miRNA probe set with 827 human, mouse, and Epstein-Barr virus miRNA was purchased from Ambion and printed onto the glass slides in the Immunogenetics Section of DTM. A complete list of miR included in our microRNA assay is available at http://nciarray.nci.nih.gov/gal_files/CCDTM-miRNA700-V3px-A.gal. After labeling, the sample and reference were cohybridized to the miRNA array at room temperature overnight. Both the processed cDNA and the miRNA array slides were scanned by using GenePix Pro 4.0 (Axon). Microarray data have been deposited with Gene Expression Omnibus under accession number GSE16936.
Data and statistical analysis
For analysis of the cDNA and miRNA array data, the raw dataset was filtered according to a standard procedure to exclude spots with minimum intensity. This filtering was arbitrarily set to an intensity parameter of 300 for cDNA expression data and 100 for miRNA array data in both fluorescence channels. Spots flagged by the analysis software and/or spots with diameters less than 25 μm for cDNA expression array and less than 10 μm for the miRNA array were excluded from the analysis.
The filtered data were normalized using Lowess Smoother and were retrieved by the BRB ArrayTool (http://linus.nci.nih.gov/BRB-ArrayTools.html) developed at the National Cancer Institute, Biometric Research Branch, Division of Cancer Treatment and Diagnosis. Hierarchical cluster analysis was conducted on the genes or miRNA by using Cluster and TreeView software.17 The relationships between the different groups of mobilized CD34+ cells were tested by applying unsupervised the Eisen hierarchical clustering method.17
For annotation of genes and functional pathways, the Database for Annotation, Visualization, and Integrated Discovery (DAVID) 2007 software (http://david.abcc.ncifcrf.gov/)18 and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway (http://www.genome.ad.jp/kegg) were used. All target prediction analysis used BRB ArrayTool mi-croRNA targets program (http://linus.nci.nih.gov/BRB-ArrayTools.html), TargetScan (http://www.targetscan.org), and miRBase Targets (http://microrna.sanger.ac.uk).
Results
PBSC mobilization and collection
The greatest quantities of CD34+ cells were isolated from plerixafor plus G-CSF–mobilized PBMC concentrates (Table 1). Similar quantities of CD34+ cells were isolated from G-CSF–mobilized and plerixafor-mobilized PBSC concentrates. Interestingly, the composition of the products differed with more mononuclear cells appearing in the plerixafor-mobilized products than either the G-CSF–mobilized or plerixafor plus G-CSF–mobilized products.
Gene expression analysis
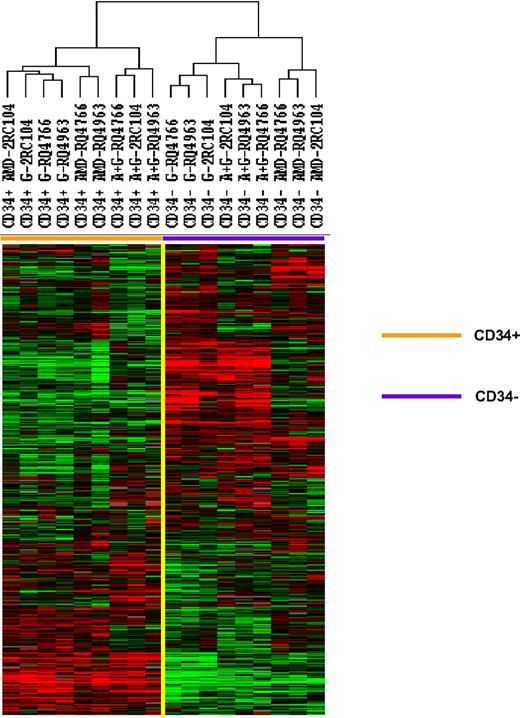
Mobilized CD34+ cells and the peripheral blood leukocytes that were not absorbed with anti-CD34 were analyzed by gene expression profiling using a microarray with 17 500 cDNA probes. The 5378 genes that were expressed in 80% of these samples and whose expression was increased 2-fold or more in at least one sample were analyzed by unsupervised hierarchical clustering of Eisen (Figure 1). The samples clustered into 2 major groups: the CD34+ group, which included all 9 CD34+ cell samples, and the peripheral blood leukocyte group, which included all 9 unabsorbed leukocyte samples. Within the CD34+ cell group, a large cluster of genes were expressed at similar levels among all CD34+ cell samples. The expression of a second cluster of genes, however, differed among CD34+ cells. In fact, the CD34+ group was made up of 3 separate clusters: one group that contained all 3 G-CSF–mobilized CD34+ cell samples and one plerixafor-mobilized CD34+ cell sample, one group that contained 2 plerixafor-mobilized CD34+ cell samples, and one that contained all 3 plerixafor plus G-CSF–mobilized CD34+ cells samples. These results suggest that CD34+ cells mobilized by G-CSF alone and plerixafor alone differ and CD34+ cells mobilized by the combination of plerixafor plus G-CSF differed from those mobilized by either G-CSF alone or by plerixafor alone.
Comparison of gene expression profiles of CD34+ cells mobilized with plerixafor, G-CSF, and plerixafor plus G-CSF with PBMCs. CD34+ cells were isolated by immunomagnetic selection from plerixafor, G-CSF, and plerixafor plus G-CSF–mobilized PBSCs collected by apheresis. The CD34+ cells and unabsorbed PBMCs were analyzed by gene expression profiling using a cDNA microarray with more than 17 000 genes. The 5378 genes that were expressed in at least 80% of the samples and were increased at least 2-fold in one sample were analyzed by hierarchical cluster of Eisen. G indicates G-CSF; A, plerixafor; A+G, plerixafor plus G-CSF.
Comparison of gene expression profiles of CD34+ cells mobilized with plerixafor, G-CSF, and plerixafor plus G-CSF with PBMCs. CD34+ cells were isolated by immunomagnetic selection from plerixafor, G-CSF, and plerixafor plus G-CSF–mobilized PBSCs collected by apheresis. The CD34+ cells and unabsorbed PBMCs were analyzed by gene expression profiling using a cDNA microarray with more than 17 000 genes. The 5378 genes that were expressed in at least 80% of the samples and were increased at least 2-fold in one sample were analyzed by hierarchical cluster of Eisen. G indicates G-CSF; A, plerixafor; A+G, plerixafor plus G-CSF.
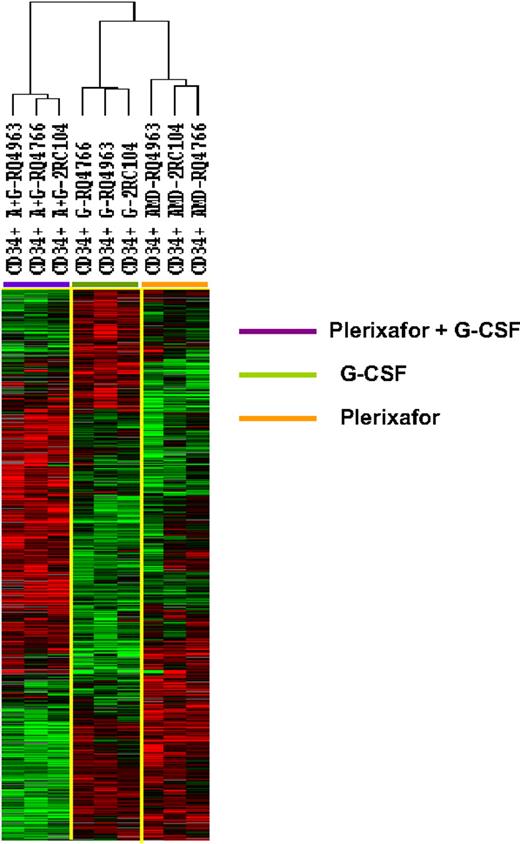
To further explore the differences among the 3 types of CD34+ cells, we identify 1097 genes that were differentially expressed among the 3 types of CD34+ cells (F tests, P < .005). Hierarchical clustering analysis of the differentially expressedgenes separated the CD34+ cells into 3 clusters: one with all 3 G-CSF–mobilized samples, one with all 3 plerixafor-mobilized samples, and one with all 3 plerixafor plus G-CSF–mobilized samples (Figure 2). Although the CD34+ cells samples were clustered into 3 separate groups, according to mobilization protocol, the transcription profiles of the plerixafor plus G-CSF–mobilized CD34+ cells were not simply a mixture of genes differently expressed by G-CSF–mobilized and plerixafor-mobilized cells. Although some genes that were up-regulated in plerixafor plus G-CSF–mobilized CD34+ cells were also up-regulated in plerixafor-mobilized or G-CSF–mobilized CD34+ cells, many genes were only up-regulated in plerixafor plus G-CSF–mobilized CD34+ cells (Figure 2). Similarly, many genes were only down-regulated in plerixafor plus G-CSF–mobilized CD34+ cells (Figure 2). These results suggest that G-CSF, plerixafor, and the combination of plerixafor plus G-CSF mobilized 3 biologically distinct types of HSCs.
Unsupervised hierarchical clustering analysis of gene differentially expressed among CD34+ cells mobilized plerixafor, G-CSF, and plerixafor plus G-CSF in 3 rhesus macaques. The CD34+ cells were analyzed by gene expression profiling using a cDNA microarray with more than 17 000 genes. Among 5378 genes that were expressed in at least 80% of the samples and were increased at least 2-fold, 1097 were found to be differentially expressed in 3 clusters of CD34+ cells (F test, P ≤ .005). The differentially expressed genes were analyzed by hierarchical cluster of Eisen. G indicates G-CSF; A, plerixafor; A+G, plerixafor plus G-CSF.
Unsupervised hierarchical clustering analysis of gene differentially expressed among CD34+ cells mobilized plerixafor, G-CSF, and plerixafor plus G-CSF in 3 rhesus macaques. The CD34+ cells were analyzed by gene expression profiling using a cDNA microarray with more than 17 000 genes. Among 5378 genes that were expressed in at least 80% of the samples and were increased at least 2-fold, 1097 were found to be differentially expressed in 3 clusters of CD34+ cells (F test, P ≤ .005). The differentially expressed genes were analyzed by hierarchical cluster of Eisen. G indicates G-CSF; A, plerixafor; A+G, plerixafor plus G-CSF.
Comparison of gene expression in G-CSF– and plerixafor-mobilized CD34+ cells
Student t tests were used to identify genes that were differentially expressed between G-CSF–mobilized CD34+ cells and plerixafor-mobilized CD34+ cells. The expression of 593 genes was greater in G-CSF–mobilized CD34+ cells than plerixafor-mobilized CD34+ cells, and the expression of 618 was greater in plerixafor-mobilized CD34+ cells (P < .05). Genes in the complement and coagulation cascades, Toll-like receptor signaling, ErbB signaling, acute myeloid leukemia, vascular endothelial growth factor signaling, and several metabolism and biosynthesis pathways were more likely to be up-regulated in G-CSF–mobilized CD34+ cells (Table 2). Up-regulated genes in plerixafor-mobilized CD34+ cells were more likely to include those in the T-cell receptor, primary immunodeficiency, TGF-beta signaling pathway, cell cycle, B-cell receptor, antigen presentation, and cell adhesion molecules signaling pathways. Genes in several pathways were up-regulated in both G-CSF– and plerixafor-mobilized CD34+ cells, including cytokine-cytokine interaction, Jak-STAT signaling pathway, MAPK signaling pathway, hematopoietic cell lineage, natural killer cell (NK)–mediated cytotoxicity, Fc epsilon RI signaling, Wnt signaling, focal adhesion, and neuroactive ligand-receptor interaction (Table 2).
KEGG pathways with the most genes up-regulated in G-CSF–mobilized CD34+ cells and plerixafor-mobilized CD34+ cells
| Pathways with genes up-regulated by G-CSF . | Pathways with genes up-regulated by plerixafor . | ||
|---|---|---|---|
| Pathway . | No. of genes in pathway . | Pathway . | No. of genes in pathway . |
| Cytokine-cytokine receptor interaction | 30 | Cytokine-cytokine receptor interaction | 29 |
| Jak-STAT signaling pathway | 25 | Calcium signaling pathway | 27 |
| MAPK signaling pathway | 24 | T-cell receptor signaling pathway | 27 |
| Focal adhesion | 22 | Hematopoietic cell lineage | 24 |
| Insulin signaling pathway | 21 | Wnt signaling pathway | 22 |
| Hematopoietic cell lineage | 20 | Primary immunodeficiency | 21 |
| Natural killer cell–mediated cytotoxicity | 19 | Cell cycle | 20 |
| Purine metabolism | 19 | MAPK signaling pathway | 20 |
| ErbB signaling pathway | 16 | Focal adhesion | 19 |
| VEGF signaling pathway | 15 | Natural killer cell mediated cytotoxicity | 19 |
| Regulation of actin cytoskeleton | 14 | TGF-beta signaling pathway | 19 |
| Toll-like receptor signaling pathway | 14 | Fc epsilon RI signaling pathway | 18 |
| Tyrosine metabolism | 14 | Jak-STAT signaling pathway | 18 |
| Acute myeloid leukemia | 13 | GnRH signaling pathway | 15 |
| Fc epsilon RI signaling pathway | 13 | Cell adhesion molecules | 14 |
| GnRH signaling pathway | 13 | Tight junction | 13 |
| Neuroactive ligand-receptor interaction | 13 | Adherens junction | 12 |
| Wnt signaling pathway | 13 | Antigen processing and presentation | 12 |
| Calcium signaling pathway | 12 | B-cell receptor signaling pathway | 12 |
| Complement and coagulation cascades | 12 | Neuroactive ligand-receptor interaction | 12 |
| Fructose and mannose metabolism | 12 | Gap junction | 11 |
| Glycine, serine, and threonine metabolism | 12 | Long-term depression | 11 |
| Leukocyte transendothelial migration | 12 | Melanogenesis | 11 |
| Melanogenesis | 12 | Regulation of actin cytoskeleton | 11 |
| Pathways with genes up-regulated by G-CSF . | Pathways with genes up-regulated by plerixafor . | ||
|---|---|---|---|
| Pathway . | No. of genes in pathway . | Pathway . | No. of genes in pathway . |
| Cytokine-cytokine receptor interaction | 30 | Cytokine-cytokine receptor interaction | 29 |
| Jak-STAT signaling pathway | 25 | Calcium signaling pathway | 27 |
| MAPK signaling pathway | 24 | T-cell receptor signaling pathway | 27 |
| Focal adhesion | 22 | Hematopoietic cell lineage | 24 |
| Insulin signaling pathway | 21 | Wnt signaling pathway | 22 |
| Hematopoietic cell lineage | 20 | Primary immunodeficiency | 21 |
| Natural killer cell–mediated cytotoxicity | 19 | Cell cycle | 20 |
| Purine metabolism | 19 | MAPK signaling pathway | 20 |
| ErbB signaling pathway | 16 | Focal adhesion | 19 |
| VEGF signaling pathway | 15 | Natural killer cell mediated cytotoxicity | 19 |
| Regulation of actin cytoskeleton | 14 | TGF-beta signaling pathway | 19 |
| Toll-like receptor signaling pathway | 14 | Fc epsilon RI signaling pathway | 18 |
| Tyrosine metabolism | 14 | Jak-STAT signaling pathway | 18 |
| Acute myeloid leukemia | 13 | GnRH signaling pathway | 15 |
| Fc epsilon RI signaling pathway | 13 | Cell adhesion molecules | 14 |
| GnRH signaling pathway | 13 | Tight junction | 13 |
| Neuroactive ligand-receptor interaction | 13 | Adherens junction | 12 |
| Wnt signaling pathway | 13 | Antigen processing and presentation | 12 |
| Calcium signaling pathway | 12 | B-cell receptor signaling pathway | 12 |
| Complement and coagulation cascades | 12 | Neuroactive ligand-receptor interaction | 12 |
| Fructose and mannose metabolism | 12 | Gap junction | 11 |
| Glycine, serine, and threonine metabolism | 12 | Long-term depression | 11 |
| Leukocyte transendothelial migration | 12 | Melanogenesis | 11 |
| Melanogenesis | 12 | Regulation of actin cytoskeleton | 11 |
Comparison of the specific genes up-regulated in G-CSF–mobilized and plerixafor-mobilized CD34+ cells revealed that the genes maximally up-regulated by G-CSF included the neutrophil genes, neutrophil cytosolic factor 4 (NCF4) and proteinase 3 (PR3), and the NK-cell gene Fc-gamma receptor IIIa (CD16a; Table 3). G-CSF–mobilized CD34+ cells were also more likely to express mononuclear phagocyte genes CX3CR1, cathepsin L (CTSL), and lymphotoxin B receptor precursor. Genes expressed by several types of leukocytes that were up-regulated in G-CSF–mobilized CD34+ cells included Fc-gamma receptor IIb, which is expressed by neutrophils and B cells; cystatin F, which is expressed by NK cells and mononuclear phagocytes; and IL-21 receptor, which is expressed by T cells, B cells, NK cells, and mononuclear phagocytes. Analysis of the expression of CTSL by quantitative real-time polymerase chain reaction (qRT-PCR) confirmed that CTSL expression was greater in G-CSF–mobilized CD34+ cells than in plerixafor-mobilized CD34+ cells (Figure 3). The level of CTSL expressed by plerixafor plus G-CSF– and G-CSF–mobilized CD34+ cells were similar.
Genes whose expression was increased the greatest in G-CSF–mobilized CD34+ cells compared with plerixafor-mobilized CD34+ cells
| Gene . | . | Fold increase* . | P . |
|---|---|---|---|
| NCF4 | Neutrophil cytosolic factor 4, 40 kDa | 9.04 | 5.30 × 10−6 |
| VNN2 | Vanin 2 | 7.57 | 7.90 × 10−6 |
| PRTN3 | Proteinase 3 (serine proteinase, neutrophil, Wegener granulomatosis autoantigen) | 7.25 | 4.49 × 10−4 |
| LTBR | Lymphotoxin-beta receptor precursor = tumor necrosis factor receptor 2 related protein = tumor necrosis factor C receptor | 7.23 | 1.33 × 10−4 |
| CST7 | Cystatin F (leukocystatin) (inhibits cathepsins) | 7.05 | 9.00 × 10−6 |
| FCGR3A | CD16 = Fc-gamma receptor IIIa | 7.00 | 2.60 × 10−5 |
| CTSL | Cathepsin L | 6.50 | 8.00 × 10−7 |
| ANXA3 | Annexin A3 | 6.17 | 1.17 × 10−4 |
| CST7 | Cystatin F (leukocystatin) | 5.83 | 5.80 × 10−6 |
| TSPYL2 | TSPY-like 2 | 4.73 | 8.58 × 10−4 |
| HSPA1B | Heat shock 70-kDa protein 1B | 4.67 | .012 |
| ANXA3 | Annexin A3 | 4.64 | 8.42 × 10−5 |
| DYSF | Dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) | 4.65 | 1.47 × 10−3 |
| CLU | Clusterin | 4.51 | 8.50 × 10−6 |
| MYCO | Myocilin, trabecular meshwork inducible glucocorticoid response | 4.23 | 1.20 × 10−3 |
| PPBP | CTAP-III = NAP2 = connective tissue activation peptide III = pro-platelet basic protein | 4.09 | 6.47 × 10−3 |
| C1QB | Complement component 1, q subcomponent, B chain | 4.00 | 4.14 × 10−3 |
| RNASE2 | Ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) | 3.99 | 7.57 × 10−4 |
| GOS2 | putative lymphocyte G0/G1 switch gene | 3.96 | 8.50 × 10−5 |
| TNFRSF1 | Tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) | 3.84 | 3.72 × 10−4 |
| CXCL2 | Chemokine (C-X-C motif) ligand | 3.65 | 6.73 × 10−4 |
| FCGR2B | Low-affinity IgG Fc receptor II-B and C isoforms | 3.61 | 1.98 × 10−4 |
| IL21R | Interleukin-21 receptor | 3.55 | 3.12 × 10−4 |
| CX3CR1 | Chemokine (C-X3-C motif) receptor 1 | 3.52 | 1.20 × 10−4 |
| SLCIA5 | Solute carrier family 1 (neutral amino acid transporter), member 5 | 3.35 | 2.43 × 10−4 |
| PTRF | Polymerase I and transcript release factor | 3.28 | 3.92 × 10−5 |
| Gene . | . | Fold increase* . | P . |
|---|---|---|---|
| NCF4 | Neutrophil cytosolic factor 4, 40 kDa | 9.04 | 5.30 × 10−6 |
| VNN2 | Vanin 2 | 7.57 | 7.90 × 10−6 |
| PRTN3 | Proteinase 3 (serine proteinase, neutrophil, Wegener granulomatosis autoantigen) | 7.25 | 4.49 × 10−4 |
| LTBR | Lymphotoxin-beta receptor precursor = tumor necrosis factor receptor 2 related protein = tumor necrosis factor C receptor | 7.23 | 1.33 × 10−4 |
| CST7 | Cystatin F (leukocystatin) (inhibits cathepsins) | 7.05 | 9.00 × 10−6 |
| FCGR3A | CD16 = Fc-gamma receptor IIIa | 7.00 | 2.60 × 10−5 |
| CTSL | Cathepsin L | 6.50 | 8.00 × 10−7 |
| ANXA3 | Annexin A3 | 6.17 | 1.17 × 10−4 |
| CST7 | Cystatin F (leukocystatin) | 5.83 | 5.80 × 10−6 |
| TSPYL2 | TSPY-like 2 | 4.73 | 8.58 × 10−4 |
| HSPA1B | Heat shock 70-kDa protein 1B | 4.67 | .012 |
| ANXA3 | Annexin A3 | 4.64 | 8.42 × 10−5 |
| DYSF | Dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) | 4.65 | 1.47 × 10−3 |
| CLU | Clusterin | 4.51 | 8.50 × 10−6 |
| MYCO | Myocilin, trabecular meshwork inducible glucocorticoid response | 4.23 | 1.20 × 10−3 |
| PPBP | CTAP-III = NAP2 = connective tissue activation peptide III = pro-platelet basic protein | 4.09 | 6.47 × 10−3 |
| C1QB | Complement component 1, q subcomponent, B chain | 4.00 | 4.14 × 10−3 |
| RNASE2 | Ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) | 3.99 | 7.57 × 10−4 |
| GOS2 | putative lymphocyte G0/G1 switch gene | 3.96 | 8.50 × 10−5 |
| TNFRSF1 | Tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) | 3.84 | 3.72 × 10−4 |
| CXCL2 | Chemokine (C-X-C motif) ligand | 3.65 | 6.73 × 10−4 |
| FCGR2B | Low-affinity IgG Fc receptor II-B and C isoforms | 3.61 | 1.98 × 10−4 |
| IL21R | Interleukin-21 receptor | 3.55 | 3.12 × 10−4 |
| CX3CR1 | Chemokine (C-X3-C motif) receptor 1 | 3.52 | 1.20 × 10−4 |
| SLCIA5 | Solute carrier family 1 (neutral amino acid transporter), member 5 | 3.35 | 2.43 × 10−4 |
| PTRF | Polymerase I and transcript release factor | 3.28 | 3.92 × 10−5 |
The fold increase for each gene is the average expression of each gene in G-CSF–mobilized CD34+ cells from 3 rhesus macaque divided by the average expression of the same gene in plerixafor-mobilized CD34+ cells from 3 monkeys.
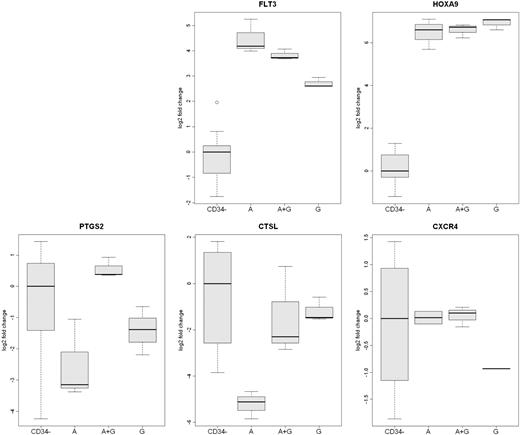
Analysis of differentially expressed mobilized CD34+ cells genes by qRT-PCR. The expression of 5 genes (FLT3, HOXA9, PTGS2, CXCR4, and CTSL) in CD34+ cells from the 3 rhesus macaque mobilized by plerixafor (A), G-CSF (G), or plerixafor plus G-CSF (A+G) were analyzed by qRT-PCR. The expression of CXCR4 was greater in plerixafor and plerixafor plus G-CSF–mobilized CD34+ cells. The expression of CTSL was greater on G-CSF– and G-CSF plus plerixafor-mobilized CD34+ cells. FLT3 expression was greatest on plerixafor-mobilized CD34+ cells, and the expression of PTGS2 was greatest on plerixafor plus G-CSF–mobilized cells. The level of expression of HOXA9 by the 3 types of CD34+ cells was similar but much greater than peripheral blood leukocytes. The expression of the 5 genes in leukocytes that were not absorbed by anti-CD34 from the 9 mobilized PBSC concentrates (CD34−) are shown as a control.
Analysis of differentially expressed mobilized CD34+ cells genes by qRT-PCR. The expression of 5 genes (FLT3, HOXA9, PTGS2, CXCR4, and CTSL) in CD34+ cells from the 3 rhesus macaque mobilized by plerixafor (A), G-CSF (G), or plerixafor plus G-CSF (A+G) were analyzed by qRT-PCR. The expression of CXCR4 was greater in plerixafor and plerixafor plus G-CSF–mobilized CD34+ cells. The expression of CTSL was greater on G-CSF– and G-CSF plus plerixafor-mobilized CD34+ cells. FLT3 expression was greatest on plerixafor-mobilized CD34+ cells, and the expression of PTGS2 was greatest on plerixafor plus G-CSF–mobilized cells. The level of expression of HOXA9 by the 3 types of CD34+ cells was similar but much greater than peripheral blood leukocytes. The expression of the 5 genes in leukocytes that were not absorbed by anti-CD34 from the 9 mobilized PBSC concentrates (CD34−) are shown as a control.
The genes differentially up-regulated most by plerixafor included several copies of CXCR4, which is expressed by hematopoietic stem cells, T cells, B cells, monocytes, and neutrophils. Other plerixafor up-regulated genes included the B-cell genes CD79A, interleukin-7 receptor, Epstein-Barr virus–induced gene 2, and Bruton agammaglobulinemia tyrosine kinase; a T-cell gene, T-cell receptor–associated transmembrane adaptor 1; and a mast cell gene, Fc receptor of IgE 1a (Table 4). In addition, the expression of the HSC marker Fms-related tyrosine kinase 3 (FLT3) was up-regulated in plerixafor-mobilized CD34+ cells. Analysis of CXCR4 and FLT3 expression by qRT-PCR confirmed that the expression of both was greater in plerixafor-mobilized CD34+ cells than G-CSF–mobilized CD34+ cells (Figure 3). These results suggest that G-CSF–mobilized CD34+ cells are more likely to include neutrophil and macrophage precursors, and plerixafor-mobilized CD34+ cells are more likely to contain T-cell, B-cell, and mast cell precursors.
Genes whose expression was increased the greatest in plerixafor-mobilized CD34+ cells compared with G-CSF–mobilized CD34+ cells
| Gene . | . | Fold increase* . | P . |
|---|---|---|---|
| ICSBP1 | Interferon consensus sequence binding protein 1 | 6.14 | 9.20 × 10−6 |
| BLNK | B-cell linker | 5.99 | 1.30 × 10−6 |
| CXCR4 | CXC chemokine receptor 4 | 5.17 | 1.17 × 10−5 |
| CXCR4 | CXC chemokine receptor 4 | 5.07 | 4.80 × 10−6 |
| CDK5R1 | Cyclin-dependent kinase 5, regulatory subunit 1 (p35) | 4.62 | 3.80 × 10−6 |
| TP53I11 | Tumor protein p53 inducible protein 11 | 4.35 | 8.40 × 10−6 |
| SMAD7 | Homologue of Mothers Against Decapentaplegic (MAD) = negative regulator of TGF-beta signaling | 4.19 | 8.90 × 10−6 |
| FLT3 | Fms-related tyrosine kinase 3 | 3.44 | 2.05 × 10−6 |
| CXCR4 | CXC chemokine receptor 4 | 3.32 | 2.49 × 10−4 |
| LDB2 | LIM domain binding 2 | 3.23 | 3.24 × 10−4 |
| COG1 | Component of oligomeric golgi complex 1 | 3.12 | 3.15 × 10−5 |
| DCN | Decorin | 3.04 | 5.96 × 10−5 |
| IL23A | Enhancer of polycomb homolog 1 (Drosophila) | 2.95 | 3.58 × 10−3 |
| AGTRL1 | Angiotensin II receptor-like 1 | 2.92 | 4.68 × 10−4 |
| SPIB | Spi-B transcription factor | 2.91 | 5.17 × 10−4 |
| EBI2 | Epstein-Barr virus–induced gene 2 (lymphocyte-specific G protein–coupled receptor) | 2.90 | 1.81 × 10−4 |
| IL7R | Interleukin-7 receptor | 2.86 | 1.81 × 10−4 |
| TRAT1 | T-cell receptor–associated transmembrane adaptor 1 | 2.83 | .014 |
| GAP43 | Growth associated protein 43 | 2.82 | 7.42 × 10−4 |
| CD79A | CD79A, BCR alpha chain = mb-1 | 2.95 | 3.58 × 10−3 |
| TIEG | TGF-beta inducible early protein | 2.75 | 1.84 × 10−4 |
| IL18R1 | Interleukin-18 receptor 1 | 2.73 | 8.62 × 10−3 |
| SMARCD | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 | 2.73 | 2.73 × 10−3 |
| BTK | Btk = Bruton agammaglobulinemia tyrosine kinase | 2.72 | 2.67 × 10−3 |
| FCER1A | Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide | 2.67 | .037 |
| KHDRBS3 | KH domain containing, RNA binding, signal transduction associated 3 | 2.63 | 3.46 × 10−3 |
| Gene . | . | Fold increase* . | P . |
|---|---|---|---|
| ICSBP1 | Interferon consensus sequence binding protein 1 | 6.14 | 9.20 × 10−6 |
| BLNK | B-cell linker | 5.99 | 1.30 × 10−6 |
| CXCR4 | CXC chemokine receptor 4 | 5.17 | 1.17 × 10−5 |
| CXCR4 | CXC chemokine receptor 4 | 5.07 | 4.80 × 10−6 |
| CDK5R1 | Cyclin-dependent kinase 5, regulatory subunit 1 (p35) | 4.62 | 3.80 × 10−6 |
| TP53I11 | Tumor protein p53 inducible protein 11 | 4.35 | 8.40 × 10−6 |
| SMAD7 | Homologue of Mothers Against Decapentaplegic (MAD) = negative regulator of TGF-beta signaling | 4.19 | 8.90 × 10−6 |
| FLT3 | Fms-related tyrosine kinase 3 | 3.44 | 2.05 × 10−6 |
| CXCR4 | CXC chemokine receptor 4 | 3.32 | 2.49 × 10−4 |
| LDB2 | LIM domain binding 2 | 3.23 | 3.24 × 10−4 |
| COG1 | Component of oligomeric golgi complex 1 | 3.12 | 3.15 × 10−5 |
| DCN | Decorin | 3.04 | 5.96 × 10−5 |
| IL23A | Enhancer of polycomb homolog 1 (Drosophila) | 2.95 | 3.58 × 10−3 |
| AGTRL1 | Angiotensin II receptor-like 1 | 2.92 | 4.68 × 10−4 |
| SPIB | Spi-B transcription factor | 2.91 | 5.17 × 10−4 |
| EBI2 | Epstein-Barr virus–induced gene 2 (lymphocyte-specific G protein–coupled receptor) | 2.90 | 1.81 × 10−4 |
| IL7R | Interleukin-7 receptor | 2.86 | 1.81 × 10−4 |
| TRAT1 | T-cell receptor–associated transmembrane adaptor 1 | 2.83 | .014 |
| GAP43 | Growth associated protein 43 | 2.82 | 7.42 × 10−4 |
| CD79A | CD79A, BCR alpha chain = mb-1 | 2.95 | 3.58 × 10−3 |
| TIEG | TGF-beta inducible early protein | 2.75 | 1.84 × 10−4 |
| IL18R1 | Interleukin-18 receptor 1 | 2.73 | 8.62 × 10−3 |
| SMARCD | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 | 2.73 | 2.73 × 10−3 |
| BTK | Btk = Bruton agammaglobulinemia tyrosine kinase | 2.72 | 2.67 × 10−3 |
| FCER1A | Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide | 2.67 | .037 |
| KHDRBS3 | KH domain containing, RNA binding, signal transduction associated 3 | 2.63 | 3.46 × 10−3 |
The fold increase for each gene is the average expression of each gene in plerixafor-mobilized CD34+ cells from 3 rhesus macaque divided by the average expression of the same gene in G-CSF–mobilized CD34+ cells from 3 monkeys.
Gene expression analysis of CD34+ cells mobilized by plerixafor plus G-CSF
To assess the nature of CD34+ cells mobilized by the combination of plerixafor plus G-CSF, we combined the gene expression results from CD34+ cells mobilized with G-CSF alone and with plerixafor alone, and compared this dataset with CD34+ cells mobilized with plerixafor plus G-CSF. A total of 1375 genes were up-regulated in plerixafor plus G-CSF–mobilized CD34+ cells compared with those mobilized with G-CSF alone and plerixafor alone, and 1449 were up-regulated in CD34+ cells regulated with G-CSF alone and plerixafor alone (t tests, P < .05).
Analysis of the pathways that contained the most genes up-regulated in CD34+ cells found that the genes up-regulated in the plerixafor plus G-CSF–mobilized CD34+ cells were more likely to belong to B-cell signaling, Fc epsilon RI signaling, Toll-like receptor, and cell cycle (Table 5). Those down-regulated in the plerixafor plus G-CSF–mobilized CD34+ cells were more likely to belong to ribosome, antigen processing and presentation, and vascular endothelial growth factor signaling pathways.
KEGG pathways with the most genes up-regulated and down-regulated in CD34+ cells mobilized by plerixafor plus G-CSF compared with those mobilized by plerixafor alone and G-CSF alone
| Pathways with genes up-regulated by plerixafor plus G-CSF . | Pathways with genes down-regulated by plerixafor plus G-CSF . | ||
|---|---|---|---|
| Pathway . | No. of up-regulated genes in pathway . | Pathway . | No. of down-regulated genes in pathway . |
| Cytokine-cytokine receptor interaction | 68 | MAPK signaling pathway | 51 |
| MAPK signaling pathway | 63 | Focal adhesion | 46 |
| Natural killer cell–mediated cytotoxicity | 41 | Oxidative phosphorylation | 40 |
| Jak-STAT signaling pathway | 39 | Calcium signaling pathway | 37 |
| Focal adhesion | 33 | Ribosome | 36 |
| Leukocyte transendothelial migration | 33 | Cytokine-cytokine receptor interaction | 33 |
| Regulation of actin cytoskeleton | 32 | Wnt signaling pathway | 33 |
| Insulin signaling pathway | 29 | Tight junction | 31 |
| Cell cycle | 28 | Jak-STAT signaling pathway | 29 |
| Gap junction | 28 | Cell adhesion molecules | 28 |
| Calcium signaling pathway | 25 | Natural killer cell–mediated cytotoxicity | 28 |
| Fc epsilon RI signaling pathway | 25 | Insulin signaling pathway | 27 |
| Oxidative phosphorylation | 25 | Axon guidance | 26 |
| Ubiquitin mediated proteolysis | 25 | ErbB signaling pathway | 25 |
| T-cell receptor signaling pathway | 24 | Small cell lung cancer | 25 |
| B-cell receptor signaling pathway | 22 | Antigen processing and presentation | 23 |
| Cell adhesion molecules | 22 | Leukocyte transendothelial migration | 23 |
| Toll-like receptor signaling pathway | 22 | Gap junction | 22 |
| Wnt signaling pathway | 22 | Hematopoietic cell lineage | 22 |
| GnRH signaling pathway | 21 | Long-term depression | 22 |
| Melanogenesis | 21 | Regulation of actin cytoskeleton | 22 |
| Tight junction | 21 | VEGF signaling pathway | 22 |
| Apoptosis | 20 | Colorectal cancer | 21 |
| Hematopoietic cell lineage | 20 | T-cell receptor signaling pathway | 21 |
| Pathogenic Escherichia coli infection—EHEC | 20 | Ubiquitin mediated proteolysis | 21 |
| Pathways with genes up-regulated by plerixafor plus G-CSF . | Pathways with genes down-regulated by plerixafor plus G-CSF . | ||
|---|---|---|---|
| Pathway . | No. of up-regulated genes in pathway . | Pathway . | No. of down-regulated genes in pathway . |
| Cytokine-cytokine receptor interaction | 68 | MAPK signaling pathway | 51 |
| MAPK signaling pathway | 63 | Focal adhesion | 46 |
| Natural killer cell–mediated cytotoxicity | 41 | Oxidative phosphorylation | 40 |
| Jak-STAT signaling pathway | 39 | Calcium signaling pathway | 37 |
| Focal adhesion | 33 | Ribosome | 36 |
| Leukocyte transendothelial migration | 33 | Cytokine-cytokine receptor interaction | 33 |
| Regulation of actin cytoskeleton | 32 | Wnt signaling pathway | 33 |
| Insulin signaling pathway | 29 | Tight junction | 31 |
| Cell cycle | 28 | Jak-STAT signaling pathway | 29 |
| Gap junction | 28 | Cell adhesion molecules | 28 |
| Calcium signaling pathway | 25 | Natural killer cell–mediated cytotoxicity | 28 |
| Fc epsilon RI signaling pathway | 25 | Insulin signaling pathway | 27 |
| Oxidative phosphorylation | 25 | Axon guidance | 26 |
| Ubiquitin mediated proteolysis | 25 | ErbB signaling pathway | 25 |
| T-cell receptor signaling pathway | 24 | Small cell lung cancer | 25 |
| B-cell receptor signaling pathway | 22 | Antigen processing and presentation | 23 |
| Cell adhesion molecules | 22 | Leukocyte transendothelial migration | 23 |
| Toll-like receptor signaling pathway | 22 | Gap junction | 22 |
| Wnt signaling pathway | 22 | Hematopoietic cell lineage | 22 |
| GnRH signaling pathway | 21 | Long-term depression | 22 |
| Melanogenesis | 21 | Regulation of actin cytoskeleton | 22 |
| Tight junction | 21 | VEGF signaling pathway | 22 |
| Apoptosis | 20 | Colorectal cancer | 21 |
| Hematopoietic cell lineage | 20 | T-cell receptor signaling pathway | 21 |
| Pathogenic Escherichia coli infection—EHEC | 20 | Ubiquitin mediated proteolysis | 21 |
Analysis of the specific genes up-regulated in plerixafor plus G-CSF–mobilized CD34+ cells found that, when compared with G-CSF alone and plerixafor alone mobilized CD34+ cells, plerixafor plus G-CSF–mobilized CD34+ cells expressed increased levels of B-cell genes: immunoglobulin alpha heavy chain constant region, Bruton agammaglobulinemia tyrosine kinase, and immunoglobulin gamma 3 heavy chain constant region (Table 6). In addition, the T-cell gene granzyme A; the HSC gene HOXA9; an NK cell gene, lectin-like NK-cell receptor (LLT1); and a gene encoding a protein found in the extracellular matrix and macrophages tissue inhibitor of metalloproteinase (TIMP3) were up-regulated (Table 6). The CD34+ cell genes up-regulated most by plerixafor plus G-CSF also included prostaglandin-endoperoxide synthase 2 (PTGS2) or cyclooxygenase 2 (COX2), an important enzyme in arachidonic acid metabolism and prostaglandin production. Analysis of PTGS2 and HOXA9 expression by qRT-PCR confirmed that the expression of PTGS2 was greater in plerixafor plus G-CSF–mobilized CD34+ cells than in those mobilized by plerixafor alone or G-CSF alone (Figure 3). The expression of HOXA9 was similar among CD34+ cells mobilized by each of the 3 regimens but was much greater than that for peripheral blood leukocytes (Figure 3).
Genes whose expression was increased the greatest in plerixafor plus G-CSF–mobilized CD34+ cells compared with those mobilized by plerixafor alone and G-CSF alone
| Gene . | Description . | Fold increase* . | Parametric P . |
|---|---|---|---|
| MGC27165 | Immunoglobulin alpha (1 or 2) heavy chain constant region | 9.04 | 2.45 × 10−4 |
| BTK | btk = Bruton agammaglobulinemia tyrosine kinase | 5.13 | < 1 × 10−7 |
| PILRB | Similar to hCG2024106 | 5.07 | 2.70 × 10−6 |
| IGHG3 | Immunoglobulin gamma 3 heavy chain constant region | 5.04 | 3.26 × 10−3 |
| IL1R2 | Interleukin 1 receptor, type II | 4.82 | 2.69 × 10−4 |
| GNA14 | G alpha 14 = guanine nucleotide binding protein alpha 14 | 4.72 | <1 × 10−7 |
| ULK2 | Unc-51-like kinase 2 (C. elegans) | 4.26 | 1.27 × 10−3 |
| BAI3 | Brain-specific angiogenesis inhibitor 3 | 4.16 | 3.92 × 10−4 |
| IL8 | Interleukin 8 | 3.98 | 2.28 × 10−4 |
| HRASLS3 | HRAS-like suppressor 3 | 3.68 | 2.24 × 10−4 |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | 3.67 | 1.61 × 10−4 |
| PPM1A | Protein phosphatase 1A (formerly 2C), magnesium-dependent, alpha isoform | 3.61 | 1.30 × 10−6 |
| CYP3A7 | Cytochrome P450, family 3, subfamily A, polypeptide 7 | 3.59 | 1.45 × 10−5 |
| CCR1 | Chemokine (C-C motif) receptor 1 | 3.58 | 1.31 × 10−3 |
| CX3CR1 | Chemokine (C-X3-C motif) receptor 1 | 3.58 | 1.39 × 10−3 |
| UBE2K | Ubiquitin-conjugating enzyme E2K (UBC1 homolog, yeast) | 3.43 | 3.00 × 10−7 |
| RPL3L | Ribosomal protein L3-like | 3.42 | 3.00 × 10−7 |
| HOXA9 | Homeobox A9 | 3.40 | 2.10 × 10−4 |
| TMSB10 | Thymosin beta 10 | 3.38 | 1.34 × 10−3 |
| ZNF644 | Zinc finger protein 644 | 3.34 | 1.74 × 10−3 |
| HIRA | HIR histone cell-cycle regulation defective homolog A (S cerevisiae) | 3.31 | 2.33 × 10−5 |
| LLT1 | Lectin-like NK-cell receptor | 3.30 | 4.62 × 10−5 |
| PRDM15 | PR domain containing 15 | 3.23 | 2.82 × 10−4 |
| TIMP3 | Tissue inhibitor of metalloproteinase 3 (Sorsby fundus dystrophy, pseudoinflammatory) | 3.19 | 2.26 × 10−4 |
| ACACA | Acetyl-coenzyme A carboxylase alpha | 3.14 | 2.70 × 10−6 |
| GZMA | Granzyme A (granzyme 1, cytotoxic T-lymphocyte–associated serine esterase 3) | 3.12 | 1.19 × 10−3 |
| ICOS | Inducible T-cell co-stimulator | 3.09 | 2.63 × 10−4 |
| CYP1B1 | Cytochrome P450, family 1, subfamily B, polypeptide 1 | 3.05 | 3.64 × 10−5 |
| LYN | Lyn = tyrosine kinase | 3.02 | 8.78 × 10−5 |
| GPA33 | Glycoprotein A33 (transmembrane) | 3.02 | 1.74 × 10−4 |
| Gene . | Description . | Fold increase* . | Parametric P . |
|---|---|---|---|
| MGC27165 | Immunoglobulin alpha (1 or 2) heavy chain constant region | 9.04 | 2.45 × 10−4 |
| BTK | btk = Bruton agammaglobulinemia tyrosine kinase | 5.13 | < 1 × 10−7 |
| PILRB | Similar to hCG2024106 | 5.07 | 2.70 × 10−6 |
| IGHG3 | Immunoglobulin gamma 3 heavy chain constant region | 5.04 | 3.26 × 10−3 |
| IL1R2 | Interleukin 1 receptor, type II | 4.82 | 2.69 × 10−4 |
| GNA14 | G alpha 14 = guanine nucleotide binding protein alpha 14 | 4.72 | <1 × 10−7 |
| ULK2 | Unc-51-like kinase 2 (C. elegans) | 4.26 | 1.27 × 10−3 |
| BAI3 | Brain-specific angiogenesis inhibitor 3 | 4.16 | 3.92 × 10−4 |
| IL8 | Interleukin 8 | 3.98 | 2.28 × 10−4 |
| HRASLS3 | HRAS-like suppressor 3 | 3.68 | 2.24 × 10−4 |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | 3.67 | 1.61 × 10−4 |
| PPM1A | Protein phosphatase 1A (formerly 2C), magnesium-dependent, alpha isoform | 3.61 | 1.30 × 10−6 |
| CYP3A7 | Cytochrome P450, family 3, subfamily A, polypeptide 7 | 3.59 | 1.45 × 10−5 |
| CCR1 | Chemokine (C-C motif) receptor 1 | 3.58 | 1.31 × 10−3 |
| CX3CR1 | Chemokine (C-X3-C motif) receptor 1 | 3.58 | 1.39 × 10−3 |
| UBE2K | Ubiquitin-conjugating enzyme E2K (UBC1 homolog, yeast) | 3.43 | 3.00 × 10−7 |
| RPL3L | Ribosomal protein L3-like | 3.42 | 3.00 × 10−7 |
| HOXA9 | Homeobox A9 | 3.40 | 2.10 × 10−4 |
| TMSB10 | Thymosin beta 10 | 3.38 | 1.34 × 10−3 |
| ZNF644 | Zinc finger protein 644 | 3.34 | 1.74 × 10−3 |
| HIRA | HIR histone cell-cycle regulation defective homolog A (S cerevisiae) | 3.31 | 2.33 × 10−5 |
| LLT1 | Lectin-like NK-cell receptor | 3.30 | 4.62 × 10−5 |
| PRDM15 | PR domain containing 15 | 3.23 | 2.82 × 10−4 |
| TIMP3 | Tissue inhibitor of metalloproteinase 3 (Sorsby fundus dystrophy, pseudoinflammatory) | 3.19 | 2.26 × 10−4 |
| ACACA | Acetyl-coenzyme A carboxylase alpha | 3.14 | 2.70 × 10−6 |
| GZMA | Granzyme A (granzyme 1, cytotoxic T-lymphocyte–associated serine esterase 3) | 3.12 | 1.19 × 10−3 |
| ICOS | Inducible T-cell co-stimulator | 3.09 | 2.63 × 10−4 |
| CYP1B1 | Cytochrome P450, family 1, subfamily B, polypeptide 1 | 3.05 | 3.64 × 10−5 |
| LYN | Lyn = tyrosine kinase | 3.02 | 8.78 × 10−5 |
| GPA33 | Glycoprotein A33 (transmembrane) | 3.02 | 1.74 × 10−4 |
The fold increase for each gene is the average expression in the plerixafor plus G-CSF–mobilized CD34+ cells from 3 rhesus divided by the average expression of the same gene in the CD34+ cells mobilized by plerixafor from 3 monkeys and CD34+ cells mobilized by G-CSF from 3 monkeys.
We also identified the genes that were down-regulated most in plerixafor plus G-CSF–mobilized CD34+ cells. Among the genes down-regulated most in plerixafor plus G-CSF–mobilized CD34+ cells were a number of ribosomal protein genes, cathepsin S (an enzyme important for antigen processing), neutrophil receptor formyl peptide receptor 1, bone morphogenic protein 2, laminin, and gamma 3 (Table 7).
Genes whose expression was decreased the greatest in plerixafor plus G-CSF–mobilized CD34+ cells compared with those mobilized by plerixafor alone and G-CSF alone
| Gene symbol . | Description . | Fold decrease* . | Parametric P . |
|---|---|---|---|
| CTSS | Cathepsin S | 13.95 | < 1 × 10−7 |
| C9orf3 | Chromosome 9 open reading frame 3 | 7.64 | 2.16 × 10−4 |
| NTRK2 | Neurotrophic tyrosine kinase, receptor, type 2 | 5.82 | < 1 × 10−7 |
| NMT1 | N-myristoyltransferase 1 | 5.64 | 2.00 × 10−7 |
| SCN1B | Sodium channel, voltage-gated, type I, beta | 5.34 | < 1 × 10−7 |
| IL16 | Interleukin 16 (lymphocyte chemoattractant factor) | 4.98 | 1.20 × 10−6 |
| CYP27B1 | Cytochrome P450, family 27, subfamily B, polypeptide 1 | 4.56 | 2.35 × 10−4 |
| KNS2 | kinesin 2 (60-70 kDa) | 4.45 | 1.18 × 10−4 |
| SMARCD2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 | 4.373 | 6.81 × 10−4 |
| CCL25 | TECK chemokine (CCL25) | 4.27 | 8.89 × 10−5 |
| PFDN6 | Prefoldin subunit 6 | 4.26 | 2.53 × 10−3 |
| EIF5A | Eukaryotic translation initiation factor 5A | 4.24 | 6.40 × 10−5 |
| TYMS | Thymidylate synthetase | 4.23 | 2.20 × 10−6 |
| OCLN | Occludin = tight junction protein | 4.21 | 2.35 × 10−4 |
| NAT2 | N-acetyltransferase 2 (arylamine N-acetyltransferase) | 4.09 | 8.14 × 10−4 |
| RPL23 | Ribosomal protein L23 | 4.04 | 6.20 × 10−6 |
| WISP2 | WNT1 inducible signaling pathway protein 2 | 4.01 | 1.20 × 10−6 |
| MVD | Mevalonate (diphospho) decarboxylase | 3.84 | 2.33 × 10−5 |
| BMP2 | Bone morphogenetic protein 2 | 3.79 | 3.03 × 10−4 |
| UXT | Ubiquitously expressed transcript | 3.78 | 2.80 × 10−6 |
| IQGAP1 | IQ motif containing GTPase activating protein 1 | 3.67 | 4.00 × 10−6 |
| KCNA5 | Potassium voltage-gated channel, shaker-related subfamily, member 5 | 3.67 | 1.80 × 10−3 |
| RPS27A | Ribosomal protein S27a = fused to ubiquitin | 3.60 | 1.00 × 10−5 |
| ECH1 | Enoyl coenzyme A hydratase 1, peroxisomal | 3.52 | 2.10 × 10−6 |
| FPR1 | Formyl peptide receptor 1 | 3.41 | 8.60 × 10−6 |
| STAP2 | Signal transducing adaptor family member 2 | 3.36 | 3.15 × 10−4 |
| ZNF525 | Zinc finger protein 525 | 3.27 | 8.30 × 10−5 |
| LAMC3 | Laminin, gamma 3 | 3.23 | 1.03 × 10−3 |
| DES | Desmin | 3.18 | 1.78 × 10−5 |
| RPS15 | Ribosomal protein S15 | 3.17 | 2.80 × 10−6 |
| Gene symbol . | Description . | Fold decrease* . | Parametric P . |
|---|---|---|---|
| CTSS | Cathepsin S | 13.95 | < 1 × 10−7 |
| C9orf3 | Chromosome 9 open reading frame 3 | 7.64 | 2.16 × 10−4 |
| NTRK2 | Neurotrophic tyrosine kinase, receptor, type 2 | 5.82 | < 1 × 10−7 |
| NMT1 | N-myristoyltransferase 1 | 5.64 | 2.00 × 10−7 |
| SCN1B | Sodium channel, voltage-gated, type I, beta | 5.34 | < 1 × 10−7 |
| IL16 | Interleukin 16 (lymphocyte chemoattractant factor) | 4.98 | 1.20 × 10−6 |
| CYP27B1 | Cytochrome P450, family 27, subfamily B, polypeptide 1 | 4.56 | 2.35 × 10−4 |
| KNS2 | kinesin 2 (60-70 kDa) | 4.45 | 1.18 × 10−4 |
| SMARCD2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 | 4.373 | 6.81 × 10−4 |
| CCL25 | TECK chemokine (CCL25) | 4.27 | 8.89 × 10−5 |
| PFDN6 | Prefoldin subunit 6 | 4.26 | 2.53 × 10−3 |
| EIF5A | Eukaryotic translation initiation factor 5A | 4.24 | 6.40 × 10−5 |
| TYMS | Thymidylate synthetase | 4.23 | 2.20 × 10−6 |
| OCLN | Occludin = tight junction protein | 4.21 | 2.35 × 10−4 |
| NAT2 | N-acetyltransferase 2 (arylamine N-acetyltransferase) | 4.09 | 8.14 × 10−4 |
| RPL23 | Ribosomal protein L23 | 4.04 | 6.20 × 10−6 |
| WISP2 | WNT1 inducible signaling pathway protein 2 | 4.01 | 1.20 × 10−6 |
| MVD | Mevalonate (diphospho) decarboxylase | 3.84 | 2.33 × 10−5 |
| BMP2 | Bone morphogenetic protein 2 | 3.79 | 3.03 × 10−4 |
| UXT | Ubiquitously expressed transcript | 3.78 | 2.80 × 10−6 |
| IQGAP1 | IQ motif containing GTPase activating protein 1 | 3.67 | 4.00 × 10−6 |
| KCNA5 | Potassium voltage-gated channel, shaker-related subfamily, member 5 | 3.67 | 1.80 × 10−3 |
| RPS27A | Ribosomal protein S27a = fused to ubiquitin | 3.60 | 1.00 × 10−5 |
| ECH1 | Enoyl coenzyme A hydratase 1, peroxisomal | 3.52 | 2.10 × 10−6 |
| FPR1 | Formyl peptide receptor 1 | 3.41 | 8.60 × 10−6 |
| STAP2 | Signal transducing adaptor family member 2 | 3.36 | 3.15 × 10−4 |
| ZNF525 | Zinc finger protein 525 | 3.27 | 8.30 × 10−5 |
| LAMC3 | Laminin, gamma 3 | 3.23 | 1.03 × 10−3 |
| DES | Desmin | 3.18 | 1.78 × 10−5 |
| RPS15 | Ribosomal protein S15 | 3.17 | 2.80 × 10−6 |
The fold decrease for each gene is the average expression in the plerixafor plus G-CSF–mobilized CD34+ cells from 3 rhesus divided by the average expression of the same gene in the CD34+ cells mobilized by plerixafor from 3 monkeys and CD34+ cells mobilized by G-CSF from 3 monkeys.
MIR expression analysis
A comparison of differentially expressed miR between plerixafor- and G-CSF–mobilized CD34+ cells found that 2 miR were up-regulated in plerixafor-mobilized CD34+ cells, miR-34c and miR-432 (Table 8). A miR, miR-126, that was previously found to be up-regulated in G-CSF–mobilized peripheral blood CD34+ cells compared with peripheral blood leukocytes19 was up-regulated in G-CSF–mobilized CD34+ cells compared with plerixafor-mobilized CD34+ cells (Table 8). The expression of miR-155, which is up-regulated during macrophage activation,20 was also greater in G-CSF–mobilized CD34+ cells. In addition, among the miR up-regulated in G-CSF–mobilized CD34+ cells were miR-21, miR-129-3p, miR-146b, miR-99a, miR-126*, and miR-10a*.
miRs that were differently expressed between plerixafor-mobilized CD34+ cells and G-CSF–mobilized CD34+ cells
| Increased in plerixafor-mobilized CD34+ cells . | Increased in G-CSF–mobilized CD34+ cells . | ||||
|---|---|---|---|---|---|
| miR . | Fold increase* . | P . | miR . | Fold increase† . | P . |
| hsa-miR-34c | 1.69 | .031 | hsa-miR-21 | 3.67 | 1.72 × 10−4 |
| hsa-miR-432* | 1.45 | .032 | hsa-miR-126* | 2.80 | 1.21 × 10−3 |
| mmu-miR-129-3p | 2.62 | 3.67 × 10−3 | |||
| hsa-miR-146b | 2.31 | 1.35 × 10−3 | |||
| hsa-miR-99a | 2.13 | .011 | |||
| hsa-miR-126* | 2.08 | 9.23 × 10−3 | |||
| hsa-miR-10a* | 1.98 | .019 | |||
| hsa-miR-424 | 1.90 | 8.57 × 10−3 | |||
| hsa-miR-126 | 1.70 | .041 | |||
| hsa-miR-155 | 1.65 | .033 | |||
| hsa-miR-335 | 1.55 | .049 | |||
| Increased in plerixafor-mobilized CD34+ cells . | Increased in G-CSF–mobilized CD34+ cells . | ||||
|---|---|---|---|---|---|
| miR . | Fold increase* . | P . | miR . | Fold increase† . | P . |
| hsa-miR-34c | 1.69 | .031 | hsa-miR-21 | 3.67 | 1.72 × 10−4 |
| hsa-miR-432* | 1.45 | .032 | hsa-miR-126* | 2.80 | 1.21 × 10−3 |
| mmu-miR-129-3p | 2.62 | 3.67 × 10−3 | |||
| hsa-miR-146b | 2.31 | 1.35 × 10−3 | |||
| hsa-miR-99a | 2.13 | .011 | |||
| hsa-miR-126* | 2.08 | 9.23 × 10−3 | |||
| hsa-miR-10a* | 1.98 | .019 | |||
| hsa-miR-424 | 1.90 | 8.57 × 10−3 | |||
| hsa-miR-126 | 1.70 | .041 | |||
| hsa-miR-155 | 1.65 | .033 | |||
| hsa-miR-335 | 1.55 | .049 | |||
The fold increase is the average expression of each microRNA in CD34+ cells from 3 rhesus macaque given plerixafor divided by the average expression of the same microRNA in 3 monkeys given G-CSF.
The fold increase is the average expression of each microRNA in CD34+ cells from 3 rhesus macaque given G-CSF divided by the average expression of the same microRNA in 3 monkeys given plerixafor.
We also identified miRs differentially expressed between CD34+ cells mobilized by plerixafor plus G-CSF and those mobilized by G-CSF alone and plerixafor alone. The levels of miR-126* and miR-142-3p were greater in plerixafor plus G-CSF–mobilized CD34+ cells than in those mobilized by either G-CSF alone (Table 9) or plerixafor alone (Table 10). The expression of miR-21 was also increased in plerixafor plus G-CSF–mobilized CD34+ cells compared with plerixafor-mobilized CD34+ cells (Table 10).
miRs that were differently expressed between plerixafor plus G-CSF–mobilized CD34+ cells and those mobilized with G-CSF alone
| Increased in plerixafor + G-CSF–mobilized CD34+ cells . | Increased in G-CSF–mobilized CD34+ cells . | ||||
|---|---|---|---|---|---|
| miR . | Fold increase* . | P . | miR . | Fold increase† . | P . |
| hsa-miR-142-3p | 3.27 | 3.34 × 10−3 | sv40-miR-S1-5p | 2.56 | 3.17 × 10−3 |
| hsa-miR-142-3p | 3.15 | 3.04 × 10−3 | hsa-miR-335 | 2.17 | .045 |
| hsa-miR-142-5p | 2.91 | 3.95 × 10−3 | hsa-mir-615 | 2.00 | .03 |
| hsa-miR-29b | 2.36 | 7.71 × 10−3 | hsa-miR-363* | 1.94 | .012 |
| hsa-miR-101 | 2.34 | 8.77 × 10−3 | mmu-miR-10b | 1.81 | .012 |
| hsa-miR-525 | 2.18 | 8.19 × 10−3 | mmu-mir-691 | 1.80 | .01 |
| hsa-mir-622 | 2.16 | .019 | hsa-miR-299-3p | 1.77 | .018 |
| hsa-miR-487b | 2.15 | 9.93 × 10−3 | mmu-miR-346 | 1.72 | 8.92 × 10−3 |
| hsa-miR-126* | 2.11 | 9.92 × 10−3 | mmu-mir-615 | 1.71 | .01 |
| hsa-miR-199b | 2.10 | .012 | hsa-miR-320 | 1.61 | .031 |
| mmu-mir-804 | 2.09 | 0.016 | hcmv-miR-UL70-3p | 1.56 | .03 |
| hsa-miR-486 | 2.08 | 3.63 × 10−3 | kshv-miR-K12-10a | 1.54 | .038 |
| hsa-miR-197 | 1.99 | 6.74 × 10−3 | hsa-miR-518f* | 1.54 | .029 |
| hsa-miR-19a | 1.98 | .01 | mmu-mir-423 | 1.51 | .044 |
| rno-miR-343 | 1.97 | .015 | hsa-mir-765 | 1.47 | .048 |
| hsa-miR-15a | 1.97 | .011 | |||
| hsa-miR-19b | 1.96 | .012 | |||
| hsa-miR-29c | 1.87 | .026 | |||
| hsa-miR-27a | 1.74 | .017 | |||
| hsa-miR-374 | 1.71 | .027 | |||
| hsa-miR-30e-5p | 1.70 | .044 | |||
| hsa-miR-29a | 1.68 | .029 | |||
| hsa-mir-662 | 1.66 | .02 | |||
| mmu-miR-489 | 1.63 | .025 | |||
| hsa-mir-421 | 1.60 | .03 | |||
| hsa-miR-20a | 1.60 | .025 | |||
| hsa-miR-106a | 1.59 | .022 | |||
| hsa-miR-203 | 1.58 | .032 | |||
| hsa-miR-20b | 1.58 | .023 | |||
| ebv-mir-BART3 | 1.57 | .022 | |||
| hsa-miR-17-5p | 1.55 | .034 | |||
| Increased in plerixafor + G-CSF–mobilized CD34+ cells . | Increased in G-CSF–mobilized CD34+ cells . | ||||
|---|---|---|---|---|---|
| miR . | Fold increase* . | P . | miR . | Fold increase† . | P . |
| hsa-miR-142-3p | 3.27 | 3.34 × 10−3 | sv40-miR-S1-5p | 2.56 | 3.17 × 10−3 |
| hsa-miR-142-3p | 3.15 | 3.04 × 10−3 | hsa-miR-335 | 2.17 | .045 |
| hsa-miR-142-5p | 2.91 | 3.95 × 10−3 | hsa-mir-615 | 2.00 | .03 |
| hsa-miR-29b | 2.36 | 7.71 × 10−3 | hsa-miR-363* | 1.94 | .012 |
| hsa-miR-101 | 2.34 | 8.77 × 10−3 | mmu-miR-10b | 1.81 | .012 |
| hsa-miR-525 | 2.18 | 8.19 × 10−3 | mmu-mir-691 | 1.80 | .01 |
| hsa-mir-622 | 2.16 | .019 | hsa-miR-299-3p | 1.77 | .018 |
| hsa-miR-487b | 2.15 | 9.93 × 10−3 | mmu-miR-346 | 1.72 | 8.92 × 10−3 |
| hsa-miR-126* | 2.11 | 9.92 × 10−3 | mmu-mir-615 | 1.71 | .01 |
| hsa-miR-199b | 2.10 | .012 | hsa-miR-320 | 1.61 | .031 |
| mmu-mir-804 | 2.09 | 0.016 | hcmv-miR-UL70-3p | 1.56 | .03 |
| hsa-miR-486 | 2.08 | 3.63 × 10−3 | kshv-miR-K12-10a | 1.54 | .038 |
| hsa-miR-197 | 1.99 | 6.74 × 10−3 | hsa-miR-518f* | 1.54 | .029 |
| hsa-miR-19a | 1.98 | .01 | mmu-mir-423 | 1.51 | .044 |
| rno-miR-343 | 1.97 | .015 | hsa-mir-765 | 1.47 | .048 |
| hsa-miR-15a | 1.97 | .011 | |||
| hsa-miR-19b | 1.96 | .012 | |||
| hsa-miR-29c | 1.87 | .026 | |||
| hsa-miR-27a | 1.74 | .017 | |||
| hsa-miR-374 | 1.71 | .027 | |||
| hsa-miR-30e-5p | 1.70 | .044 | |||
| hsa-miR-29a | 1.68 | .029 | |||
| hsa-mir-662 | 1.66 | .02 | |||
| mmu-miR-489 | 1.63 | .025 | |||
| hsa-mir-421 | 1.60 | .03 | |||
| hsa-miR-20a | 1.60 | .025 | |||
| hsa-miR-106a | 1.59 | .022 | |||
| hsa-miR-203 | 1.58 | .032 | |||
| hsa-miR-20b | 1.58 | .023 | |||
| ebv-mir-BART3 | 1.57 | .022 | |||
| hsa-miR-17-5p | 1.55 | .034 | |||
The fold increase is the average expression of each microRNA in CD34+ cells from 3 rhesus macaque given plerixafor plus G-CSF divided by the average expression of the same microRNA in CD34+ cells from 3 monkeys given G-CSF.
The fold increase is the average expression of each microRNA CD34+ cells from 3 rhesus given G-CSF divided by the average expression of the same microRNA in CD34+ cells from 3 monkeys given plerixafor plus G-CSF.
miRs that were differently expressed between plerixafor plus G-CSF–mobilized CD34+ cells and those mobilized with plerixafor alone
| Increased in plerixafor + G-CSF–mobilized CD34+ cells . | Increased in plerixafor-mobilized CD34+ cells . | ||||
|---|---|---|---|---|---|
| miR . | Fold increase* . | P . | miR . | Fold increase† . | P . |
| hsa-miR-21 | 5.98 | 4.29 × 10−5 | hsa-mir-615 | 2.06 | .0305 |
| hsa-miR-126* | 3.91 | 1.6 × 10−3 | mmu-miR-329 | 2.01 | .0210 |
| hsa-miR-126 | 3.34 | 2.90 × 10−4 | sv40-miR-S1-5p | 1.88 | .0129 |
| hsa-miR-142-3p | 2.88 | 8.98 × 10−4 | kshv-miR-K12-1 | 1.79 | .0143 |
| hsa-miR-142-3p | 2.75 | 1.03 × 10−3 | kshv-miR-K12-10a | 1.78 | .0103 |
| hsa-miR-29b | 2.57 | 2.92 × 10−3 | mmu-mir-709 | 1.69 | .0235 |
| hsa-miR-142-5p | 2.56 | 1.22 × 10−3 | rno-miR-327 | 1.68 | .0155 |
| hsa-miR-101 | 2.54 | 3.01 × 10−3 | hsa-miR-518f* | 1.63 | .0312 |
| hsa-miR-29c | 2.44 | 8.59 × 10−3 | hsa-mir-612 | 1.62 | .0353 |
| hsa-miR-345 | 2.39 | 8.68 × 10−3 | hcmv-miR-UL70-3p | 1.58 | .0457 |
| hsa-miR-424 | 2.34 | .011 | mmu-mir-705 | 1.53 | .0365 |
| hsa-miR-27a | 2.21 | 9.84 × 10−3 | ebv-mir-BART6 | 1.51 | .0475 |
| hsa-miR-148a | 2.21 | .044 | hsa-mir-557 | 1.51 | .0404 |
| hsa-miR-29a | 2.15 | 5.57 × 10−3 | hsa-miR-363* | 1.50 | .0440 |
| mmu-mir-804 | 2.09 | .01 | |||
| hsa-miR-146b | 2.07 | 5.72 × 10−3 | |||
| hsa-miR-19a | 2.07 | 3.23 × 10−3 | |||
| hsa-miR-24 | 2.03 | .014 | |||
| hsa-miR-19b | 1.93 | 4.96 × 10−3 | |||
| hsa-miR-146a | 1.90 | .021 | |||
| hsa-miR-374 | 1.72 | .025 | |||
| hsa-miR-487b | 1.71 | .043 | |||
| hsa-miR-486 | 1.71 | .034 | |||
| hsa-miR-15a | 1.71 | .029 | |||
| hsa-miR-18a | 1.69 | .025 | |||
| rno-miR-374 | 1.62 | .03 | |||
| hsa-miR-30e-5p | 1.62 | .04 | |||
| hsa-miR-20a | 1.59 | .04 | |||
| hsa-miR-106a | 1.53 | .043 | |||
| Increased in plerixafor + G-CSF–mobilized CD34+ cells . | Increased in plerixafor-mobilized CD34+ cells . | ||||
|---|---|---|---|---|---|
| miR . | Fold increase* . | P . | miR . | Fold increase† . | P . |
| hsa-miR-21 | 5.98 | 4.29 × 10−5 | hsa-mir-615 | 2.06 | .0305 |
| hsa-miR-126* | 3.91 | 1.6 × 10−3 | mmu-miR-329 | 2.01 | .0210 |
| hsa-miR-126 | 3.34 | 2.90 × 10−4 | sv40-miR-S1-5p | 1.88 | .0129 |
| hsa-miR-142-3p | 2.88 | 8.98 × 10−4 | kshv-miR-K12-1 | 1.79 | .0143 |
| hsa-miR-142-3p | 2.75 | 1.03 × 10−3 | kshv-miR-K12-10a | 1.78 | .0103 |
| hsa-miR-29b | 2.57 | 2.92 × 10−3 | mmu-mir-709 | 1.69 | .0235 |
| hsa-miR-142-5p | 2.56 | 1.22 × 10−3 | rno-miR-327 | 1.68 | .0155 |
| hsa-miR-101 | 2.54 | 3.01 × 10−3 | hsa-miR-518f* | 1.63 | .0312 |
| hsa-miR-29c | 2.44 | 8.59 × 10−3 | hsa-mir-612 | 1.62 | .0353 |
| hsa-miR-345 | 2.39 | 8.68 × 10−3 | hcmv-miR-UL70-3p | 1.58 | .0457 |
| hsa-miR-424 | 2.34 | .011 | mmu-mir-705 | 1.53 | .0365 |
| hsa-miR-27a | 2.21 | 9.84 × 10−3 | ebv-mir-BART6 | 1.51 | .0475 |
| hsa-miR-148a | 2.21 | .044 | hsa-mir-557 | 1.51 | .0404 |
| hsa-miR-29a | 2.15 | 5.57 × 10−3 | hsa-miR-363* | 1.50 | .0440 |
| mmu-mir-804 | 2.09 | .01 | |||
| hsa-miR-146b | 2.07 | 5.72 × 10−3 | |||
| hsa-miR-19a | 2.07 | 3.23 × 10−3 | |||
| hsa-miR-24 | 2.03 | .014 | |||
| hsa-miR-19b | 1.93 | 4.96 × 10−3 | |||
| hsa-miR-146a | 1.90 | .021 | |||
| hsa-miR-374 | 1.72 | .025 | |||
| hsa-miR-487b | 1.71 | .043 | |||
| hsa-miR-486 | 1.71 | .034 | |||
| hsa-miR-15a | 1.71 | .029 | |||
| hsa-miR-18a | 1.69 | .025 | |||
| rno-miR-374 | 1.62 | .03 | |||
| hsa-miR-30e-5p | 1.62 | .04 | |||
| hsa-miR-20a | 1.59 | .04 | |||
| hsa-miR-106a | 1.53 | .043 | |||
The fold increase in the average expression of each microRNA in CD34+ cells from 3 rhesus macaque given plerixafor plus G-CSF divided by the average expression of the same microRNA in CD34+ cells from 3 monkeys given plerixafor.
The fold increase in the average expression of each microRNA in CD34+ cells from in 3 rhesus macaque given plerixafor divided by the average expression of the same microRNA in CD34+ cells from monkeys given plerixafor plus G-CSF.
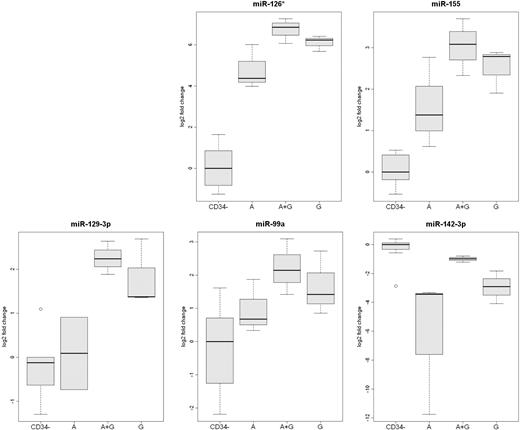
The expression of miR-126*, miR-155, miR-142-3p, miR-129-3p, and miR-99a were also measured by qRT-RCR (Figure 4). This analysis confirmed that the expression of miR-126* and miR-142-3p was greater in plerixafor plus G-CSF–mobilized CD34+ cells than in the other 2 types of CD34+ cells. In addition, the analysis confirmed that the expression of miR-99a, miR-129-3p, and miR-155 was greater in G-CSF–mobilized CD34+ cells than plerixafor-mobilized CD34+ cells.
Analysis of differentially expressed mobilized CD34+ cell miRs by qRT-PCR. The expression of miR-126*, miR-129-3p, miR-142-3p, miR-99, and miR-155 was analyzed by qRT-PCR in CD34+ cells from 3 rhesus macaque mobilized by plerixafor (A), G-CSF (G), or plerixafor plus G-CSF (A+G). The expression of the 5 microRNAs in leukocytes that were not absorbed by anti-CD34 from the 9 mobilized PBSC concentrates (CD34−) are shown as a control.
Analysis of differentially expressed mobilized CD34+ cell miRs by qRT-PCR. The expression of miR-126*, miR-129-3p, miR-142-3p, miR-99, and miR-155 was analyzed by qRT-PCR in CD34+ cells from 3 rhesus macaque mobilized by plerixafor (A), G-CSF (G), or plerixafor plus G-CSF (A+G). The expression of the 5 microRNAs in leukocytes that were not absorbed by anti-CD34 from the 9 mobilized PBSC concentrates (CD34−) are shown as a control.
Discussion
Plerixafor has recently been approved by the US Food and Drug Administration for use with G-CSF to mobilize PBSCs for autologous transplantation. Because plerixafor is now clinically available, it will also likely be used to mobilize stem cells in the 1% to 5% of allogeneic donors who mobilize HSCs poorly in response to G-CSF.21,22 The number of CD34+ cells mobilized by the combination of plerixafor and G-CSF is significantly greater than either agent alone. Because the success of allogeneic transplants is highly dependent on rapid myeloid engraftment and immune reconstitution in the transplant recipient, it is important to determine whether the critical in vivo biological activity or potency of HSCs mobilized by plerixafor and plerixafor plus G-CSF is similar to those mobilized by G-CSF. We used a surrogate assay for potency testing, global gene and microRNA expression analysis, to compare CD34+ cells mobilized by plerixafor, G-CSF, and the combination of the 2 agents.
Although many of the same genes were up-regulated in CD34+ cells mobilized by either plerixafor or G-CSF, the expression of a large number of genes also differed between CD34+ cells mobilized by these 2 agents. G-CSF–mobilized CD34+ cells were more likely to express neutrophil and mononuclear phagocyte genes than those mobilized by plerixafor. In addition, G-CSF–mobilized CD34+ cells also expressed high levels of miR-155, which is expressed in maturing dendritic cells.20 In contrast, plerixafor-mobilized CD34+ cells were more likely to express B-, T-, and mast cell genes. These results suggest that G-CSF mobilizes a greater proportion of HSCs that are committed toward myeloid differentiation, whereas those mobilized by plerixafor are more likely to be committed toward B, T, and mast cells.
Surprisingly, the combination of plerixafor plus G-CSF mobilized a population of CD34+ cells that were distinct from CD34+ cells mobilized by either agent alone. CD34+ cells mobilized by plerixafor plus G-CSF were more likely to express B-cell and T-cell genes than G-CSF–mobilized and plerixafor-mobilized CD34+ cells. In addition, plerixafor plus G-CSF–mobilized CD34+ cells were also remarkable for their increased expression of miR142-3p and miR-142-5p. miR-142-3p and miR-142-5p are highly expressed in T cells,23 and miR-142-3p has been found to be increased in childhood B-cell precursor acute lymphoblastic leukemia.24 The results suggest plerixafor plus G-CSF mobilizes a greater proportion of B- and T-cell precursors than either G-CSF or plerixafor alone.
It is not clear if plerixafor, G-CSF, or their combination mobilizes a more primitive HSC. CD34+ cells mobilized by plerixafor expressed greater levels of CXCR4 and FLT3 than G-CSF–mobilized CD34+ cells. Increased expression of CXCR4 in the plerixafor-mobilized population is not surprising and is consistent with the nature of the mobilization protocol, that is, interfering with the binding of CXCR4 and CXCL12. Thus, CD34+ cells mobilized with plerixafor alone or in combination with G-CSF should be expressing CXCR4. FLT3 is a member of the receptor tyrosine kinase family and plays an important role in hematopoietic homeostasis.25 It is expressed on both common lymphoid and multipotent progenitors. Higher FLT3 expression was observed in those CD34+ cells mobilized with plerixafor alone or with the combination plerixafor plus G-CSF than with G-CSF alone. These increases in expression were confirmed by qRT-PCR analysis. We also found that the expression of COX2 was up-regulated in plerixafor plus G-CSF–mobilized CD34+ cells. COX2 is an inducible enzyme that metabolizes arachidonic acid to a number of prostaglandins, including prostaglandin E2 (PGE2). PGE2 has been found to be important in hematopoiesis. In zebrafish PGE2 increases HSC numbers, and in mice it enhances spleen colony-forming units and the frequency of long-term repopulating cells.26 PEG2 also has been found to enhance the expression of CXCR4 on mouse CD34+ cells.27 This suggests that greater quantities of these progenitors are among plerixafor-mobilized and plerixafor plus G-CSF–mobilized CD34+ cells than G-CSF–mobilized CD34+ cells.
G-CSF–mobilized CD34+ cells, however, have increased levels of miR-126, miR-126*, and miR-10a* compared with plerixafor-mobilized CD34+ cells. miR-126 and miR-10a have previously been found to be expressed by CD34+ cells.19 In addition to being expressed by HSCs, miR-126 is also expressed by megakaryocytes and endothelial cells and is important in angiogenesis.28,29 miR-126* and miR-10a* are mirror images of miR-126 and miR-10a, respectively, and they are derived from the same pre-miR stem loop, so it is not surprising that both miR-126 and miR-126* were expressed in G-CSF–mobilized CD34+ cells. These 2 miR, however, target different genes, and they may play a different role in HSC function. We found that the levels of miR-126 and miR-126* were greater in plerixafor plus G-CSF–mobilized CD34+ cells than in the other types of CD34+ cells. Another miR whose expression was increased in G-CSF–mobilized CD34+ cells was miR-99a, which is also expressed by megakaryocytes.29 These results suggest that G-CSF–mobilized CD34+ cells may contain a wider range of lineage progenitors. Transplantation studies will be required to determine whether plerixafor, G-CSF, or the combination of these agents mobilizes a more primitive HSC.
The finding that plerixafor, G-CSF, and plerixafor plus G-CSF mobilize different types of CD34+ cells is consistent with other studies that found that different agents mobilized different populations of HSCs. In a mouse model, Pitchford and colleagues have found that G-CSF mobilized greater quantities of hematopoietic progenitor cells than plerixafor but plerixafor mobilized greater quantities of endothelial progenitor cells than G-CSF.30 They also found that the combination of G-CSF plus plerixafor mobilized greater quantities of hematopoietic progenitors than either agent alone, but not endothelial progenitors.
The results of this study have several implications for clinical HSC transplantation with mobilized PBSCs. Because plerixafor, G-CSF, and plerixafor plus G-CSF mobilize different subpopulations of CD34+ cells, the potency of PBSCs mobilized by these agents may differ. The rate of engraftment and profile of transplant-associated comorbidities may differ between recipients of allogeneic transplants with plerixafor-mobilized PBSCs and those who receive G-CSF–mobilized PBSCs. In addition, the dose of CD34+ cells required for successful engraftment may be different for plerixafor- and plerixafor plus G-CSF–mobilized PBSC concentrates. A preliminary study of allogeneic transplantations involving 20 HLA-compatible siblings has shown that plerixafor-mobilized PBSCs resulted in prompt neutrophil and platelet engraftment although a relatively low median dose of CD34+ cells was given, 2.9 × 106 per kilogram of recipient weight.4 This same study found that plerixafor-mobilized PBSCs contained greater quantities of CD3+, CD4+, and CD8+ cells than G-CSF–mobilized PBSCs, but the rates of acute and chronic graft versus host disease that were similar to those of historic controls who received G-CSF–mobilized PBSCs at the same institution.4 Our data suggest that if similar quantities of CD34+ cell are administered, those mobilized with G-CSF may result in faster neutrophil recovery and those mobilized with plerixafor or plerixafor plus G-CSF may have more rapid B-cell and T-cell recovery. Larger controlled studies will be required to assess similarities and differences in rates of engraftment, acute and chronic graft-versus-host disease, and immune reconstitution between plerixafor- and G-CSF–mobilized PBSCs. Clinical correlates will be required to determine the minimum and optimal CD34+ cell doses of plerixafor-mobilized and plerixafor plus G-CSF–mobilized PBSCs required for successful transplantations and to determine whether the rate and sequence of immune recovery differs among the mobilization regimens.
In conclusion, the composition of CD34+ cells in PBSC concentrates is dependent on the mobilization regimen, and the composition of CD34+ cells mobilized by a combination of agents is not a simple mixture of the cells mobilized by single agents. Unique expression profile signatures appear between populations. G-CSF, plerixafor, and plerixafor plus G-CSF mobilized different populations of CD34+ cells, but further in vitro and clinical studies are needed to compare the potency of those cells mobilized by these agents.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was funded by the Hematology Branch, National Heart, Lung, and Blood Institute, National Institutes of Health, and the Department of Transfusion Medicine, Clinical Center, National Institutes of Health.
National Institutes of Health
Authorship
Contribution: R.E.D. designed the study, analyzed data, and wrote the manuscript; P.J. helped design the study, performed studies, analyzed data, and wrote the manuscript; A.C.B., M.E.M., J.R., and E.W. performed studies and analyzed data; and D.F.S. helped design the study, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: A drug used in this study, plerixafor, was provided by Genzyme. The authors declare no competing financial interests.
Correspondence: David Stroncek, MD, Department of Transfusion Medicine, Clinical Center, National Institutes of Health, 10 Center Dr, Bldg 10, Rm 1C711, Bethesda, MD 20892-1184; e-mail: dstroncek@cc.nih.gov.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal