Abstract
In imatinib-treated chronic myeloid leukemia (CML), secondary drug resistance is often caused by mutations in the BCR-ABL kinase domain (KD). As alternative therapies are available for imatinib resistance, early identification of mutations may prevent disease progression. Because most patients are routinely monitored by BCR-ABL quantitative polymerase chain reaction (PCR), it is important to define the optimal increase in BCR-ABL that should trigger mutation testing. Expert panels have provisionally recommended a 10-fold BCR-ABL increase as the trigger for mutation screening, acknowledging the lack of consensus. To address this question, we monitored 150 CML patients by quantitative PCR and DNA sequencing. Thirty-five different mutations were identified in 53 patients, and, during 22.5 months (median) of follow-up after sequencing, mutations were significantly predictive of shorter progression-free survival. An unbiased receiver operating characteristic analysis identified a 2.6-fold increase in BCR-ABL RNA as the optimal cutoff for predicting a concomitant KD mutation, with a sensitivity of 77% (94% if including subsequent samples). The 2.6-fold threshold approximated the analytic precision limit of our PCR assay. In contrast, transcript rise cutoffs of 5-fold or greater had poor diagnostic sensitivity and no significant association with mutations. We conclude that the currently recommended 10-fold threshold to trigger mutation screening is insensitive and not universally applicable.
Introduction
The most common mechanism of secondary (acquired) imatinib resistance in patients with chronic myeloid leukemia (CML) is point mutations in the BCR-ABL kinase domain (KD) that impair imatinib binding to its target. To date, more than 50 different KD mutations have been described that confer variable degrees of imatinib resistance (reviewed in Apperley1 and O'Hare et al2 ). The second-line BCR-ABL inhibitors nilotinib and dasatinib have shown activity in patients with secondary imatinib resistance, including patients with KD mutations. Current consensus recommendations are to perform mutation analysis if patients fail to achieve certain milestones of response, or experience loss of response.3-5 Because most patients are routinely monitored by BCR-ABL real-time quantitative reverse-transcription polymerase chain reaction (RQ-PCR), it is necessary to determine the optimal increase in BCR-ABL RNA that should trigger mutation testing, while minimizing both false-positive and false-negative test results. One study suggested a 2-fold rise of BCR-ABL RNA as an appropriate threshold,6 but this was not confirmed in another study.7 Recognizing the lack of a consensus, expert panels and the National Comprehensive Cancer Network (NCCN) guidelines have provisionally recommended mutation screening in cases with a 10-fold or greater increase of BCR-ABL RNA.4,5 To address this issue in an unbiased fashion, we have applied a receiver operating characteristic (ROC) analysis to determine the threshold BCR-ABL rise that optimizes KD mutation detection in a cohort of patients treated at Oregon Health & Science University (OHSU).
Methods
Patients
The 150 study subjects represent all consented patients with imatinib-treated CML followed at our institution that were serially monitored with BCR-ABL RQ-PCR and later, as part of this retrospective study, underwent successful KD mutation analysis of residual archival samples by DNA sequencing (average, 2.2 sequenced samples per patient; range, 1-8 samples). The majority of these patients (n = 98; 65%) were diagnosed more than 6 months before initiating imatinib therapy (Table 1), and 92% of these had received previous interferon-alpha treatment. Mutation screening was undertaken in 101 of these patients because BCR-ABL RNA had increased at least 3-fold between successive samples during treatment (deemed an “RQ-PCR relapse”). In these 101 patients, the best molecular response before the RQ-PCR relapse was roughly evenly distributed among 4 response categories, with a greater than 3 log-drop in 22 patients, a 2 to 3 log-drop in 25 patients, a 1 to 2 log-drop in 33 patients, and a less than 1 log-drop in 21 patients. Of the 194 total samples with RQ-PCR relapse, 148 (76%) had residual samples available for sequencing. The 49 other patients were randomly selected for retrospective sequencing of residual archival samples from 137 patients who had never experienced a greater than 3-fold rise in BCR-ABL RNA during treatment.
Demographic characteristics of study subjects
| . | RQ-PCR rise greater than 3-fold, n = 101 . | No RQ-PCR rise greater than 3-fold, n = 49 . | P . | All patients, N = 150 . |
|---|---|---|---|---|
| Median age, y (range) | 51.1 (16-79) | 47.8 (23-74) | .45 | 50.6 (16-79) |
| Sex, % male | 56 | 47 | .15 | 53 |
| Median months from leukemia diagnosis to imatinib start (range) | 21 (0-161) | 6 (0-155) | .03 | 17 (0-161) |
| Patients with “early” CML diagnosis, fewer than 6 mo before imatinib, % | 26 | 53 | .002 | 35 |
| Baseline diagnosis of chronic-phase CML/ accelerated-phase/blast crisis, % | 79/20/1 | 90/10/0 | .31 | 82/17/1 |
| Median months of follow-up, after imatinib (range) | 48 (11-80) | 40 (7-75) | 46 (7-80) | |
| Median no. of laboratory monitoring visits (range) | 9 (2-21) | 7 (2-18) | .76 | 8 (2-21) |
| Median months between monitoring visits (range) | 3.9 (1.6-8.3) | 4.0 (2.3-7.8) | .66 | 3.9 (1.6-8.3) |
| CCR ever achieved, % | 58 | 82 | .003 | 66 |
| MCR (< 35% Ph) never achieved, % | 24 | 10 | .051 | 19 |
| Imatinib treatment failure, % (primary resistance)* | 60 | 37 | .11 | 53 |
| MMR ever achieved, % | 39 | 57 | .019 | 45 |
| Median RQ-PCR log-drop at time of DNA sequencing (range) | 0.77 (−1.0 to 2.6) | 1.8 (−0.4 to 5.2) | < .001 | 1.0 (−1.0 to 5.2) |
| CCR (MCR) at time of sequencing, % | 28 (49) | 58 (75) | .007 | 37 (57) |
| Complete hematologic response at time of sequencing, % | 80 | 90 | .17 | 83 |
| Kinase domain mutation, no. (%) | 43 (43) | 10 (20) | .010 | 53 (35) |
| . | RQ-PCR rise greater than 3-fold, n = 101 . | No RQ-PCR rise greater than 3-fold, n = 49 . | P . | All patients, N = 150 . |
|---|---|---|---|---|
| Median age, y (range) | 51.1 (16-79) | 47.8 (23-74) | .45 | 50.6 (16-79) |
| Sex, % male | 56 | 47 | .15 | 53 |
| Median months from leukemia diagnosis to imatinib start (range) | 21 (0-161) | 6 (0-155) | .03 | 17 (0-161) |
| Patients with “early” CML diagnosis, fewer than 6 mo before imatinib, % | 26 | 53 | .002 | 35 |
| Baseline diagnosis of chronic-phase CML/ accelerated-phase/blast crisis, % | 79/20/1 | 90/10/0 | .31 | 82/17/1 |
| Median months of follow-up, after imatinib (range) | 48 (11-80) | 40 (7-75) | 46 (7-80) | |
| Median no. of laboratory monitoring visits (range) | 9 (2-21) | 7 (2-18) | .76 | 8 (2-21) |
| Median months between monitoring visits (range) | 3.9 (1.6-8.3) | 4.0 (2.3-7.8) | .66 | 3.9 (1.6-8.3) |
| CCR ever achieved, % | 58 | 82 | .003 | 66 |
| MCR (< 35% Ph) never achieved, % | 24 | 10 | .051 | 19 |
| Imatinib treatment failure, % (primary resistance)* | 60 | 37 | .11 | 53 |
| MMR ever achieved, % | 39 | 57 | .019 | 45 |
| Median RQ-PCR log-drop at time of DNA sequencing (range) | 0.77 (−1.0 to 2.6) | 1.8 (−0.4 to 5.2) | < .001 | 1.0 (−1.0 to 5.2) |
| CCR (MCR) at time of sequencing, % | 28 (49) | 58 (75) | .007 | 37 (57) |
| Complete hematologic response at time of sequencing, % | 80 | 90 | .17 | 83 |
| Kinase domain mutation, no. (%) | 43 (43) | 10 (20) | .010 | 53 (35) |
RQ-PCR indicates real-time quantitative reverse-transcription polymerase chain reaction; CML, chronic myeloid leukemia; CCR, complete cytogenetic response; MCR, major cytogenetic response; and MMR, major molecular response.
Defined, per European LeukemiaNet (ELN) criteria,3 as failure to reach MCR (< 35% Ph-positivity) by 12 months or CCR by 18 months.
In this cohort selectively enriched for patients with rising BCR-ABL RNA levels, approximately half of the patients (53%) had primary imatinib resistance (Table 1), that is, they met the criteria for failure as defined by the European LeukemiaNet (ELN) recommendations3 (less than a major cytogenetic response [< 35% Ph-positivity] by 12 months or less than a complete cytogenetic response [CCR] by 18 months). In contrast, secondary (also known as “acquired”) imatinib resistance was defined as a loss of response, after an initial response, and, in this paper, was equivalent to “disease progression.” Disease progression (secondary resistance) criteria were defined as per our previous studies,8,9 and were identical to those from the International Randomized Study of Interferon versus STI571 (IRIS) imatinib treatment trial.10 These criteria included progression to accelerated or blastic phase, loss of a complete hematologic response, or loss of a complete cytogenetic response (CCR). As our prior studies,8,9 (but not IRIS),10 were restricted to patients who had achieved a CCR, an additional disease progression criterion for the present study was the loss of a major cytogenetic response—as defined in the IRIS study10 as an increase in Ph-positive bone marrow cells in metaphase by at least 30 percentage points.
The time from imatinib initiation to the initial mutation screen was no different (P > .2) for the patients with (average, 27 months) versus those without (average, 24 months) a 3-fold rise in BCR-ABL RNA. We chose 3-fold as our initial threshold, as we have previously found that a 3.2-fold (half-log) increase in BCR-ABL RNA is predictive of disease progression—as defined by hematologic and cytogenetic (not molecular) criteria.8 Furthermore, our BCR-ABL RQ-PCR assay can, with 95% confidence, distinguish serial changes in BCR-ABL RNA of greater than or equal to 2.9-fold.8 For comparison, if we had chosen a higher 5-fold or 10-fold threshold for defining RQ-PCR relapse, as has been recommended by expert consensus groups,4,5 the fraction of our patients fulfilling RQ-PCR relapse criteria would have decreased from 67% (3-fold) to 50% (5-fold) or 29% (10-fold). As the mutation analyses were performed retrospectively, no treatment alterations were made based on the results, and imatinib was the only tyrosine kinase inhibitor used during follow-up. As per our previous study, the dose of imatinib was initially 400 mg/day in patients with chronic-phase CML, and 600 mg/day in those with accelerated phase.8 However, because in some patients imatinib dose information was not comprehensively recorded at every visit throughout follow-up, we cannot definitively rule out transient imatinib dose reductions (or noncompliance) as contributing to the rise in RQ-PCR levels in some patients. The study was approved by the Institutional Review Board at OHSU, with informed consent provided according to the Declaration of Helsinki.
Disease monitoring
Patients were monitored with complete blood counts, bone marrow morphology, and bone marrow cytogenetics. Real-time quantitative RT-PCR (RQ-PCR) for BCR-ABL RNA relative to the G6PDH reference RNA was performed as previously described8,9 (3.9-month median interval between visits; Table 1). To determine the relative reduction of transcripts compared with a standardized baseline value,4,11 the BCR-ABL to G6PDH transcript ratio was compared with the laboratory-specific baseline transcript ratio and expressed as a “log-drop” from baseline (on a base-10 log scale), as described.8,9 Screening for mutations in the ABL KD was performed by bidirectional direct DNA sequencing of BCR-ABL RT-PCR products, as previously described, with an estimated detection sensitivity of 20%.8
Statistics
Continuous variables were compared with the Mann-Whitney (Wilcoxon) rank sum test (for 2 groups) or the nonparametric Kruskal-Wallis test (for 3 or more groups). Categoric variables were compared with the χ2 or Fisher exact test, and the Mantel-Haenszel test was used to adjust for baseline molecular and cytogenetic response levels. A Cox proportional hazard regression model was used to determine the relative hazard ratio (for disease progression) and define the time-dependent disease progression risk. The Kaplan-Meier method was used to determine the cumulative probability of progression-free survival. Differences in progression-free survival were compared using the log-rank test. An ROC curve analysis was used to determine the threshold value of rising BCR-ABL RNA levels that optimally predicted a concomitant BCR-ABL KD point mutation.12 All reported P values are 2-sided, and P values less than .05 were considered significant. The Statview software program (SAS) was used for all statistical calculations, except for the ROC curve analysis, which was performed with SPSS (SPSS Inc).
Results
Kinase domain mutations
Table 1 summarizes the key demographic features of patients with and without a 3-fold RQ-PCR rise. RQ-PCR relapses were significantly more frequent in patients with longer lag times to imatinib treatment, those failing to achieve a complete cytogenetic response (CCR) or major molecular response (MMR, 3 log reduction in BCR-ABL RNA), and those with lesser degrees of cytogenetic or molecular responses at the time of sequencing. Thirty-five different KD mutations, 5 of which were novel, were detected in 53 patients (35%), a median of 28 months (range, 3-57 months) after imatinib initiation (Table 2). Forty-three of these mutation-positive patients belonged to the higher-risk group of 101 subjects with a 3-fold RQ-PCR rise (P = .01). Mutations were no more likely to develop, however, in patients with, versus those without, prior primary imatinib resistance (P > .8). The average number of mutation-positive samples was 1.9 (maximum 6), and 8 patients had more than 1 mutation type at some time during treatment (2 with 4, 1 with 3, and 5 with 2 different mutations). Fourteen samples (from the same 8 patients) had more than 1 detectable mutation type in the same sample (1 of which had 3 different mutations). In 7 patients (5 of whom had more than 1 detectable mutation at some time during treatment), a total of 11 mutant BCR-ABL clones were present only transiently (M244V [twice; 1 as a minor clone with a peak height on the sequencing trace exceeded by another allele], Y253H [twice; both as minor clones], L273M, E282G, M351T, F359I [minor clone], L387M [minor clone], S417Y [minor clone], E453K [minor clone]), and were undetectable in subsequent specimens. Of these 7 patients with transient mutations, 5 subsequently experienced disease progression, 1 with different mutations (E453K and S417Y, both as minor clones) than were previously detected (M244V). The most frequent mutation site was the ATP-binding loop (17 patients; Table 2). Other common mutations were M244V (8 patients), M351T (3 patients), F359V/I (6 patients), H396P/R (3 patients), and F486S (3 patients). The T315I mutation was found in only 2 patients, both of whom had no subsequent follow-up cytogenetic data to assess disease progression.
Distribution of kinase domain mutations in 53 patients
| Mutation . | No. of patients . | Imatinib sensitivity . |
|---|---|---|
| M244V | 8 | Intermediate |
| L248V | 1 | Intermediate |
| G250E | 4 | Intermediate |
| G251D | 1 | NA |
| Q252H | 2 | Intermediate |
| Y253F | 3 | Insensitive |
| Y253H | 3 | Insensitive |
| E255K | 4 | Insensitive |
| L273M | 1 | NA |
| E279Z | 1 | NA* |
| E282G | 2 | NA |
| E292V | 1 | NA |
| T315I | 2 | Insensitive |
| R328M | 1 | NA* |
| M351T | 3 | Sensitive |
| E352D | 1 | NA* |
| E352G | 1 | NA |
| E355G | 1 | Intermediate |
| F359I | 2 | NA |
| F359V | 4 | Intermediate |
| L364I | 1 | Sensitive |
| V379I | 2 | Intermediate |
| F382L | 1 | NA |
| L387M | 2 | Sensitive |
| M388L | 1 | Sensitive |
| H396P | 1 | Sensitive |
| H396R | 2 | Intermediate |
| S417Y | 1 | NA |
| I418V | 1 | NA |
| E450G | 1 | NA |
| E453D | 1 | NA* |
| E453K | 1 | NA |
| E459G | 1 | NA |
| D482V | 1 | NA* |
| F486S | 3 | Intermediate |
| Mutation . | No. of patients . | Imatinib sensitivity . |
|---|---|---|
| M244V | 8 | Intermediate |
| L248V | 1 | Intermediate |
| G250E | 4 | Intermediate |
| G251D | 1 | NA |
| Q252H | 2 | Intermediate |
| Y253F | 3 | Insensitive |
| Y253H | 3 | Insensitive |
| E255K | 4 | Insensitive |
| L273M | 1 | NA |
| E279Z | 1 | NA* |
| E282G | 2 | NA |
| E292V | 1 | NA |
| T315I | 2 | Insensitive |
| R328M | 1 | NA* |
| M351T | 3 | Sensitive |
| E352D | 1 | NA* |
| E352G | 1 | NA |
| E355G | 1 | Intermediate |
| F359I | 2 | NA |
| F359V | 4 | Intermediate |
| L364I | 1 | Sensitive |
| V379I | 2 | Intermediate |
| F382L | 1 | NA |
| L387M | 2 | Sensitive |
| M388L | 1 | Sensitive |
| H396P | 1 | Sensitive |
| H396R | 2 | Intermediate |
| S417Y | 1 | NA |
| I418V | 1 | NA |
| E450G | 1 | NA |
| E453D | 1 | NA* |
| E453K | 1 | NA |
| E459G | 1 | NA |
| D482V | 1 | NA* |
| F486S | 3 | Intermediate |
NA indicates mutation previously described, but in vitro imatinib sensitivity not determined; and NA*, mutation not previously described.
The number of patients sums to 66 (not 53) due to the presence of multiple mutations in some patients.
In vitro imatinib sensitivity according to proliferation assays of cells transfected with each BCR-ABL variant.24 As per our previous thresholds,2 “sensitive” is defined as an IC50 of 1000 nM or less imatinib, and “insensitive” as an IC50 of more than 3000 nM. The imatinib sensitivity data are from O'Hare et al,24 except L248V,25 E355G,26 L364I,27 M388L,25 and F486S.28
Defining the optimal transcript rise threshold to trigger mutation screening
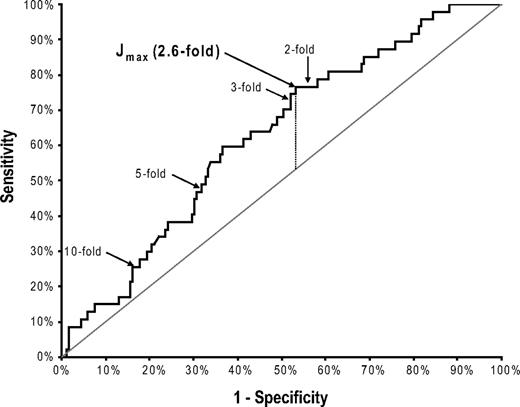
Eighteen of the 150 patients were excluded from this analysis because sequencing was performed on a sample for which no numeric PCR rise could be concomitantly determined due to a negative or weakly positive (below the accurate linear quantitation range) PCR result in the preceding baseline sample. The maximal transcript level that theoretically could have been present in the preceding sample in these excluded cases (as assessed by the reference gene level) was a 4.4 log-drop (median). Of the 18 excluded patients, 17 never experienced an RQ-PCR relapse (in 167 evaluated samples), and 6 of 18 had a kinase domain mutation. As these 18 patients were specifically excluded from this analysis because of their low residual disease levels, they had, expectedly, better cytogenetic and molecular responses than the 132 included patients. All other variables shown in Table 1, however, were no different in the included versus excluded patients, including the postimatinib follow-up time, the number of laboratory monitoring visits, and the prevalence and first appearance of kinase domain mutations. Of the 132 included patients with a more moderate response level (median baseline BCR-ABL = 1.1 log-drop; range, −1.2 to 3.8 log-drop), 47 had a KD mutation, and 35 of these had a concomitant 3-fold or greater rise in BCR-ABL RNA. A 3-fold transcript level rise thus had a 74% diagnostic sensitivity (95% CI, 60%-86%) for predicting mutations. However, an optimal threshold for sequencing will consider both specificity and sensitivity. Thus, to more rigorously define the specific increase in BCR-ABL RNA that optimally predicts a concomitant KD mutation, we performed an ROC analysis of this relationship.12 The resulting ROC curve of sensitivity versus (1 − specificity), with an area under the curve (AUC) of 0.63 (95% CI, 0.54-0.71), confirmed that an increase in BCR-ABL transcripts is a significant predictor of a simultaneous KD mutation (Figure 1). In comparison, an AUC of 1.0 would represent perfect diagnostic performance (100% sensitive and specific).
ROC curve for optimally predicting a kinase domain mutation by a rise in BCR-ABL RNA. The quantitative increase in BCR-ABL RNA levels was determined on 233 samples (from 132 patients) with a readable kinase domain (KD) DNA sequence, and a numeric BCR-ABL RNA level on both the sequenced sample and the immediately prior sample. Sensitivity was defined as the number of mutation-bearing samples with a transcript level rise above a moving (fold-change) cutoff threshold divided by the total number of samples with a mutation. Specificity was defined as the number of wild-type samples with a transcript level rise below the same cutoff threshold divided by the total number of samples without a mutation. The Youden index (J) is the vertical distance from each point on the receiver operating characteristic (ROC) curve to the diagonal “chance” line (from 0,0 to 1,1). The maximal J value (Jmax, vertical dotted line), defining the optimal cutoff threshold (2.6-fold transcript level rise) for predicting a concomitant mutation, is denoted, as are the ROC points associated with a 2-, 3-, 5- and 10-fold transcript level rise.
ROC curve for optimally predicting a kinase domain mutation by a rise in BCR-ABL RNA. The quantitative increase in BCR-ABL RNA levels was determined on 233 samples (from 132 patients) with a readable kinase domain (KD) DNA sequence, and a numeric BCR-ABL RNA level on both the sequenced sample and the immediately prior sample. Sensitivity was defined as the number of mutation-bearing samples with a transcript level rise above a moving (fold-change) cutoff threshold divided by the total number of samples with a mutation. Specificity was defined as the number of wild-type samples with a transcript level rise below the same cutoff threshold divided by the total number of samples without a mutation. The Youden index (J) is the vertical distance from each point on the receiver operating characteristic (ROC) curve to the diagonal “chance” line (from 0,0 to 1,1). The maximal J value (Jmax, vertical dotted line), defining the optimal cutoff threshold (2.6-fold transcript level rise) for predicting a concomitant mutation, is denoted, as are the ROC points associated with a 2-, 3-, 5- and 10-fold transcript level rise.
A consensus statistical method to determine the optimal cutoff value for any predictive laboratory test is the Youden index (J),13,14 which maximizes the sum of test sensitivity plus test specificity. J can be visually determined as the point on the ROC curve with the maximal vertical distance from the diagonal “chance line” (from [0,0] to [1,1]), which represents the ROC curve of a biomarker with no significant diagnostic utility. Using this algorithm, the optimal cutoff value that best predicted the presence of a concomitant KD mutation was a 2.6-fold (0.42 log) transcript level increase (Figure 1). The baseline BCR-ABL RNA level, before this 2.6-fold rise, was a 1.8 log-drop (median; range, −0.65 to 3.8 log-drop). At this optimized cutoff level, of the 47 patients with a point mutation, 36 had a rising RQ-PCR value of at least 2.6-fold (in the same sample), defining a diagnostic sensitivity of 77% (95% CI, 62%-88%) for a single RQ-PCR test. There was a significantly increased risk (odds ratio = 2.9) of finding a concomitant point mutation for samples with (vs without) a greater than 2.6-fold RQ-PCR rise (P = .005). This mutation-predictive capability of a 2.6-fold RQ-PCR rise was not significantly affected by the baseline transcript level at which the rise began—with a common odds ratio of 4.1 (P = .003) after adjustment for the heterogeneous levels of baseline BCR-ABL RNA. The mutation-predictive capability of a 2.6-fold RQ-PCR rise was also not significantly affected by the cytogenetic response level at the time of sequencing—with a common odds ratio of 4.1 (P = .002) after adjustment for the cytogenetic response.
Eight of 11 patients with a point mutation but no concomitant 2.6-fold RQ-PCR increase had a more than 2.6-fold transcript level rise in a subsequent sample, drawn 6.2 months (median) after the initial mutation discovery. In comparison, disease progression occurred 10.4 months (median) after mutation discovery (Figure 2). In this retrospective study, the choice as to which samples to sequence was not based on any real-time knowledge of patient-specific RQ-PCR or prior sequencing results. Thus, among all patients resequenced after a prior mutation analysis, only a minority (64/150 = 43%) had a 2.6-fold RQ-PCR rise at the time of resequencing. Further proof of the absence of any significant selection bias for resequenced samples was the finding that there was no significant difference in the prevalence of a prior KD mutation among resequenced samples with (31% with a mutation) versus without (47% with a mutation) a 2.6-fold RQ-PCR relapse. Of the 47 patients with a mutation, a total of 44 thus had a 2.6-fold RQ-PCR rise, either at the same time as the initial mutation discovery (n = 36), or at a later time (n = 8). In 1 of the 8 mutation-positive patients without an initial 2.6-fold RQ-PCR rise, the mutation that was initially discovered (M244V) was transient, and was later replaced by 2 new variants (S417Y and E453K).
The presence of a kinase domain mutation predicts shorter progression-free survival after sequencing. The cumulative rate of progression-free survival (Kaplan-Meier method) is shown for patients with, versus patients without, a detectable KD mutation.
The presence of a kinase domain mutation predicts shorter progression-free survival after sequencing. The cumulative rate of progression-free survival (Kaplan-Meier method) is shown for patients with, versus patients without, a detectable KD mutation.
Table 3 shows the diagnostic sensitivities, specificities, negative predictive values, and odds ratios for predicting a concomitant KD mutation at various rising RQ-PCR cutoff thresholds, including the optimal cutoff value of a 2.6-fold increase in BCR-ABL RNA. All cutoff values between a 2- and 4.5-fold increase were significantly predictive of a concomitant mutation, with 2.6-fold being the optimal cutoff level by several criteria (J, negative predictive value, and odds ratio). In contrast, cutoff rises of 5-fold or greater had poor diagnostic sensitivity (missing that majority of mutation cases with lower levels of transcript rises) and were not significantly predictive of mutations.
An increase in BCR-ABL RNA above a variable cutoff threshold predicts the presence of a concomitant KD mutation
| RQ-PCR increase, fold change . | Sensitivity, % (95% CI) . | Specificity, % . | Negative predictive value, % (95% CI) . | Odds ratio (95% CI) . | P . |
|---|---|---|---|---|---|
| 2.0 | 77 (62-88) | 44 | 88 (80-94) | 2.6 (1.2-5.4) | .01 |
| 2.5 | 77 (62-88) | 46 | 89 (81-94) | 2.8 (1.4-5.9) | .005 |
| 2.6 | 77 (62-88) | 47 | 89 (81-94) | 2.9 (1.4-6.0) | .005 |
| 2.6* (include subsequent sample) | 94 (82-99) | 47 | 97 (91-99) | 13 (3.9-43) | < .001 |
| 3.0 | 74 (60-86) | 48 | 88 (80-94) | 2.7 (1.3-5.5) | .008 |
| 3.5 | 64 (49-77) | 54 | 86 (78-91) | 2.1 (1.1-4.1) | .03 |
| 4.0 | 60 (44-74) | 60 | 85 (78-91) | 2.2 (1.1-4.2) | .02 |
| 4.5 | 55 (40-70) | 64 | 85 (78-90) | 2.2 (1.2-4.2) | .02 |
| 5.0 | 47 (32-62) | 68 | 84 (77-89) | 1.9 (1.0-3.6) | .06 |
| 10 | 26 (14-40) | 83 | 82 (75-87) | 1.7 (0.8-3.7) | .2 |
| RQ-PCR increase, fold change . | Sensitivity, % (95% CI) . | Specificity, % . | Negative predictive value, % (95% CI) . | Odds ratio (95% CI) . | P . |
|---|---|---|---|---|---|
| 2.0 | 77 (62-88) | 44 | 88 (80-94) | 2.6 (1.2-5.4) | .01 |
| 2.5 | 77 (62-88) | 46 | 89 (81-94) | 2.8 (1.4-5.9) | .005 |
| 2.6 | 77 (62-88) | 47 | 89 (81-94) | 2.9 (1.4-6.0) | .005 |
| 2.6* (include subsequent sample) | 94 (82-99) | 47 | 97 (91-99) | 13 (3.9-43) | < .001 |
| 3.0 | 74 (60-86) | 48 | 88 (80-94) | 2.7 (1.3-5.5) | .008 |
| 3.5 | 64 (49-77) | 54 | 86 (78-91) | 2.1 (1.1-4.1) | .03 |
| 4.0 | 60 (44-74) | 60 | 85 (78-91) | 2.2 (1.1-4.2) | .02 |
| 4.5 | 55 (40-70) | 64 | 85 (78-90) | 2.2 (1.2-4.2) | .02 |
| 5.0 | 47 (32-62) | 68 | 84 (77-89) | 1.9 (1.0-3.6) | .06 |
| 10 | 26 (14-40) | 83 | 82 (75-87) | 1.7 (0.8-3.7) | .2 |
Except for the indicated row, table includes only those patients with successful RQ-PCR and sequencing performed on the same sample. If samples after the initial genotype determination are included, at the optimal 2.6-fold cutoff, the sensitivity, negative predictive value, and odds ratio increase.
Kinase domain mutations predict a shorter progression-free survival
Forty-nine of the 150 patients (19 of whom had a mutation) were ineligible for an analysis of the effects of a kinase domain mutation on progression-free survival because they had no progression-free follow-up time after the initial genotype determination. These exclusions were due to either (1) a less than minor cytogenetic response (> 65% Ph positivity) that could, by definition, never fulfill the progression criteria (> 30% increase in Ph positivity) for a subsequent loss of a major cytogenetic response (n = 24), or (2) the lack of any cytogenetic follow-up data (n = 25). The postgenotype follow-up was 22.5 months (median) in the remaining 101 patients eligible for the survival analysis (74/101 with a 3-fold RQ-PCR relapse). Sixty (59%) of these 101 eligible patients subsequently progressed, including 4 who progressed to accelerated phase or blast crisis. Thirty-four of these 101 patients had a KD mutation, 29 (85%) of whom progressed, and 67 had no detectable mutation, 31 (46%) of whom progressed. The progression-free survival was significantly shorter (P < .001) for the patients with mutations (median, 10.4 months after the mutation screen) compared with those without (median, 32.4 months), and the hazard ratio was 3.2 (95% CI, 1.9-5.4; Figure 2). In a subset analysis in which patients with imatinib treatment failure (primary resistance) were excluded, the presence of a kinase domain mutation remained a significant risk factor for subsequent disease progression (n = 54; hazard ratio = 3.2; 95% CI, 1.5-7.2; P = .004). In a landmark analysis addressing the same question, 41 patients had both a mutation analysis performed by 30 months after the initiation of imatinib therapy and disease progression (or last follow-up) occurring after 30 months. Excluded from the landmark analysis were 52 patients with sequencing performed only after 30 months (19 with a mutation), and patients with disease progression (n = 43; 16 with a mutation) or last follow-up (n = 14; 3 with a mutation) occurring before 30 months. Of the 15 landmark-eligible patients with a point mutation detected at 30 months, 14 subsequently progressed (median, 34 months after imatinib initiation). In comparison, of the 26 patients without a mutation at 30 months, 12 subsequently progressed, and the progression-free survival was significantly prolonged (median, 52 months after imatinib initiation; P < .001; Figure 3).
The presence of a kinase domain mutation predicts shorter progression-free survival after imatinib initiation. The cumulative rate of progression-free survival (Kaplan-Meier method) is shown for patients with versus patients without, a detectable KD mutation, detected by 30 months after initiation of imatinib therapy. This 30-month landmark analysis excludes patients with sequencing performed after 30 months, and those with disease progression or last follow-up occurring before 30 months.
The presence of a kinase domain mutation predicts shorter progression-free survival after imatinib initiation. The cumulative rate of progression-free survival (Kaplan-Meier method) is shown for patients with versus patients without, a detectable KD mutation, detected by 30 months after initiation of imatinib therapy. This 30-month landmark analysis excludes patients with sequencing performed after 30 months, and those with disease progression or last follow-up occurring before 30 months.
Progression-free survival was not significantly different in the 10 patients with a P-loop mutation (median, 9.4 months after mutation detection), 8 of whom progressed, compared with 24 patients with other mutation types (median, 10.4 months; P > .8). With the exception of 1 patient with an L248V mutation (with disease progression 28 months after mutation discovery), all disease progression events occurred within 2 standard deviations of the mean lag time from mutation detection to disease progression (10.2 ± 5.8 months), and without any significant difference according to mutation type (Figure 4). The adverse prognostic significance of a KD mutation was independently maintained when adjusted for the variables presented in Table 1. Other than the presence of a KD mutation (hazard ratio = 2.5; 95% CI, 1.4-4.4; multivariable P = .001), the only variable independently predictive of a significantly higher risk of disease progression was the level of BCR-ABL RNA at the time of the mutation analysis (hazard ratio = 2.1; 95% CI, 1.5-3.0; multivariable P < .001). In this multivariable analysis, the lag time to the initiation of imatinib therapy, previous imatinib treatment failure (primary resistance), and the achievement of a complete hematologic, major cytogenetic, or complete cytogenetic response were not independently predictive of disease progression risk.
Time to disease progression after mutation discovery. The time from the initial discovery of a KD mutation to subsequent disease progression (y-axis) is plotted against the location of the mutation (kinase domain amino acid number; x-axis). The 37 data points reflect the presence of multiple mutations in some of the 29 patients with disease progression after a mutation was discovered. Shaded bars depict average values for each mutation location. Underlined codons represent the mutation location of 5 patients with no subsequent disease progression during follow-up. For these 5 patients, the follow-up time from mutation discovery to the last monitoring visit is indicated in parentheses.
Time to disease progression after mutation discovery. The time from the initial discovery of a KD mutation to subsequent disease progression (y-axis) is plotted against the location of the mutation (kinase domain amino acid number; x-axis). The 37 data points reflect the presence of multiple mutations in some of the 29 patients with disease progression after a mutation was discovered. Shaded bars depict average values for each mutation location. Underlined codons represent the mutation location of 5 patients with no subsequent disease progression during follow-up. For these 5 patients, the follow-up time from mutation discovery to the last monitoring visit is indicated in parentheses.
Discussion
KD mutations are the most common and best characterized mechanism of secondary imatinib resistance, and as yet, the only mechanism for which diagnostic tests are routinely available. To maximize the diagnostic yield of the mutation test, however, it is critical to define the optimal threshold rise in BCR-ABL RNA that should trigger mutation analysis. Reasoning that the optimal threshold for transcript level rises would maximize both test sensitivity (to capture as many patients with mutations as possible) and test specificity (to avoid unnecessary sequencing), we applied an unbiased ROC analysis to our data set of 150 patients on regular BCR-ABL RQ-PCR monitoring (for a median of 46 months). We found that a 2.6-fold rise in BCR-ABL RNA is the optimal cutoff for predicting the presence of a concomitant KD mutation. This 2.6-fold threshold detected a KD mutation with a diagnostic sensitivity of 77% for transcript levels drawn at the same time as the earliest mutation discovery. This estimate of diagnostic sensitivity may, however, be an underestimate, given that, in this retrospective study in which genotyping was performed in patients both with and without an RQ-PCR rise, mutations were discovered in 11 patients without a concomitant transcript rise—8 of whom had a transcript rise exceeding 2.6-fold in a sample drawn after the initial mutation discovery. If these 8 mutation-positive patients were considered to have had a sufficient transcript rise (at any time during monitoring), the diagnostic sensitivity of this biomarker would be 94%. At this 2.6-fold threshold, the negative predictive value was 97%, implying a small 3% risk of failing to detect a deleterious point mutation when the BCR-ABL RNA increase does not reach this optimized cutoff level.
The practical recommendation from this data is to perform mutation screening whenever the BCR-ABL RNA level increases more than 2.6-fold (0.42 log) from its previous value. Furthermore, as this relationship retains significance regardless of the baseline response level, sequencing is indicated in any patient with a 2.6-fold rise, and not just those with CCR. Applying a similar transcript rise cutoff of 2.0-fold (occurring at any time relative to sequencing), Branford et al established quite comparable assay performance characteristics—a diagnostic sensitivity of 97% and a negative predictive value of 99% for predicting KD mutations.6 The consistent 2- to 3-fold mutation-screening trigger established by each of 2 independent studies suggests that the currently recommended 5- to 10-fold cutoff may be set too high. Consistent with this, in our laboratory, cutoff rises of 5- or 10-fold thus had no significant association with mutation detection and had poor diagnostic sensitivity, with 53% to 74% of mutations being missed. The implication is that all patients with RQ-PCR rises greater than 2.6-fold should be screened for mutations, including those with a 5- to 10-fold rise. However, given the widespread variability and poor interlaboratory standardization of BCR-ABL RQ-PCR technical methods,4 the optimal cutoff for mutation analysis should, ideally, be determined by each individual laboratory. The choice of the RQ-PCR reference gene would be predicted to be less critical for standardization of the optimal BCR-ABL RNA level rise that should trigger mutation screening—as any reference gene–specific assay bias would be eliminated in the determination of the RQ-PCR difference between successive samples. Although any significant rise of BCR-ABL that is not due to treatment noncompliance mandates a re-evaluation of the therapeutic strategy, knowledge of the precise mutation type can help guide therapy. For example, because F317L BCR-ABL exhibits intermediate sensitivity to dasatinib and high sensitivity to nilotinib, the latter would be the treatment of choice.2 Most importantly, if T315I was detected, then none of the currently available second-line BCR-ABL inhibitors would be indicated.
In our laboratory, the 2.6-fold mutation-screening threshold determined by ROC analysis is consistent with the analytic precision of our assay at low levels of residual disease, where the 95% confidence interval for RQ-PCR is 2.9-fold.8 Thus, a small minority (∼ 5%) of observed PCR rises of more than 2.6-fold are, by definition, unavoidable “analytic” false-positives that would trigger an unnecessary (wild type) sequence analysis, which, although costly, would have minimal clinical impact. In contrast, a false-negative transcript rise, as would predominate with an insensitive 5- to 10-fold cutoff, would not trigger a reflex mutation screen (even in the presence of a mutation), and potentially delay beneficial adjustments to the therapeutic strategy.
Consistent with published results, we find that the presence of a KD mutation is predictive of future disease progression.15-17 Nevertheless, a minority (15%) of patients with a detectable KD mutation did not progress, perhaps due to the reduced “fitness” of some clones or the presence of the mutation in more differentiated cells incapable of sustaining malignant hematopoiesis.2 Although 57% of our patients with disease progression had a detectable KD mutation, other mechanisms of resistance—BCR-ABL overexpression, clonal evolution, impaired drug bioavailability, and dosage/compliance issues—may have contributed to the expanding disease burden of a significant minority of patients.
Similar to other reports,1,4,17 we observed a diverse spectrum of KD mutations. Only 2 of our patients, however, had the pan-resistant T315I mutation, a considerably lower frequency than has been reported in some,1,4 but not other,17 studies. This relative paucity of T315I mutations could reflect the fact that more than 80% of our patients were in chronic phase, whereas T315I is more common in patients with advanced disease18 and those being treated with a second-generation tyrosine kinase inhibitor such as dasatinib or nilotinib.19-21 None of the patients in our study, however, received any tyrosine kinase inhibitor other than imatinib during their entire follow-up. Several of the KD mutations in our study have not been previously reported, and await functional validation of their ability to confer imatinib resistance. Contrary to some reports,16,17,22,23 but not others,15 progression-free survival was similar for patients with P-loop mutations compared with patients with a mutation elsewhere in the KD. However, within our relatively small cohort, we cannot exclude that some individual P-loop mutations may indeed confer an adverse prognosis. Across the entire KD sequence, we failed to observe any discernable prognostic impact associated with mutation localization.
In summary, despite the limitations of this study—its retrospective nature and the heterogeneity of the patient population and their response to imatinib—we show that increasing levels of BCR-ABL RNA can be used to sensitively predict the presence of a KD mutation in CML patients treated with imatinib. Using an unbiased ROC analysis, a 2.6-fold transcript level increase offered the optimal diagnostic yield for mutations. Thus, to institute alternative therapies as soon as possible in patients with evolving imatinib resistance, mutation screening should be considered whenever transcript level increases exceed this threshold. Conversely, foregoing an expensive mutation screen on a patient without a minimal 2.6-fold BCR-ABL rise will only rarely (in 3% of cases) miss a mutation with potential therapeutic consequences.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported, in part, by National Institutes of Health grant R21 CA095203 (R.D.P.), National Heart, Lung, and Blood Institute (NHLBI) grant HL082978-01 (M.W.N.D.), and the Leukemia & Lymphoma Society 7393-06: Specialized Center of Research (M.W.N.D.). M.W.N.D. is a Scholar in Clinical Research of the Leukemia & Lymphoma Society.
National Institutes of Health
Authorship
Contribution: R.D.P. designed the research, collated and analyzed the data, and wrote the paper; S.G.W. and J.L. performed the research and managed the database; M.J.M. managed the study patients; and M.W.N.D. managed the study patients and designed and oversaw the research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Richard D. Press, Department of Pathology, L113, Oregon Health & Science University, 3181 SW Sam Jackson Park Rd, Portland, OR 97201; e-mail: pressr@ohsu.edu.