Abstract
The prognosis of fludarabine (F)–refractory chronic lymphocytic leukemia (CLL) is very poor, and underlying mechanisms are only partly understood. To assess the contribution of p53 abnormalities to F-refractory CLL, we studied TP53 mutations in the CLL2H trial (subcutaneous alemtuzumab; n = 99). We found TP53 mutations in 37% of patients. Twelve of 67 (18%) patients without the 17p deletion showed a TP53 mutation and 50% showed evidence of uniparental disomy. A total of 75% of cases with TP53 mutation (without 17p−) showed clonal evolution/expansion. TP53 mutations had no impact on overall survival (P = .48). CLL with the 17p deletion or TP53 mutation showed very low miR-34a expression. To investigate the mechanisms underlying refractory CLL beyond p53, we studied cases without 17p−/TP53 mutation in detail. In several paired samples before and after F-refractory disease, no change in p21/p53 induction was observed after DNA damage. Although TP53 mutations and 17p deletions are found in a high proportion of F-refractory CLL, more than half of the cases cannot be explained by p53 defects (deletion or mutation), and alternative mechanisms need to be investigated. Alemtuzumab is effective irrespective of genetic high-risk subgroups with TP53 mutations. These clinical trials are registered at www.clinicaltrials.gov as #NCT00274976.
Introduction
Independent of the treatment chosen, none of the randomized first-line trials has as yet shown improved overall survival (OS) resulting from a specific therapy in chronic lymphocytic leukemia (CLL).1-3 Indeed, when the most important molecular prognostic markers are included in multivariate models with the treatment modalities, biologic disease characteristics (thymidine kinase, β2-microglobulin, unmutated VH status, 11q and 17p deletion) appear more important in determining outcome than initial treatment choice, even if single-center series suggest that outcome has improved over time with intensification of treatment.2,4-7 One way to improve treatment results may thus be the early tailoring of treatment according to the biologic disease profile. These patients may be best treated with agents that act differently from chemotherapy as, for example, antibodies and targeted therapies.
The group of patients with a deletion of 17p13 have been shown repeatedly to respond particularly poorly to chemotherapy.2,4,8 The reason for drug resistance of CLL with the 17p deletion is probably caused by the inactivation of p53 by mutation of the remaining TP53 allele, which is found in the majority of cases. Although TP53 mutations have been known to occur in CLL since the early 1990s, their precise prognostic role, in particular when not accompanied by 17p deletion, has been less clear.8-15
In recent studies, TP53 mutation in the absence of 17p deletion was found in 4% to 5%.16-18 Survival was equally poor for patients with deletion 17p plus TP53 mutation, TP53 mutation only, and 17p deletion only. Other studies have suggested an intermediate prognosis of CLL with TP53 mutation in the absence of the 17p deletion.19 TP53 mutation was shown to be an independent predictor of poor survival or progression-free survival irrespective of 17p deletion.16-18
In addition to loss or mutation of TP53, other principal components of the DNA damage pathway have been reported to be deregulated by mutation or deletion.20-22 The functionality of the ATM/p53 pathway has been monitored by exposing CLL cells in vitro to DNA damage (eg, by irradiation) and assessing the inducibility of p21/p53, and more recent techniques include the activation of p53 by nutlins.23-25 Resistant CLL cells have also been shown to exhibit increased DNA repair capacity and deregulated nonhomologous end-joining.26,27 However, irrespective of the method used, only a proportion of drug-resistant cases is explained by the currently identified factors, such as p53 and ATM. This is in part because few studies have specifically assessed the biology of F-refractory CLL in large and homogeneously treated cohorts.28,29 Because p53 loss by 17p deletion and/or TP53 mutation are the best documented causes for refractoriness to fludarabine or other chemotherapy, defects of other parts of the p53 pathway might underlie refractory disease. We have recently shown that a miR component of the p53 pathway is impaired not only in CLL cases with the 17p deletion but also in cases with F-refractory disease in the absence of 17p deletion, suggesting that this is indeed the case.30
In the present study, we analyzed the contribution of TP53 mutations and 17p deletion to F-refractory CLL in a multicenter treatment trial population with mature follow-up and detailed molecular characterization. The aim of the study was to precisely define the contribution of p53 defects to refractory CLL. In addition, we wanted to identify a cohort of patients with refractory disease that may not be explained by our current molecular profiling as a prerequisite to identify new mechanisms causing refractory CLL.
Methods
Study design and treatment schedule
Details on the clinical trial were reported separately.31 Briefly, CLL2H was a multicenter, open-label, phase 2 trial. Patients were eligible if they had a diagnosis of CLL requiring treatment (Binet-C or “active disease”) and had been refractory to a fludarabine-containing regimen (defined as no complete response/partial response [CR/PR] according to NCI criteria or disease progression within 6 months). Eligible patients were staged and registered, and samples were sent for central reference diagnostics before therapy. Patients were treated with subcutaneous alemtuzumab 3 × 30 mg weekly for up to 12 weeks (n = 103; #NCT00274976). The study was conducted with informed consent obtained in accordance with the Declaration of Helsinki and was approved by the Institutional Review Boards of all participating institutions.
For 41 patients, multiple separate samples (median 3; range, 2-7 samples) taken over a 148- to 3283-day interval (median, 938 days) were available for sequential analyses. We obtained material at diagnosis (n = 9), before first-line treatment (n = 22), before second-line treatment (n = 13), and during follow-up after alemtuzumab therapy in CLL2H (n = 16), respectively.
TP53 deletion and VH mutation status analyses
Fluorescence in situ hybridization analysis and VH sequencing were performed in all cases as previously described.8,32-34 To define the cutoff level for the diagnosis of TP53 deletion, hybridization experiments of blood specimens from probands were performed. The cutoff level was defined by the mean plus 3 SDs of the frequency of control cells exhibiting only one TP53 signal. A germline homology of 98% was used as the cutoff between VH mutated and VH unmutated cases.
TP53 mutation analyses by DHPLC
WAVE technology was used to identify samples containing mutations in exons 2 to 11 (coding region of p53; n = 99).16,35 WAVE analysis is based on temperature-dependent differences in column-retention time of polymerase chain reaction (PCR) products generated from homoduplex (wild-type) and heteroduplex (mutated) DNA, resulting in the presence of distorted or additional peaks if sequence variations are present. Each test sample was mixed with a known wild-type DNA control (20%). Assay conditions were optimized for analysis of each PCR fragment with samples previously characterized by DNA sequencing.16 All samples were denatured and cooled slowly to room temperature before WAVE analysis to maximize heteroduplex formation (95°C for 2 minutes, then decrease 1°C every 40 seconds to 45°C for 30 minutes). Oligonucleotide primers and denaturing high-performance liquid chromatography (DHPLC) conditions were chosen as previously described with small modifications (available on request).16,35
TP53 sequence analysis
We analyzed all samples with an aberrant DHPLC finding by automated fluorescent sequencing using the Big Dye Terminator Kit and ABI 3100 sequencer (Applied Biosystems; exons 2-11). The primers were designed to cover all coding exons and intron-exon boundaries (available on request). Cases with the 17p deletion in which no aberration was identified by DHPLC were sequenced in all coding exons to confirm the absence of a mutation.
RNA isolation and real-time reverse transcription-PCR
RNA was isolated using the miRVana PARIS Kit (Ambion). Expression of hsa-miR-34a was analyzed using the real-time reverse-transcribed (RT)–PCR Detection Kit and specific primer sets (Ambion) and SYBR Green Rox Mix (ABgene). RT reactions were performed according to the manufacturer's protocol. Small nuclear RNA U6 was used for normalization. RT-PCR was run on the ABI Prism 7700 Sequence Detector (Applied Biosystems). Relative expression was calculated using the 2−ΔΔCT method taking one patient sample with a very low expression as a reference unless otherwise stated.30,36
p53-p21 expression analyses by flow cytometry
Cells were harvested after in vitro culture 24 hours after irradiation (or unirradiated control), washed with phosphate-buffered saline, and stained with CD19-PC7 antibody (Beckman Coulter). Cells were then fixed in 2% cold paraformaldehyde and overnight in 80% ethanol at −20°C. Fixed cells were washed with bovine serum albumin in phosphate-buffered saline to prevent unspecific binding and then stained with p53-phycoerythrin antibody (clone DO-7; BD Biosciences) and p21-fluorescein isothiocyanate (Calbiochem) or the corresponding isotype controls. After incubation at 4°C for 1 hour, cells were analyzed on the FACSCalibur and data were analyzed using the CellQuest Pro software.
Statistical analysis
Endpoints were progression-free survival, time to treatment failure (TTTF), and OS from the time of CLL2H treatment start. Survival time distributions were plotted using Kaplan-Meier estimates. Median duration of follow-up was calculated according to the method of Korn. Treatment failure was defined as death, progression, or new treatment.
Group-wise comparisons of distributions of clinical, laboratory, and genetic data were performed with the Fisher exact test (categorical variables). All tests were 2-sided. An effect was considered statistically significant at P less than .05. The proportional hazards regression model of Cox was used to identify prognostic variables by multivariate analysis on 82 patients with all parameters available. All statistical analyses were performed with the statistical software environment R, Version 2.7.1, using the R package Design, Version 2.0-12.
Results
TP53 mutations and molecular genetics of F-refractory CLL
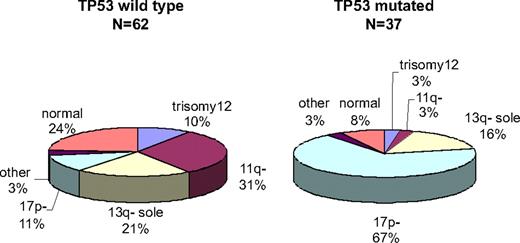
We detected 38 TP53 mutations in 37 of 99 (37%) patients with F-refractory CLL in the CLL2H trial. There was a high concordance rate between 17p deletion and mutation, with 25 of 32 patients with the 17p deletion also showing a mutation of the remaining allele (78%; Figure 1). The 7 cases with the 17p deletion in which no mutation was detected by DHPLC were all sequenced directly to confirm the results. In addition, 2 cases with the 17p deletion clone size less than 50% by fluorescence in situ hybridization were studied with a resequencing chip (Roche Molecular Systems) confirming the absence of a mutation. The assays used were not designed to detect large intragenic deletions.
Genomic profile of F-refractory CLL stratified by TP53 mutation. F-refractory CLL cases with (n = 37) and without (n = 62) TP53 mutation show a different hierarchical genetic profile. Cases with the 17p deletion are overrepresented in the mutation group (P < .001), whereas cases with the 11q deletion are overrepresented in the group without the TP53 mutation.
Genomic profile of F-refractory CLL stratified by TP53 mutation. F-refractory CLL cases with (n = 37) and without (n = 62) TP53 mutation show a different hierarchical genetic profile. Cases with the 17p deletion are overrepresented in the mutation group (P < .001), whereas cases with the 11q deletion are overrepresented in the group without the TP53 mutation.
Table 1 shows the characteristics of the whole CLL2H patient cohort in which TP53 mutations were analyzed stratified by TP53 mutation status. Of note, 12 of 67 (18%) patients without the 17p deletion showed a TP53 mutation. TP53 mutations in the absence of the 17p deletion were found in patients with deletion 13q single (as a sole aberration; n = 6), no aberration (n = 2), trisomy 12 (n = 1), but only 1 of 20 patients with deletion 11q without 17p− (1 of 12 with TP53 mutation vs 19 of 55 without TP53 mutation; P < .001; Figure 1). We found no association between the presence of TP53 mutations and the number of prior lines of treatment (data not shown).
Characteristics of patients analyzed for TP53 mutations within the CLL2H trial
| Hierarchical categorization of genomic aberrations . | Total (n = 99) . | WT TP53 (n = 62) . | Mutated TP53 (n = 37) . | P . | |||
|---|---|---|---|---|---|---|---|
| n . | % . | n . | % . | n . | % . | ||
| Trisomy 12 | 7 | 7 | 6 | 10 | 1 | 3 | .25 |
| 11q− | 20 | 20 | 19 | 31 | 1 | 3 | < .001 |
| 13q− sole | 19 | 19 | 13 | 21 | 6 | 16 | .6 |
| 17p− | 32 | 32 | 7 | 11 | 25 | 68 | < .001 |
| Other | 3 | 3 | 2 | 3 | 1 | 3 | 1 |
| Normal | 18 | 18 | 15 | 24 | 3 | 8 | .06 |
| VH mutation status | |||||||
| Mutated | 22 | 22 | 12 | 19 | 10 | 27 | |
| Unmutated | 70 | 71 | 44 | 71 | 26 | 70 | .6 |
| NA | 7 | 7 | 6 | 10 | 1 | 3 | |
| Response | |||||||
| CR | 4 | 4 | 3 | 5 | 1 | 3 | 1 |
| PR | 30 | 30 | 22 | 35 | 8 | 22 | .25 |
| SD | 37 | 37 | 24 | 39 | 13 | 35 | 1 |
| PD | 24 | 24 | 12 | 19 | 12 | 32 | .14 |
| NA | 4 | 4 | 1 | 2 | 3 | 8 | |
| Hierarchical categorization of genomic aberrations . | Total (n = 99) . | WT TP53 (n = 62) . | Mutated TP53 (n = 37) . | P . | |||
|---|---|---|---|---|---|---|---|
| n . | % . | n . | % . | n . | % . | ||
| Trisomy 12 | 7 | 7 | 6 | 10 | 1 | 3 | .25 |
| 11q− | 20 | 20 | 19 | 31 | 1 | 3 | < .001 |
| 13q− sole | 19 | 19 | 13 | 21 | 6 | 16 | .6 |
| 17p− | 32 | 32 | 7 | 11 | 25 | 68 | < .001 |
| Other | 3 | 3 | 2 | 3 | 1 | 3 | 1 |
| Normal | 18 | 18 | 15 | 24 | 3 | 8 | .06 |
| VH mutation status | |||||||
| Mutated | 22 | 22 | 12 | 19 | 10 | 27 | |
| Unmutated | 70 | 71 | 44 | 71 | 26 | 70 | .6 |
| NA | 7 | 7 | 6 | 10 | 1 | 3 | |
| Response | |||||||
| CR | 4 | 4 | 3 | 5 | 1 | 3 | 1 |
| PR | 30 | 30 | 22 | 35 | 8 | 22 | .25 |
| SD | 37 | 37 | 24 | 39 | 13 | 35 | 1 |
| PD | 24 | 24 | 12 | 19 | 12 | 32 | .14 |
| NA | 4 | 4 | 1 | 2 | 3 | 8 | |
WT indicates wild type; CR, complete remission; PR, partial remission; SD, stable disease; PD, progressive disease; and NA, not available.
The cohort showed an incidence of 17p deletion and/or TP53 mutation of 44% (44 of 99). Of the remaining 55 patients, 19 had an 11q deletion, leaving 36 patients (36%) in whom no adverse genomic marker (TP53 mutation, 17p or 11q deletion) was found despite refractory disease.
TP53 mutation profile in F-refractory CLL
TP53 mutations were mainly missense mutations, including some of the known hot spots (eg, codon 173, 220, 248, and 273), but we also observed a high incidence of frameshift mutations (supplemental Table 1; supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Of note, there were 2 mutations (p.Y220C, n = 3; and p.R209KfsX6, n = 4) that accounted for 18% of the overall mutation incidence (supplemental Table 1). The p.Y220C mutation is of particular interest because recent data suggest that the Y220C mutant may be a “druggable” target for developing novel anticancer drugs based on protein stabilization.37 Transitions (n = 16) were more common than transversions (n = 9), but transitions at cytosine-phosphate-guanosine (CpG) were very uncommon (n = 4), which is in keeping with the profile that has been reported for CLL (Table 2, supplemental Table 1, supplemental Figure 1).38
Characterization of TP53 mutations not accompanied by 17p deletion
| Sample no. . | Mutation details . | CLL genetics . | Clonal expansion . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation (amino acid) . | Exon . | UPD (mutated allele > 50%) . | WAF1 . | Cytogenetics . | IGVH mutation status . | Initial clone size (mutated allele), % . | Clone size at CLL2H entry (mutated allele), % . | Clonal evolution/expansion . | Time interval (clonal evolution/expansion), mo . | Treatment during clonal evolution/expansion . | |
| 2H-27 | p.K132E | 5 | NA | 0.6 | 13q14− | UM | 0 | 25 | Yes | 10 | FC |
| 2H-38 | p.W146CfsX251 | 5 | No | 13q14− | M | NA | 5 | No* | 23 | Alemtuzumab | |
| 2H-92 | p.V173M | 5 | Yes | 10.5 | 13q14− | UM | 0 | 70 | Yes | 98 | DexaBeam, TBI/Cy + auto-SCT, FC, R-FC, CHOP |
| 2H-25 | p.R209KfsX6 | 6 | Yes | +12q13, 13q14− | UM | 75 | 90 | Yes | 5 | CHOP, FC | |
| P121 | p.R209KfsX6 | 6 | Yes | 13q14 bi | M | NA | 80 | NA | NA | ||
| 2H-60 | p.Y220C | 6 | Yes | 1.2 | Normal | UM | NA | 90 | NA | NA | |
| UL-86 | p.N239S | 7 | No | 14.9 | 13q14 bi | M | NA | 10 | Yes | 41 | Alemtuzumab† |
| 2H-41 | p.S241F | 7 | No | 0 | Normal | UM | 0 | 50 | Yes | 61 | FC |
| 2H-106 | p.C242W | 7 | No | 14.2 | 11q23− | UM | 0 | 5 | Yes | 54 | FC, DexaBeam, TBI/Cy + auto-SCT, FC |
| 2H-17 | p.L252SfsX93 | 7 | Yes | 6q21−, 13q14− | UM | NA | 100 | NA | NA | ||
| 2H-18 | p.R273L | 8 | Yes | 0.9 | 13q14 bi | M | NA | 70 | NA | NA | |
| 2H-76 | p.R290H | 8 | No | 67.3 | Normal | UM | 50 | 40 | No | 11 | FC |
| Sample no. . | Mutation details . | CLL genetics . | Clonal expansion . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation (amino acid) . | Exon . | UPD (mutated allele > 50%) . | WAF1 . | Cytogenetics . | IGVH mutation status . | Initial clone size (mutated allele), % . | Clone size at CLL2H entry (mutated allele), % . | Clonal evolution/expansion . | Time interval (clonal evolution/expansion), mo . | Treatment during clonal evolution/expansion . | |
| 2H-27 | p.K132E | 5 | NA | 0.6 | 13q14− | UM | 0 | 25 | Yes | 10 | FC |
| 2H-38 | p.W146CfsX251 | 5 | No | 13q14− | M | NA | 5 | No* | 23 | Alemtuzumab | |
| 2H-92 | p.V173M | 5 | Yes | 10.5 | 13q14− | UM | 0 | 70 | Yes | 98 | DexaBeam, TBI/Cy + auto-SCT, FC, R-FC, CHOP |
| 2H-25 | p.R209KfsX6 | 6 | Yes | +12q13, 13q14− | UM | 75 | 90 | Yes | 5 | CHOP, FC | |
| P121 | p.R209KfsX6 | 6 | Yes | 13q14 bi | M | NA | 80 | NA | NA | ||
| 2H-60 | p.Y220C | 6 | Yes | 1.2 | Normal | UM | NA | 90 | NA | NA | |
| UL-86 | p.N239S | 7 | No | 14.9 | 13q14 bi | M | NA | 10 | Yes | 41 | Alemtuzumab† |
| 2H-41 | p.S241F | 7 | No | 0 | Normal | UM | 0 | 50 | Yes | 61 | FC |
| 2H-106 | p.C242W | 7 | No | 14.2 | 11q23− | UM | 0 | 5 | Yes | 54 | FC, DexaBeam, TBI/Cy + auto-SCT, FC |
| 2H-17 | p.L252SfsX93 | 7 | Yes | 6q21−, 13q14− | UM | NA | 100 | NA | NA | ||
| 2H-18 | p.R273L | 8 | Yes | 0.9 | 13q14 bi | M | NA | 70 | NA | NA | |
| 2H-76 | p.R290H | 8 | No | 67.3 | Normal | UM | 50 | 40 | No | 11 | FC |
The table summarizes genetic background of CLL cases with TP53 mutation, types of TP53 mutations including the predicted residual activity of p53 toward transcriptional targets (WAF1), and details on clonal expansion. UPD was assessed by analyzing clone size of the wild-type and mutant allele. Mutant allele in excess of 50% was considered evidence for UPD. We also summarize the results of follow-up investigations to assess clonal expansion/evolution and treatment during this process.
TBI indicates total body irradiation; and SCT, stem cell transplantation.
Case 2H-38 showed disappearance of the mutation after treatment with alemtuzumab as confirmed by DHPLC and sequencing.
Case UL-86 showed clonal expansion after initial reduction in clone size after alemtuzumab therapy.
TP53 mutations in the absence of 17p deletion
The prognostic impact of TP53 mutations in CLL when not accompanied by the 17p deletion has only recently been shown.6,16-18 We therefore analyzed incidence, clone size, and clonal evolution/expansion of these cases in detail in this F-refractory cohort. The demonstration of increasing clone size with or without selective pressure during (chemo-) therapy can be considered evidence for the biologic significance of TP53 mutations. The findings are summarized in Table 2. Sequential samples were available in 8 of 12 cases with a median interval of 32 months (range, 5-98 months) between the first and last sample investigated.
We observed increasing clone size of the mutated allele in 6 of 8 cases (Table 2). All 6 of these patients received combination chemotherapy: fludarabine and cyclophosphamide (FC; n = 6); cytoxan, hydroxyrubicin, oncovin, and prednisone (CHOP; n = 2); and autologous stem cell transplantation (n = 2; Table 2). There was one case with clonal expansion at the time of disease progression after initial disappearance of the mutation (at the time point of PR) after alemtuzumab treatment (Table 2). There were 2 cases with no evidence of clonal expansion and in one case the TP53 mutation was no longer detected after therapy with alemtuzumab. One of the cases lacking clonal evolution had a very low allele frequency (5%; Table 2, case 2H-38). The second case had a mutation in which the remaining activity of the mutated p53 can be expected to be comparatively high, which might explain the finding (Table 2, case 2H-76).
When we analyzed the ratio of mutated and unmutated allele in cases without the 17p deletion, we found that 6 of 12 cases had more than 50% mutated allele, which can be explained by uniparental disomy (UPD). Therefore, at least 50% of the cases with TP53 mutation in the absence of 17p− had a homozygous TP53 mutation caused by UPD. In these cases, the biologic impact can be considered to be equivalent to cases with the 17p deletion.
Mir-34a expression in F-refractory CLL
We studied the miR-34a (a microRNA component of the p53 pathway) expression in several cases (n = 35). The cases with F-refractory disease had a significantly lower baseline miR-34a expression than a control cohort of CLL cases without refractory disease, TP53 loss, or mutation (median miR-34a expression, 8.4; range, 0.3-115.8; vs 40.3; range, 6.7-337.8; P < .001).30 We found very low levels of miR-34a in cases with the 17p deletion and TP53 mutation (median, 0.9; range, 0.3-8.43), but also in cases with the TP53 mutation in the absence of the 17p deletion (median, 9.5; range, 1.05-17.75). Interestingly, cases with the 17p deletion and absence of TP53 mutation also showed levels similar to these 2 groups, suggesting that other mechanisms of TP53 inactivation (eg, intronic mutations) might be present in these cases (Figure 2). In contrast, the median expression of the other cases was higher (median, 24.6; range, 0.59-115.8). A comparison of the cases without TP53 mutation or 17p deletion showed a somewhat lower miR-34a expression (median, 24.6; 0.6-115.8) than in patients without refractory disease taken from a prior cohort (without TP53 loss/mutation; median, 40.3; P = .1). Although this difference was not statistically significant, we have previously used a distinction based on baseline and dynamic miR-34a expression, which could explain the findings.30
Deregulated miR component of the p53 pathway in refractory CLL. The expression of miR-34a in CLL is tightly linked to the presence of TP53 mutations or 17p deletion, but also to refractory disease. When separating subgroups based on the presence of TP53 mutation or 17p deletion, all subgroups with TP53 alteration show strikingly similar miR-34a expression. In cases with F-refractory CLL (without TP53 mutation or 17p−), miR-34a levels are on average higher, but a subset of cases show expression levels similar to TP53 mutation/17p− cases. Bar represents mean expression.
Deregulated miR component of the p53 pathway in refractory CLL. The expression of miR-34a in CLL is tightly linked to the presence of TP53 mutations or 17p deletion, but also to refractory disease. When separating subgroups based on the presence of TP53 mutation or 17p deletion, all subgroups with TP53 alteration show strikingly similar miR-34a expression. In cases with F-refractory CLL (without TP53 mutation or 17p−), miR-34a levels are on average higher, but a subset of cases show expression levels similar to TP53 mutation/17p− cases. Bar represents mean expression.
Mechanisms underlying F-refractory CLL in the absence of 17p deletion or TP53 mutation
In an effort to better understand the mechanisms underlying refractory disease in the absence of TP53 mutation or 17p deletion, we investigated serial samples with viably frozen cells regarding their response to DNA damage after irradiation. To this end, we assessed p21 and p53 induction after irradiation in 13 cases (17p− or TP53 mutation n = 5; no evidence of TP53 loss or mutation n = 8). The cases with TP53 mutation showed a typical profile (type A with high baseline p53 expression [missense mutations] and type B with low p53 baseline and failure to up-regulate p21 and p53 [frameshift mutations]).23 Of the cases without TP53 loss or mutation, half showed an abnormal p53 and/or p21 induction, including 2 cases with the 11q deletion, but also 2 with no aberration or trisomy 12. The other half showed low p53 baseline as well as a normal p21 and p53 induction, suggesting that this pathway is intact.
We also studied sequential samples obtained during the disease course to assess any potential alteration in the response to DNA damage coinciding with the development of F-refractory disease. For 5 patients (without 17p− or TP53 mutation), we studied samples before fludarabine/FC treatment and at the time of refractory disease. Interestingly, we did not observe significant changes in the p21/p53 response. This suggests that in vitro analysis of p53/p21 induction will only detect a proportion of refractory cases and other mechanisms must explain refractory disease in the remaining patients (Figure 3).
Functional assessment of p21/p53 induction after DNA damage may be normal in cases with F-refractory disease without the 17p deletion or TP53 mutation. (A) Normal p21 and p53 induction before and after development of F-refractory disease in CLL cells without the 17p deletion or TP53 mutation. The pattern of p21 and p53 expression/induction did not change with the occurrence of F-refractory disease: black represents expression without 5 Gy irradiation; gray, expression after 5 Gy irradiation. Both samples show a low baseline level and normal up-regulation of p21 and p53 after irradiation. (B) For comparison, an example of type A defect with high baseline p53 levels caused by a mutation of TP53. After induction of DNA damage, p21 is not up-regulated.
Functional assessment of p21/p53 induction after DNA damage may be normal in cases with F-refractory disease without the 17p deletion or TP53 mutation. (A) Normal p21 and p53 induction before and after development of F-refractory disease in CLL cells without the 17p deletion or TP53 mutation. The pattern of p21 and p53 expression/induction did not change with the occurrence of F-refractory disease: black represents expression without 5 Gy irradiation; gray, expression after 5 Gy irradiation. Both samples show a low baseline level and normal up-regulation of p21 and p53 after irradiation. (B) For comparison, an example of type A defect with high baseline p53 levels caused by a mutation of TP53. After induction of DNA damage, p21 is not up-regulated.
Clinical impact of TP53 mutations in F-refractory CLL treated with alemtuzumab
The presence or absence of TP53 mutations showed no significant impact on OS (P = .48), suggesting that alemtuzumab is active irrespective of TP53 mutation status (median OS, 21.3 months vs 13.6 months [TP53 mutation]). The TTTF was very similar in the groups with or without TP53 mutation (5.82 vs 4.85 months; P = .8), suggesting that the somewhat longer OS could be the result of higher sensitivity toward next therapy after CLL2H treatment in cases without TP53 mutation (Figure 4). When we separated groups according to 17p deletion, TP53 mutation without the 17p deletion, and patients with none of these changes, we found no impact of the genetic profile with respect to OS or TTTF (Figure 4).
TP53 mutations do not influence clinical endpoints in F-refractory CLL treated with alemtuzumab. (A) OS of patients with F-refractory CLL stratified by TP53 and 17p status (n = 99). The median survival for patients with and without TP53 mutation was 13.6 months versus 21.3 months, respectively. The groups with the 17p deletion, TP53 mutation without 17p−, and all other patients also showed similar OS (18.6 [17p−], 19.9 [TP53 mutation, no 17p−], and 19.1 months [neither TP53 mutation nor 17p−]). (B) The TTTF was very similar for patients with or without the TP53 mutation (median time to treatment failure, 5.8 vs 4.6 months) and irrespective of 17p status (median TTTF, 5.0 [17p−], 11.1 [no 17p−/TP53 mutation], and 5.3 months [neither]).
TP53 mutations do not influence clinical endpoints in F-refractory CLL treated with alemtuzumab. (A) OS of patients with F-refractory CLL stratified by TP53 and 17p status (n = 99). The median survival for patients with and without TP53 mutation was 13.6 months versus 21.3 months, respectively. The groups with the 17p deletion, TP53 mutation without 17p−, and all other patients also showed similar OS (18.6 [17p−], 19.9 [TP53 mutation, no 17p−], and 19.1 months [neither TP53 mutation nor 17p−]). (B) The TTTF was very similar for patients with or without the TP53 mutation (median time to treatment failure, 5.8 vs 4.6 months) and irrespective of 17p status (median TTTF, 5.0 [17p−], 11.1 [no 17p−/TP53 mutation], and 5.3 months [neither]).
We also tested the prognostic impact of TP53 mutations in a multivariate model, including other important factors (age, performance status, β2MG, and thymidine kinase). The TP53 mutation had no impact on OS (hazard ratio = 1.11; range, 0.68-1.84; Table 3). The overall response (CR and PR) rate for patients with TP53 mutation was 9 of 37 (24%) compared with 25 of 62 (40%; P = .19; Table 1).
Cox regression on overall survival
| . | P . | Hazard ratio . | 95% CI . |
|---|---|---|---|
| Thymidine kinase | .003 | 1.5 | 1.14-1.97 |
| Age (increments of 10 y) | .008 | 1.46 | 1.11-1.94 |
| ECOG > 1 | .03 | 2.02 | 1.06-3.84 |
| β2MG | .07 | 1.09 | 0.99-1.19 |
| WBC | .35 | 0.92 | 0.78-1.09 |
| TP53 mutation | .69 | 1.11 | 0.68-1.84 |
| . | P . | Hazard ratio . | 95% CI . |
|---|---|---|---|
| Thymidine kinase | .003 | 1.5 | 1.14-1.97 |
| Age (increments of 10 y) | .008 | 1.46 | 1.11-1.94 |
| ECOG > 1 | .03 | 2.02 | 1.06-3.84 |
| β2MG | .07 | 1.09 | 0.99-1.19 |
| WBC | .35 | 0.92 | 0.78-1.09 |
| TP53 mutation | .69 | 1.11 | 0.68-1.84 |
WBC indicates white blood count; CI, confidence interval; and β2MG, β2-microglobulin.
Discussion
The study presented here addressed 2 main questions: (1) what is the contribution of the p53 pathway defects to F-refractory CLL and (2) what are the clinical implications of TP53 mutations in the setting of refractory CLL when treated with alemtuzumab? We addressed these key questions in a large homogeneous, multicenter trial population with detailed clinical and genetic characterization as well as long follow-up.
As further evidence for the association of TP53 mutations with refractoriness to chemotherapy, we found a high incidence of TP53 abnormalities (deletion and TP53 mutation) in this cohort. Because patients were treated with a substance that acts independently of the p53 pathway, we did not observe the same prognostic impact that would be observed in a different clinical situation. In this cohort, thymidine kinase, age, and performance score were independent predictors for outcome. Part of the disappearance of the prognostic impact of TP53 mutation could also be mediated by the “selective pressure” of the cohort in which all patients had documented refractory disease. The lack of predictive power of the known genetic factors of poor prognosis has previously been documented after treatment with alemtuzumab in a smaller study of 36 patients reported by Lozanski et al.39 The question of whether the results are applicable to other clinical situations as, for example, first-line treatment with alemtuzumab remains to be determined.40 Although the comparison of OS was not significant, there was a shorter median OS of patients with TP53 mutation (13.6 months vs 21.3 months). This was in contrast with the almost identical TTTF. When we analyzed the survival from the next line of treatment after alemtuzumab, again survival was short for patients with the TP53 mutation (6.5 vs 12 months), suggesting that part of the effect may relate to the failure to respond to subsequent treatment.
Our study adds further evidence to the association of TP53 mutation (without 17p−) and poor prognosis. We show definite evidence of clonal expansion documented within single cases (5 of 8) under selective pressure of chemotherapy. In addition, the overall incidence of 18% of TP53 mutation without the 17p deletion, compared with 3% to 5% that has been reported in cohorts of unselected patients or patients with a first-line treatment indication, supports its biologic role. Interestingly, we observed evidence for UPD on 17p in at least 50% of cases with the TP53 mutation only. UPD leads to the duplication of the mutated TP53 allele, and a similar biologic and clinical consequence to cases with the 17p deletion (and TP53 mutation) can be assumed. UPD at 17p has been described in CLL and is associated with mutated TP53.6,41 The frequency of this phenomenon is currently unknown. This is particularly important because a detailed analysis with the aim to detect UPD in subclones is challenging. For the current study, we can only state that the incidence is at least 50%, although the incidence may potentially be higher. In our analysis of cases from the first-line treatment situation, the incidence was much lower (3 of 13), suggesting that UPD at 17p is associated with advanced disease.6
Although only a limited number of patients with TP53 mutation in the absence of 17p deletion could be studied sequentially, we observed some heterogeneity that should be cautiously interpreted because the time interval was variable and follow-up samples were not systematically collected. Nonetheless, we not only found evidence for clonal expansion with disappearance and reemergence after alemtuzumab treatment, but also a case in which the mutation disappeared after treatment (although only originally detectable with low allele frequency). The observations suggest that different factors may influence clonal evolution of TP53 mutation-bearing CLL (without 17p−).
As expected, most mutations were located in the DNA-binding domain and led to impaired transcriptional activity of p53. There were 2 mutations (p.Y220C [n = 3] and p.R209KfsX6 [n = 4]) that accounted for 18% of the overall mutation incidence, but a detailed analysis of the mutation profile would necessitate a much larger study. The p.Y220C mutation is of particular interest because recent data suggest that the Y220C mutant may be a “druggable” target for developing novel anticancer drugs based on protein stabilization.37
p53 has recently been shown to target miR-34a and miR-34b/c located in the chromosomal regions 1p36 and 11q23 (the critical region of deletion 11q in CLL), respectively.42 Importantly, the miR-34 family and particularly miR-34a have been shown to mediate some of the functional consequences of p53 activation, including apoptosis, cell cycle arrest, and senescence in different cell lines and primary cells.42-44 Currently, CDK 4&6, CCND1, cMET, Cyclin E2, MycN, and BCL2 are among candidate targets of the miR-34 family.42
We and others have previously shown that CLL with the 17p deletion shows a marked down-regulation of miR-34a.30,45,46 In addition, we established an association of low miR-34a expression and induction with both TP53 mutation and F-refractory CLL.30 Our current findings support these results. We found significantly lower miR-34a levels of the cohort compared with nonrefractory control CLL cases. Particularly striking were the similar expression levels of cases with TP53 loss or mutation irrespective of the particular aberration. These findings suggest that the aberrations may be more alike than different, and there is a growing evidence that the prognostic impact of 17p deletion is mediated solely by virtue of inactivating p53.16,18 Of particular interest was the finding that miR-34a levels were also low in a subset of cases without TP53 loss or mutations. Although the difference did not reach statistical significance, in our previous study we had used baseline and dynamic miR-34a expression, which may have served as a better separator and explain this. Still, the current findings place miR-34a among few differentially expressed genes in CLL with TP53 loss or mutation even in steady state. It has previously been shown that gene expression signatures of cases with TP53 loss/mutation only vary on exposure of the cells to stress.47,48
One of the most important future questions that can be derived from our current study is from where the refractoriness of cases without TP53 mutation or 17p deletion stems. We have addressed this important question by focusing on cases without 17p− and TP53 mutation with a more detailed analysis of dynamic p21/p53 expression after DNA damage. Although we observed abnormal induction in 4 of 8 cases (including 2 with the 11q deletion), we also found normal response to irradiation in the remaining 50%. Similarly, the response to irradiation did not change during the emergence of F-refractory disease. This suggests that, at least for a significant proportion of cases without TP53 mutation or 17p deletion, the p21/p53 induction assay may not detect abnormalities that identify refractory CLL. This finding is important because efforts are underway to assess the value of p21/p53 induction assay in earlier treatment situations. In addition to these practical considerations, the findings also suggest that impaired p21/p53 inductions are not a prerequisite for the emergence of refractory CLL. This is of particular interest because a growing number of assays aimed at detecting p53 dysfunction are being assessed as “surrogate markers” of poor prognosis.49 Based on our results, not all cases with F-refractory disease will exhibit impaired activation of p53 and its target.
The cohort with refractory disease without classic high-risk genetic profile (TP53 mutation, 17p−, 11q−) makes up 36% of the current study population. In our effort to identify new mechanisms underlying refractory CLL, this cohort will be particularly suited to discover new mechanisms of resistance beyond p53 defects.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by the Else Kröner Fresenius Stiftung (P20/07//A11/07), Bayer-Schering Pharma, Amgen, Global CLL Research Foundation, and German José Carreras Leukemia-Foundation (R06/28v).
Authorship
Contribution: T.Z. designed research, performed research, analyzed data, and wrote the paper; S.H., T.D., J.M., D.W. A.B., A.S., S.G., and D.M. performed research and analyzed data; R.B. and M.H. analyzed data; H.D. designed the research and analyzed data; and S.S. designed the research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: T.Z. has received honoraria for lectures from Bayer Schering Pharma. S.S. has received honoraria for lectures and advisory board attendance from Bayer Schering Pharma. The trial was supported by Bayer Schering Pharma and Amgen. The remaining authors declare no competing financial interests.
Correspondence: Stephan Stilgenbauer, Department of Internal Medicine III, University of Ulm, Albert-Einstein-Allee 23, 89081 Ulm, Germany; e-mail: stephan.stilgenbauer@uniklinik-ulm.de.



![Figure 4. TP53 mutations do not influence clinical endpoints in F-refractory CLL treated with alemtuzumab. (A) OS of patients with F-refractory CLL stratified by TP53 and 17p status (n = 99). The median survival for patients with and without TP53 mutation was 13.6 months versus 21.3 months, respectively. The groups with the 17p deletion, TP53 mutation without 17p−, and all other patients also showed similar OS (18.6 [17p−], 19.9 [TP53 mutation, no 17p−], and 19.1 months [neither TP53 mutation nor 17p−]). (B) The TTTF was very similar for patients with or without the TP53 mutation (median time to treatment failure, 5.8 vs 4.6 months) and irrespective of 17p status (median TTTF, 5.0 [17p−], 11.1 [no 17p−/TP53 mutation], and 5.3 months [neither]).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/13/10.1182_blood-2009-05-224071/4/m_zh89990942460004.jpeg?Expires=1767755628&Signature=Q4BagaL-LZ5WHg0maDx~UeaSVmQ88KRxDee-xAUmJ06ZSmUetPuAP2oU9QqoEYZjwUKBCTt7XaEqi62BMe4eFBy0GId4vpB1Ezm0BU2m8lOok3Kjf7ArcnjHawk6sazAU6Os~cO2DnNUPxV9YezaXKnlCLqdOzAPeeoMyFYZSrrCU4Vk4F5hS7V~JhHNEeQtOJSKIfVXHBgukAYewVhCyKKaIf9AI-P03HpBY5C3cIN86OIlAy4dS91a-V2kMriQpgVPcplYarhidfuZ9zdc8JaXOS~SkaD7TGa-WNXcZ8JF~c8yYuGmxvlv0BqaufJGoHQNzhQsfcpaHZc6HJibLw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)