Abstract
Acute lymphoblastic leukemia (ALL) is currently treated with an intense regimen of chemotherapy yielding cure rates near 80%. However, additional changes using available drugs are unlikely to provide significant improvement in survival. New therapies are warranted given the risk of severe therapy-associated toxicities including infertility, organ damage, and secondary malignancy. Here, we report ectopic expression of the receptor tyrosine kinase Mer in pediatric B-cell ALL. Inhibition of Mer prevented Erk 1/2 activation, increased the sensitivity of B-ALL cells to cytotoxic agents in vitro by promoting apoptosis, and delayed disease onset in a mouse model of leukemia. In addition, we discovered cross-talk between the Mer and mammalian target of rapamycin (mTOR) signaling pathways. Our results identify Mer as a novel therapeutic target in ALL and suggest that inhibitors of Mer will interact synergistically with currently used therapies. This strategy may allow for dose reduction resulting in decreased toxicity and increased survival rates. Mer is aberrantly expressed in numerous other malignancies suggesting that this approach may have broad applications.
Introduction
Cancer is the leading cause of disease-related deaths among children 1 to 14 years of age, and acute lymphoblastic leukemia (ALL) is the most common malignancy in children. ALL manifests as overproduction of white blood cells in the bone marrow and is classified according to the immunophenotype of the malignant cells.1,2 Most pediatric leukemias are of the B-precursor subtypes with fewer cases exhibiting mature B- and T-cell immunophenotypes.3 ALL can be further classified by cytogenetic analysis of chromosome number and specific chromosomal translocations and these cytogenetic subgroups can be distinguished by differential gene expression profiles.4 One of the chromosomal rearrangements commonly found in pre-B ALL is the t(1;19),5 resulting in fusion of 2 transcription factors, E2A and PBX1.6 Specifically, the N-terminal transactivation domain of E2A is fused to the C-terminal DNA-binding domain of PBX1. The resulting chimeric protein interferes with patterns of gene expression normally regulated by the native proteins.
Historically, E2A-PBX1 positivity was a poor prognostic indicator relative to other ALL biologic subsets.7 Although modifications to the standard of care including an intensive reinduction course and prolonged maintenance therapy have dramatically increased cure rates,8 significant risk of both short- and long-term toxicities (neurocognitive sequelae, auditory complications, cardiovascular dysfunction, gastrointestinal/hepatic dysfunction, growth delay, secondary malignancies, and infertility) persist. In pediatric cancer survivors, the incidence of severe late effects is approximately 25%.9,10 Furthermore, a recent study found that survival rates for children with relapsed ALL remain poor with contemporary treatment protocols.11 Consequently, candidate novel agents will be most attractive if they increase the efficacy and/or reduce the toxicity of current chemotherapy regimens. One means to accomplish this is to develop new biologically targeted agents that interact synergistically with standard leukemia chemotherapy, permitting dose reduction.
Molecularly targeted agents currently in use for treatment of leukemia include the tyrosine kinase inhibitors imatinib and dasatinib for the treatment of chronic myelogenous leukemia and Philadelphia chromosome–positive (Ph+) ALL.12,13 In addition, lestaurtinib is a FLT3 tyrosine kinase inhibitor currently in clinical trials for the treatment of pediatric acute myeloid leukemia (AML) and MLL-rearranged ALL.14,15 Rituximab, a monoclonal antibody against CD20, is also being used as a therapy for CD20+ leukemia/lymphoma.16 The early success of these agents in the treatment of pediatric leukemia supports the concept of targeted biologic therapies as a means to increase efficacy of standard chemotherapy while decreasing associated patient toxicity. However, the biologic agents developed to date are useful only in specific subsets of pediatric leukemia. Thus, novel targets must be identified to yield less toxic therapies for E2A-PBX1+ B-ALL as well as other subtypes of pediatric leukemia.
Within the hematopoietic lineages, the receptor tyrosine kinase (RTK) Mer (also known as MerTK, Nyk, and Tyro12) is expressed in dendritic cells, monocytes/macrophages, NK cells, NKT cells, megakaryocytes, and platelets.17 However, Mer is not expressed in normal lymphocytes.18-20 In studies of T-cell ALL, we have previously shown that ectopic expression of Mer contributes to the development of lymphoblastic leukemia and lymphoma.19,20 Mer RNA expression has also been demonstrated in E2A-PBX1+ B-ALL.4 Mer is known to activate antiapoptotic signaling proteins, including Akt and Erk 1/2.21-23 Furthermore, a recent microarray study identified gas6, a ligand for Mer, as a gene which promotes survival of HEK-293 cells under conditions of serum withdrawal.24 These data led us to hypothesize that abnormal expression and activation of Mer provide a survival advantage for leukemia cells. Furthermore, inhibition of Mer may enhance the sensitivity of leukemia cells to cytotoxic agents.
Here, we present data demonstrating aberrant expression of the Mer protein in E2A-PBX1+ and other cytogenetic subgroups of B-ALL. Using both in vitro and in vivo models, we provide compelling evidence that Mer inhibition attenuates survival signaling, makes cells more susceptible to death after serum withdrawal or chemotherapy treatment, and significantly delays the onset of disease in a xenograft mouse model of leukemia. Thus, Mer is a novel therapeutic target for treatment of B-cell ALL. In addition, therapies targeting Mer may synergize with standard chemotherapy and thereby allow dose reduction and decrease toxicity.
Methods
Patient samples and cell culture
Diagnostic bone marrow or peripheral blood samples were obtained from the Children's Oncology Group (COG) repository of B-cell ALL patient samples. Additional samples were obtained from the tissue bank at The Children's Hospital. Patient samples were collected after written informed consent obtained in accordance with the Declaration of Helsinki and supplied in a deidentified manner. All experiments involving patient samples conformed to the relevant regulatory standards as approved by the Colorado Multiple Institutional Review Board (COMIRB). The REH, RCH-ACV, UOCB1, Hal01, Nalm6, and RS411 cell lines were generously provided by Lia Gore, MD (University of Colorado Denver and The Children's Hospital). The 697 cell line was obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ). All cell lines were maintained in RPMI medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin. Multiplex polymerase chain reaction (PCR) DNA profiling confirmed the identity of the 697 and REH cell lines.
Animals
Wild-type (C57Bl/6) and NOD-SCID mice were purchased from Jackson Laboratories. MerKO mice were kindly provided by Drs Glenn Matsushima and H.S. Earp (University of North Carolina). MerTg mice were generated in our laboratory and have been previously described.20 All experiments involving animals conformed to the relevant regulatory standards as approved by the University of Colorado Institutional Animal Use and Care Committee.
Flow cytometric analysis of cell surface proteins
Single-cell suspensions of normal human bone marrow cells, B-cell lymphoblasts, or mouse bone marrow cells were washed in phosphate-buffered saline (PBS) containing 2% FBS. Cells were stained in 50 μL antibody (supplemental Materials and Methods, available on the Blood website; see the Supplemental Materials link at the top of the online article) solution at 4°C for 30 minutes. Stained cells were washed with PBS-FBS and resuspended in PBS. Fluorescence was detected and analyzed using a FC 500 flow cytometer with CXP data analysis software (Beckman Coulter).
Western blotting
Whole-cell lysates of human B-ALL cells were prepared from patient samples or cultured cells in lysis buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 10 mM EDTA, 10% glycerol, and 1% Triton X-100) supplemented with phosphatase inhibitors (1 mM Na3VO4 and 0.1 mM Na2MoO4) and protease inhibitors (Complete mini, Roche Molecular Biochemicals). Samples were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and immunoblotting (see supplemental Materials and Methods for additional details). Mer knockdown in 697 cells (see Figure 2B) was quantified by densitometric analysis of semiquantitative Western blots using AlphaEase software (Alpha Innotech) as previously described.25
Real-time quantitative reverse transcriptase–PCR
Total RNA was isolated from patient samples using a spin column purification method (RNeasy Plus Mini Kit; QIAGEN). Real-time reverse transcriptase (RT)–PCR analysis was performed in the presence of fluorogenic probes using TaqMan Universal PCR Master Mix and No AmpErase UNG (Applied Biosystems); see supplemental Materials and Methods for details. Threshold cycle values were normalized to the 18S rRNA internal control and analysis performed as previously described.26 A 2-fold difference in RNA concentration per cycle was assumed for calculation of fold-change values.
Lentiviral shRNA vectors
Lentiviral vectors (pLKO.1) containing shRNA sequences targeting Mer (shMer1, Oligo ID: TRCN0000000862 and shMer4, Oligo ID: TRCN0000000865) or GFP (shControl, catalog no. RHS4459) were obtained from Open Biosystems. The GFP vector does not express the GFP protein but rather expresses a shRNA molecule that targets GFP. The GFP shRNA molecule engages the endogenous RNAi machinery. However, because human cells do not express GFP, the GFP construct serves as a nonsilencing control. Details of lentiviral particle production and target cell transduction can be found in supplemental Materials and Methods.
Functional assay of apoptosis
REH or 697 cells were plated at 2 × 105 cells/mL in 24-well plates and cultured overnight before treatment with the indicated agents for 48 hours. Alternatively, 697 cells were plated at 4-10 × 105 cells/mL in the absence or presence of serum for 24 hours. Cells were collected by centrifugation at 240g for 5 minutes. Resultant cell pellets were resuspended in PBS containing 1 μM YO-PRO-1 (Invitrogen) and 1.5 μM propidium iodide (Invitrogen) and incubated on ice for 20 to 30 minutes. Fluorescence was detected and analyzed using a FC 500 flow cytometer with CXP data analysis software (Beckman Coulter).
Functional assay of metabolically active cells
Preliminary experiments were performed to determine the maximum densities of 697 and REH cells such that nutrients would not be limiting for proliferation. Cells were plated in 96-well plates (2 × 105 cells/mL for 697 cells and 3 × 105 cells/mL for REH cells) and cultured overnight before addition of cytotoxic agents or vehicle only for an additional 48 hours. 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT; Sigma-Aldrich) reagent was added to a final concentration of 0.65 mg/mL and cells were cultured for another 4 hours. Solubilization solution (2×, 10% sodium dodecyl sulfate in 0.01 M HCl) was added and plates were incubated at 37°C overnight. Optical density was determined at 562 nm with a reference wavelength of 650 nm. Relative cell numbers were calculated by subtraction of background absorbance and normalization to untreated controls.
Xenograft mouse model of leukemia
NOD-SCID mice were injected intravenously with 697 cells or PBS as vehicle control. Animals were monitored daily and killed upon physical signs of leukemia onset (weight loss, abnormal posture, decreased activity, and/or hind limb paralysis) or at 15 weeks after injection. Upon sacrifice, whole blood and bone marrow were harvested for flow cytometric analysis of human cell-surface proteins CD19 and CD45. Leukemic engraftment was defined by physical presentation of symptoms as defined above and the presence of at least 5% human (CD19+/CD45+) cells in the bone marrow or peripheral blood. Flow cytometric analysis was also used to confirm the absence of Mer expression in CD19+/CD45+ ALL cells harvested from the blood or bone marrow of mice which were injected with Mer knockdown 697 cells. Similarly, the presence of Mer expression was confirmed in CD19+/CD45+ ALL cells collected from mice injected with control 697 cells. Kaplan-Meier survival curves were constructed. None of the animals that received PBS developed leukemia.
Statistical analysis
Where indicated, results were tested for statistical significance using Prism (Version 5.01; GraphPad Software). The specific statistical analyses performed are listed in the figure legends. Results were considered significant at P values less than .05.
Results
Mer tyrosine kinase is ectopically expressed in pediatric B-ALL
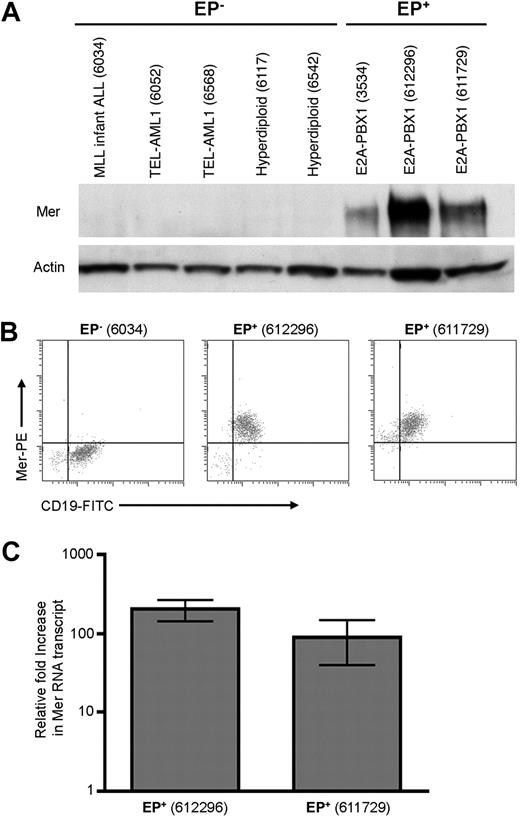
Our laboratory has previously reported that Mer is not expressed in normal mature T and B lymphocytes.18,20 Furthermore, Mer mRNA and protein are not detected in T cells at any developmental stage.18,19 Because the E2A-PBX1 fusion protein is found in B-precursor ALL,27 we used flow cytometry to demonstrate that Mer is not expressed on the surface of B cells at any stage of development (supplemental Figure 1). We next sought to verify the ectopic expression of Mer mRNA and investigate the expression of Mer protein in B-ALL. Samples from newly diagnosed patients with B-cell ALL were evaluated for expression of Mer mRNA and protein via RT-PCR, Western blot, and flow cytometric analyses. Sixteen E2A-PBX1+ B-ALL and twelve E2A-PBX1− B-ALL patient samples were examined (supplemental Table 1). All E2A-PBX1+ samples expressed Mer protein (Figure 1A-B; supplemental Table 1; and data not shown). In contrast, all but one E2A-PBX1− sample were negative for Mer protein expression. 4 E2A-PBX1+ and 6 E2A-PBX1− samples were analyzed by quantitative RT-PCR. Expression of Mer mRNA correlated with Mer protein expression in all cases (supplemental Table 1). The E2A-PBX1+ samples exhibited a 10- to 205-fold increase in Mer mRNA transcript relative to 5 E2A-PBX1− samples which expressed negligible levels of Mer mRNA (Figure 1C and data not shown). Although 11 of 12 samples obtained from newly diagnosed E2A-PBX1− patients did not express Mer, expression of Mer was detected in 7 of 7 human B-cell precursor leukemia cell lines tested, irrespective of E2A-PBX1 fusion protein expression. Specifically, Mer expression was detected in all 5 E2A-PBX1− cell lines tested and 3 of these 5 expressed Mer at levels comparable with E2A-PBX1+ cell lines (supplemental Figure 2).
Mer is expressed in diagnostic samples derived from patients with E2A-PBX1+ B-ALL. Twenty-eight diagnostic bone marrow samples from patients with B-cell ALL were analyzed by Western blot (A) and/or flow cytometry (B) for the expression of Mer protein. A subset of these samples was also analyzed by quantitative real-time RT-PCR (C) for the presence of Mer mRNA transcript. (A) Western blot analysis of Mer protein (∼ 180 kDa) in representative samples of non–E2A-PBX1 (EP−) B-ALL (lanes 1-5) and E2A-PBX1+ (EP+) B-ALL (lanes 6-8). Patient sample ID is shown in parentheses for comparison to the complete dataset listed in supplemental Table 1. The membrane was stripped and reprobed with anti-Actin (∼ 43 kDa) antibody to confirm similar loading of total protein. (B) Representative flow cytometry profiles for 1 EP− B-ALL sample (6034) and 2 EP+ B-ALL samples (612296 and 611729) are shown. All 3 samples expressed the CD19 B-lineage marker, whereas only the EP+ samples exhibited Mer staining. (C) Quantitative real time RT-PCR for 2 representative EP+ B-ALL samples exhibiting 100- to 200-fold increase in Mer transcript relative to the mean expression in 5 EP− B-ALL samples which express negligible amounts of Mer mRNA. Mean values and standard errors were derived from 3 independent experiments performed in duplicate.
Mer is expressed in diagnostic samples derived from patients with E2A-PBX1+ B-ALL. Twenty-eight diagnostic bone marrow samples from patients with B-cell ALL were analyzed by Western blot (A) and/or flow cytometry (B) for the expression of Mer protein. A subset of these samples was also analyzed by quantitative real-time RT-PCR (C) for the presence of Mer mRNA transcript. (A) Western blot analysis of Mer protein (∼ 180 kDa) in representative samples of non–E2A-PBX1 (EP−) B-ALL (lanes 1-5) and E2A-PBX1+ (EP+) B-ALL (lanes 6-8). Patient sample ID is shown in parentheses for comparison to the complete dataset listed in supplemental Table 1. The membrane was stripped and reprobed with anti-Actin (∼ 43 kDa) antibody to confirm similar loading of total protein. (B) Representative flow cytometry profiles for 1 EP− B-ALL sample (6034) and 2 EP+ B-ALL samples (612296 and 611729) are shown. All 3 samples expressed the CD19 B-lineage marker, whereas only the EP+ samples exhibited Mer staining. (C) Quantitative real time RT-PCR for 2 representative EP+ B-ALL samples exhibiting 100- to 200-fold increase in Mer transcript relative to the mean expression in 5 EP− B-ALL samples which express negligible amounts of Mer mRNA. Mean values and standard errors were derived from 3 independent experiments performed in duplicate.
Mer expression provides a survival advantage for leukemia cells
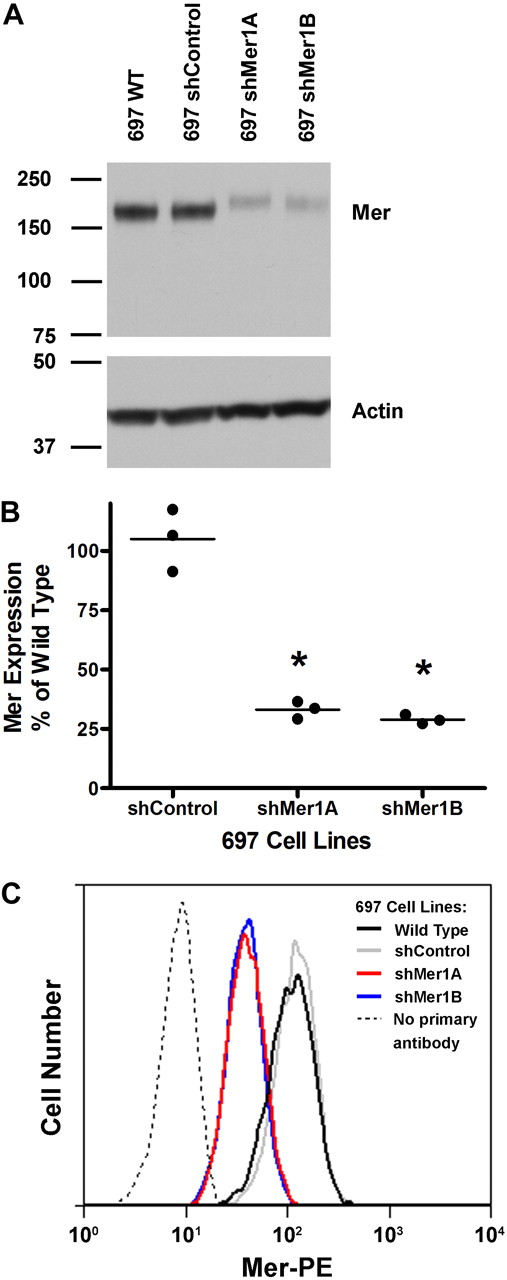
Two Mer-expressing B-ALL cell lines, 697 (E2A-PBX1+) and REH (E2A-PBX1−), were selected for further investigation. The REH cell line carries the t(12;21) chromosomal translocation which results in expression of the TEL-AML1 fusion protein and occurs in approximately 22% of pediatric ALL cases.28 Two different lentiviral short hairpin RNA (shRNA) contructs were used to inhibit Mer expression in the 697 or REH cell lines. Two clonal populations of each shRNA-expressing cell line (697 shMer1A, 697 shMer1B, REH shMer4A, and REH shMer4B) were derived by limiting dilution. Western blot analysis indicated significant knockdown of Mer protein expression in 697 shMer1A (66.9% ± 2.1%, mean ± SEM) and 697 shMer1B (71.0% ± 1.1%) cells relative to wild-type (WT) 697 cells and 697 cells transduced with nonsilencing control (shControl) shRNA (Figure 2A-B). In addition, flow cytometric analysis demonstrated that expression of Mer on the surface of 697 Mer knockdown cells was decreased relative to control cells (Figure 2C). REH shMer4A and REH shMer4B also exhibited robust (∼ 60%) knockdown of Mer protein levels compared with REH WT cells and REH shControl cells (supplemental Figure 3).
Abrogation of Mer expression in the E2A-PBX1+ human B-ALL cell line 697. Cells were infected with lentiviral particles containing short hairpin RNA (shRNA) constructs targeting Mer (shMer1A, shMer1B) or GFP (shControl) as a nonsilencing control. Mer knockdown was confirmed by Western blot analysis of whole-cell lysates (A-B) and flow cytometry (C). (A) Equal amounts of total protein were analyzed to qualitatively demonstrate reduced expression of Mer (∼ 180 kDa). The blot was stripped and reprobed with anti-Actin antibody (∼ 43 kDa) to confirm similar loading of protein. (B) Densitometric analysis of semiquantitative Western blots demonstrated significant knockdown in the clonal lines. *P < .001 vs shControl, 1-way ANOVA followed by Tukey multiple comparison test. No significant differences between shControl and wild-type were observed. Three independent measurements and mean are displayed. (C) Control and Mer knockdown 697 cells were stained with phycoerythrin-conjugated (PE) anti-Mer antibody or PE secondary antibody as an isotype control (no primary antibody). Representative histograms from 3 independent experiments are shown.
Abrogation of Mer expression in the E2A-PBX1+ human B-ALL cell line 697. Cells were infected with lentiviral particles containing short hairpin RNA (shRNA) constructs targeting Mer (shMer1A, shMer1B) or GFP (shControl) as a nonsilencing control. Mer knockdown was confirmed by Western blot analysis of whole-cell lysates (A-B) and flow cytometry (C). (A) Equal amounts of total protein were analyzed to qualitatively demonstrate reduced expression of Mer (∼ 180 kDa). The blot was stripped and reprobed with anti-Actin antibody (∼ 43 kDa) to confirm similar loading of protein. (B) Densitometric analysis of semiquantitative Western blots demonstrated significant knockdown in the clonal lines. *P < .001 vs shControl, 1-way ANOVA followed by Tukey multiple comparison test. No significant differences between shControl and wild-type were observed. Three independent measurements and mean are displayed. (C) Control and Mer knockdown 697 cells were stained with phycoerythrin-conjugated (PE) anti-Mer antibody or PE secondary antibody as an isotype control (no primary antibody). Representative histograms from 3 independent experiments are shown.
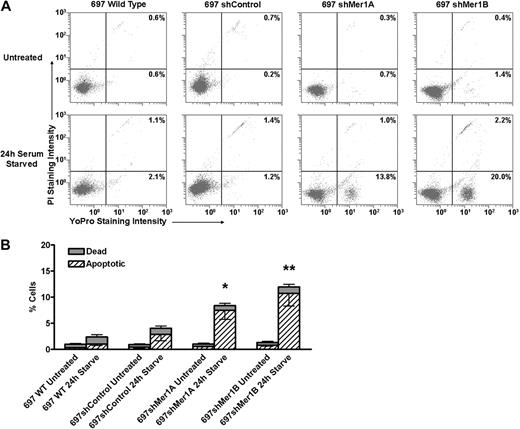
To test our hypothesis that expression of Mer provides a survival advantage for leukemia cells, we incubated control (WT and shControl) and Mer knockdown (shMer1A, shMer1B) 697 cells in media with or without 10% FBS for 24 hours. Cells were stained with YO-PRO-1 and propidium iodide to permit flow cytometric quantitation of apoptotic and dead cells (Figure 3A). No significant differences between WT and shControl cells were observed. In the presence of serum, no significant differences in the mean percentage of apoptotic (0.3%-0.8%) or dead (0.5%-0.6%) cells were observed between WT, shControl, or Mer knockdown cells (Figure 3B). In the absence of serum, both control and Mer knockdown cells exhibited an increase in the mean percentages of apoptotic and dead cells relative to untreated cells. However, the mean percentage of apoptotic cells was significantly higher for shMer1A (7.5%, P < .05) and shMer1B (10.7%, P < .001) cells compared with control (1.0%%-2.8%) cells (Figure 3B). These data support our hypothesis that expression of Mer provides a survival advantage for leukemia cells.
Mer knockdown confers susceptibility to apoptosis induced by serum starvation. Wild-type, shControl, and Mer knockdown (shMer1A, shMer1B) 697 cells were incubated in the presence (Untreated) or absence (24-hour serum-starved) of serum for 24 hours. Apoptotic and dead cells were identified by flow cytometric analysis of cells stained with YO-PRO-1 and propidium iodide. (A) Representative flow cytometric profiles are shown. Cells undergoing apoptosis are stained with YO-PRO-1 but are impermeable to propidium iodide. Dead cells and cells in late apoptosis are permeable to both dyes. Viable cells are not stained by either dye. The percentages of apoptotic (bottom right quadrants) and dead cells (top right quadrants) are shown. (B) Cumulative data demonstrate a significant accumulation of apoptotic cells when Mer knockdown cells are serum-starved for 24 hours (n = 5-6; *P < .05, **P < .001 vs shControl, 2-way repeated measures ANOVA followed by Bonferroni posttests). No significant differences between WT and shControl cells were observed. Mean values and standard errors derived from at least 3 independent experiments are shown.
Mer knockdown confers susceptibility to apoptosis induced by serum starvation. Wild-type, shControl, and Mer knockdown (shMer1A, shMer1B) 697 cells were incubated in the presence (Untreated) or absence (24-hour serum-starved) of serum for 24 hours. Apoptotic and dead cells were identified by flow cytometric analysis of cells stained with YO-PRO-1 and propidium iodide. (A) Representative flow cytometric profiles are shown. Cells undergoing apoptosis are stained with YO-PRO-1 but are impermeable to propidium iodide. Dead cells and cells in late apoptosis are permeable to both dyes. Viable cells are not stained by either dye. The percentages of apoptotic (bottom right quadrants) and dead cells (top right quadrants) are shown. (B) Cumulative data demonstrate a significant accumulation of apoptotic cells when Mer knockdown cells are serum-starved for 24 hours (n = 5-6; *P < .05, **P < .001 vs shControl, 2-way repeated measures ANOVA followed by Bonferroni posttests). No significant differences between WT and shControl cells were observed. Mean values and standard errors derived from at least 3 independent experiments are shown.
Inhibition of Mer increases the chemosensitivity of leukemia cells by promoting apoptosis
Given the observed increase in apoptosis after Mer inhibition and serum starvation, we investigated whether inhibition of Mer would enhance the sensitivity of leukemia cells to cytotoxic agents. Control and Mer knockdown 697 cells were treated with chemotherapeutic agents 6-mercaptopurine (6-MP), methotrexate (MTX), vincristine (VCR), etoposide (VP-16), or doxorubicin (DOXO) for 48 hours at clinically relevant concentrations.29-32 Relative cell numbers were determined and demonstrated that inhibition of Mer substantially increased the sensitivity of 697 cells to cytotoxic agents (Figure 4A-E). Specifically, control 697 cells were resistant to 6-MP treatment in the high micromolar range of doses tested (Figure 4A). However, the Mer knockdown lines demonstrated remarkable sensitivity to 6-MP with IC50 values in the 2- to 3-μM range (Figure 4A; Table 1). In addition, Mer inhibition resulted in a dramatic and significant increase in sensitivity to treatment with MTX, VCR, VP-16, and DOXO (Figure 4B-E) as indicated by 2- to 5-fold reductions in IC50 values for Mer shRNA-expressing cells compared with control cells (P < .01, Table 1).
Inhibition of Mer expression results in increased chemosensitivity in 697 cells. Wild-type, shControl, and Mer knockdown (shMer1A, shMer1B) 697 cells were treated with the indicated concentrations of 6-mercaptopurine (6-MP), methotrexate (MTX), vincristine (VCR), etoposide (VP-16), or doxorubicin (DOXO) for 48 hours, and relative cell numbers were determined. Mean values and standard errors from at least 3 independent experiments performed in triplicate are shown. IC50 values were determined by nonlinear regression of data from at least 3 independent experiments performed in triplicate (see Table 1).
Inhibition of Mer expression results in increased chemosensitivity in 697 cells. Wild-type, shControl, and Mer knockdown (shMer1A, shMer1B) 697 cells were treated with the indicated concentrations of 6-mercaptopurine (6-MP), methotrexate (MTX), vincristine (VCR), etoposide (VP-16), or doxorubicin (DOXO) for 48 hours, and relative cell numbers were determined. Mean values and standard errors from at least 3 independent experiments performed in triplicate are shown. IC50 values were determined by nonlinear regression of data from at least 3 independent experiments performed in triplicate (see Table 1).
IC50 values determined by nonlinear regression of MTT assay data (see Figure 4)
| . | 697 Human B-ALL cell lines . | |||
|---|---|---|---|---|
| Wild-type . | shControl . | shMer1A . | shMer1B . | |
| 6-MP, μmol/L | ||||
| IC50 | 64 | > 64 | 2.86 | 3.24 |
| 99% CI | ND | ND | 2.2-3.7 | 2.5-4.2 |
| P < .01 | ND | ND | ND | |
| MTX, nmol/L | ||||
| IC50 | 27.5 | 27.8 | 12.3 | 11.8 |
| 99% CI | 23.8-31.7 | 24.4-31.6 | 10.3-14.7 | 10.6-13.1 |
| P < .01 | NS | * | * | |
| VCR, nmol/L | ||||
| IC50 | 1.04 | 1.02 | 0.54 | 0.46 |
| 99% CI | 0.84-1.29 | 0.84-1.23 | 0.46-0.63 | 0.37-0.55 |
| P < .01 | NS | * | * | |
| VP-16, nmol/L | ||||
| IC50 | 181 | 173 | 54.0 | 60.3 |
| 99% CI | 124-264 | 124-242 | 43.5-66.8 | 49.8-73.0 |
| P < .01 | NS | * | * | |
| DOXO, nmol/L | ||||
| IC50 | 16.7 | 11.1 | 2.83 | 3.26 |
| 99% CI | 12.4-22.4 | 7.17-17.2 | 2.20-3.64 | 2.44-4.35 |
| P < .01 | NS | * | * | |
| . | 697 Human B-ALL cell lines . | |||
|---|---|---|---|---|
| Wild-type . | shControl . | shMer1A . | shMer1B . | |
| 6-MP, μmol/L | ||||
| IC50 | 64 | > 64 | 2.86 | 3.24 |
| 99% CI | ND | ND | 2.2-3.7 | 2.5-4.2 |
| P < .01 | ND | ND | ND | |
| MTX, nmol/L | ||||
| IC50 | 27.5 | 27.8 | 12.3 | 11.8 |
| 99% CI | 23.8-31.7 | 24.4-31.6 | 10.3-14.7 | 10.6-13.1 |
| P < .01 | NS | * | * | |
| VCR, nmol/L | ||||
| IC50 | 1.04 | 1.02 | 0.54 | 0.46 |
| 99% CI | 0.84-1.29 | 0.84-1.23 | 0.46-0.63 | 0.37-0.55 |
| P < .01 | NS | * | * | |
| VP-16, nmol/L | ||||
| IC50 | 181 | 173 | 54.0 | 60.3 |
| 99% CI | 124-264 | 124-242 | 43.5-66.8 | 49.8-73.0 |
| P < .01 | NS | * | * | |
| DOXO, nmol/L | ||||
| IC50 | 16.7 | 11.1 | 2.83 | 3.26 |
| 99% CI | 12.4-22.4 | 7.17-17.2 | 2.20-3.64 | 2.44-4.35 |
| P < .01 | NS | * | * | |
Cumulative data from at least 3 independent experiments performed in triplicate were analyzed.
CI indicates confidence interval; DOXO, doxorubicin; 6-MP, 6-mercaptopurine; MTX, methotrexate; ND, not determined; NS, not significant; VCR, vincristine; and VP-16, etoposide.
Significance vs shControl is indicated by nonoverlapping 99% CIs. Significance could not be statistically evaluated for 6-MP–treated cells because an IC50 was not reached for wild-type or shControl cells.
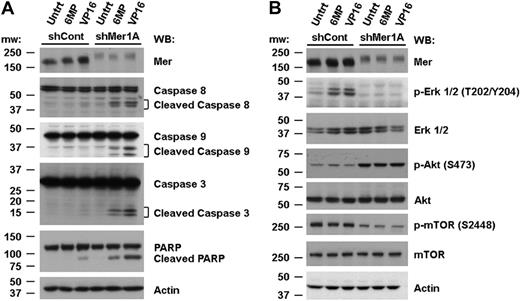
The data presented in Figure 4 were acquired via an assay that measures the relative number of metabolically active cells and thus, can reflect changes in both cell proliferation and cell death. To more specifically evaluate the role of apoptotic cell death in Mer-mediated chemoresistance, we performed flow cytometric analysis of cells stained with YO-PRO-1 and propidium iodide (Figure 5A). shControl and shMer1A 697 cells were treated with 6-MP, VP-16, or medium alone for 48 hours. Three concentrations of each drug were applied based on the dose-response curves generated in previous experiments. Cumulative analysis of 3 independent experiments (Figure 5B) revealed a dramatic dose-dependent increase in the mean percentage of apoptotic cells when shMer1A cells were treated with 6-MP (8.6%-20.2%, P < .05) or VP-16 (11.2%-26.2%, P < .001) relative to untreated cells (0.1%-0.5%) or shControl cells treated with chemotherapeutic agents (0.1%-1.4%). A dose-dependent increase in the mean percentage of dead cells was also observed for shMer1A cells treated with 6-MP (1.7%-7.2%, P < .001) or VP-16 (6.6%-66.0%, P < .05) relative to untreated cells (0.4%-0.5%) or shControl cells incubated with 6-MP (0.4%-0.9%) or VP-16 (4.5%-22.4%). Increased induction of apoptosis in shMer 697 cells treated with chemotherapy was also evident at the biochemical level. Mer shRNA-expressing cells exhibited increased cleavage of the apoptotic effectors PARP and caspases 3, 8, and 9 (Figure 6A).
Inhibition of Mer expression results in increased induction of apoptotic cell death in response to treatment with chemotherapeutic agents. Cultures of shControl or shMer1A 697 cells were exposed to 6-MP, VP-16, or medium only for 48 hours. Apoptotic and dead cells were identified by flow cytometric analysis of cells stained with YO-PRO-1 and propidium iodide as in Figure 3. (A) Representative flow cytometry profiles are shown. The percentages of apoptotic and dead cells are indicated in bottom right and top right quadrants, respectively. (B) Mean values and standard errors derived from 3 independent experiments are shown. Results were tested for significance using 2-way ANOVA and Bonferroni posttests to compare shControl and shMer1A cells at equivalent drug concentrations. Significant differences in apoptotic cell populations are indicated by asterisks (*P < .05, **P < .001). Significant differences in the number of dead cells were also observed (†P < .05, ‡P < .001).
Inhibition of Mer expression results in increased induction of apoptotic cell death in response to treatment with chemotherapeutic agents. Cultures of shControl or shMer1A 697 cells were exposed to 6-MP, VP-16, or medium only for 48 hours. Apoptotic and dead cells were identified by flow cytometric analysis of cells stained with YO-PRO-1 and propidium iodide as in Figure 3. (A) Representative flow cytometry profiles are shown. The percentages of apoptotic and dead cells are indicated in bottom right and top right quadrants, respectively. (B) Mean values and standard errors derived from 3 independent experiments are shown. Results were tested for significance using 2-way ANOVA and Bonferroni posttests to compare shControl and shMer1A cells at equivalent drug concentrations. Significant differences in apoptotic cell populations are indicated by asterisks (*P < .05, **P < .001). Significant differences in the number of dead cells were also observed (†P < .05, ‡P < .001).
Knockdown of Mer inhibits survival signaling and promotes caspase cleavage. Control (shCont) and Mer knockdown (shMer1A) 697 cells were exposed to 10 μM 6-MP, 150 nM VP-16, or medium only (Untrt) for 24 hours (A) or 40-60 minutes (B). Whole-cell lysates were prepared and expression of the indicated proteins (p- denotes a phosphorylated protein) was determined by Western blot analysis. Blots representative of 3 independent experiments are shown.
Knockdown of Mer inhibits survival signaling and promotes caspase cleavage. Control (shCont) and Mer knockdown (shMer1A) 697 cells were exposed to 10 μM 6-MP, 150 nM VP-16, or medium only (Untrt) for 24 hours (A) or 40-60 minutes (B). Whole-cell lysates were prepared and expression of the indicated proteins (p- denotes a phosphorylated protein) was determined by Western blot analysis. Blots representative of 3 independent experiments are shown.
Similar experiments conducted with REH cell lines demonstrated that Mer knockdown increases the sensitivity of REH cells to MTX. Mer shRNA-expressing REH cells exhibited significantly lower IC50 values in response to treatment with MTX (P < .05; supplemental Figure 4A-B). Inhibition of Mer also resulted in significantly greater accumulation of apoptotic (13.3%-15.4%, P < .05) and dead (32.3%-40.5%, P < .001) cells in comparison to control cells (5.2%-6.1% apoptotic, 14.1%-23.0% dead) in response to treatment with 30 nM MTX (supplemental Figure 5A-B). Importantly, the REH Mer knockdown lines were generated using a different shRNA construct than the 697 Mer knockdown lines. Thus, these results suggest that the observed increase in chemosensitivity of human leukemia cells is due specifically to inhibition of Mer and is not an off-target effect of the shRNA. Collectively, the data generated using 2 distinct cell lines expressing 2 distinct Mer shRNAs indicate that Mer functions to decrease sensitivity of B-ALL cells to treatment with cytotoxic agents and suggest that targeted inhibitors of Mer will interact synergistically with standard leukemia chemotherapy.
Inhibition of Mer expression leads to reduced phosphorylation of Erk 1/2 and mTOR
Previous reports demonstrated that Mer stimulation leads to phosphorylation of Akt and Erk 1/2, resulting in reduced apoptosis.21,23 In this study, Western blot analysis was used to compare activation of these and other signaling proteins in shControl versus shMer1A 697 cells after treatment with chemotherapeutic agents. In WT cells, treatment with 6-MP or VP-16 results in activation of Erk 1/2. In contrast, when Mer is inhibited, phosphorylation of Erk 1/2 (T202/Y204) is completely abolished (Figure 6B). Unexpectedly, phosphorylation of Akt (S473) is increased in Mer knockdown cells relative to shControl cells, even in the absence of treatment with cytotoxic agents. One possible explanation for this result is compensatory up-regulation of other proteins, such as the Mer-related RTKs Axl and Tyro3, which can activate Akt. Consistent with this possibility, a recent study demonstrated that chemotherapeutic agents induced Axl mRNA expression in AML.33 However, neither Axl nor Tyro3 proteins are expressed in shControl or shMer1A 697 cells (supplemental Figure 6A-B). Furthermore, treatment of these cells with 6-MP or VP-16 for 24 hours does not induce expression of Axl or Tyro3. An alternative explanation for the increased Akt phosphorylation in Mer knockdown cells is reduced feedback inhibition of Akt via mammalian target of rapamycin (mTOR) complex 1 (mTORC1).34,35 Consistent with this idea, we observed reduced phosphorylation of mTOR (S2448) concomitant with increased Akt phosphorylation (Figure 6B). These data indicate that inhibition of Mer leads to loss of Erk 1/2 and mTOR signaling in 697 cells and suggest a biochemical mechanism for the increased induction of apoptosis in response to chemotherapeutic agents in shMer-expressing 697 cells.
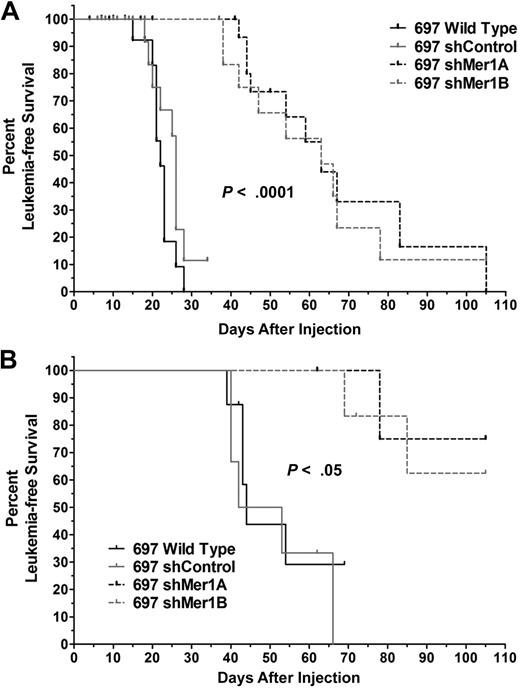
Inhibition of Mer expression delays onset of leukemia in a mouse xenograft model
To validate Mer as a novel therapeutic target in an in vivo setting, we evaluated the oncogenicity of control and Mer knockdown 697 cells in a mouse xenograft model of leukemia. Sublethally irradiated NOD-SCID mice were injected with the indicated cell lines (Figure 7A). Animals were monitored daily and killed upon physical signs of leukemia onset or at 105 days postinjection (DPI). Median survival of animals injected with control cells was 23 to 26 days. Median survival of animals in both Mer knockdown groups was 63 days. Kaplan-Meier survival curves (Figure 7A) demonstrate that Mer inhibition significantly delayed the onset of disease (P < 10−5). When unirradiated mice were injected with 10-fold fewer leukemia cells, Mer inhibition again significantly delayed the onset of disease (P < .05; Figure 7B). In this context, median survival of animals injected with control cells was 44 to 47.5 days, and overall survival was 0% at 69 DPI. Median survival of animals in both Mer knockdown groups was more than 105 DPI, the end point of the study. Importantly, of the eleven mice that received Mer knockdown cells, 6 had no physical symptoms of leukemia at the end of the study (105 DPI). Upon sacrifice, it was discovered that 3 of these 6 animals had more than 5% human cells in the bone marrow. However, leukemia cells were not detected in the remaining 3 mice. The normal lifespan of these animals prevents extension of these studies beyond 105 DPI and thus, it was not possible to evaluate whether these animals would remain leukemia-free over a longer period. These data indicate that Mer tyrosine kinase is critical for leukemogenesis and that inhibition of Mer slows the progression of disease in vivo.
Mer inhibition significantly delays the onset of disease and improves leukemia-free survival in a mouse xenograft model of human leukemia. (A) Sub-lethally irradiated NOD-SCID mice (10-26 animals per group) were injected with 5 × 106 cells of the indicated cell lines. (B) Nonirradiated NOD-SCID mice (5-8 animals per group) were injected with 5 × 105 cells of the indicated cell lines. Ticks on the Kaplan-Meier survival curves indicate censored subjects (mice for which samples could not be obtained or did not have leukemia at time of death). Comparison of survival curves revealed a significant difference in leukemia-free survival with Mer inhibition (wild-type or shControl vs shMer1A or shMer1B, log-rank test).
Mer inhibition significantly delays the onset of disease and improves leukemia-free survival in a mouse xenograft model of human leukemia. (A) Sub-lethally irradiated NOD-SCID mice (10-26 animals per group) were injected with 5 × 106 cells of the indicated cell lines. (B) Nonirradiated NOD-SCID mice (5-8 animals per group) were injected with 5 × 105 cells of the indicated cell lines. Ticks on the Kaplan-Meier survival curves indicate censored subjects (mice for which samples could not be obtained or did not have leukemia at time of death). Comparison of survival curves revealed a significant difference in leukemia-free survival with Mer inhibition (wild-type or shControl vs shMer1A or shMer1B, log-rank test).
Discussion
In this paper, we demonstrate that Mer is ectopically expressed in B-ALL. In samples from newly diagnosed B-ALL patients, Mer mRNA4 and protein expression is associated with a specific cytogenetic subtype of B-ALL defined by the presence of the fusion protein E2A-PBX1. However, Mer protein expression was detected in both E2A-PBX1+ and E2A-PBX1− human B-ALL cell lines. While it is possible that there is selective pressure for expression of Mer in vitro, this is unlikely given that some human leukemia cells lines are negative for expression of Mer even after several months of continuous culture (L.B. and D.K.G., unpublished data, February 2008). Of the 5 E2A-PBX1− cell lines that expressed Mer, 4 were established from patients at relapse and one from a patient who failed induction therapy,36-38 suggesting that Mer expression may be more prevalent in chemoresistant and/or recurrent B-ALL. Consistent with this idea, overexpression of Mer and/or the closely related RTK Axl are poor prognostic indicators in other human cancers.39-41 We are currently investigating the potential link between Mer and chemoresistant or recurrent B-ALL in an ongoing clinical study. Taken together, our data indicate that Mer expression is associated with the E2A-PBX1 cytogenetic subgroup in newly diagnosed B-ALL but also suggest that Mer expression may be more widespread among various cytogenetic subtypes in chemoresistant or recurrent B-ALL. The present findings suggest that targeted inhibition of Mer is an attractive therapeutic strategy in B-ALL where a Mer inhibitor could be administered after flow cytometric detection of Mer positivity.
The functional significance of this study is highlighted by our finding that abrogation of Mer expression increases the sensitivity of B-ALL cells to multiple chemotherapeutic agents. Importantly, 6-MP, MTX, VCR, and DOXO are used in standard pediatric ALL chemotherapy regimens and VP-16 is a major component of relapse therapy. The increased sensitivity of Mer shRNA-expressing B-ALL cells to chemotherapeutic agents was mediated, at least in part, by an increase in activation of apoptotic signaling pathways and increased induction of apoptosis. Interestingly, Mer inhibition combined with cytotoxic agents resulted in cleavage of both caspases 8 and 9, suggesting a role for Mer in regulation of apoptosis via both intrinsic and extrinsic pathways. Although Mer knockdown B-ALL cells exhibit no significant differences in proliferation or survival in the absence of an apoptotic stimulus in vitro (R.M.A.L. and D.K.G., unpublished data, April 2008), inhibition of Mer was sufficient to significantly delay onset and/or progression of disease in a mouse model of human leukemia, indicating an important role for Mer in leukemogenesis in vivo. The apparent discrepancy between these findings may be explained by our results demonstrating that serum starvation alone provides enough stress to induce significantly more cell death in Mer knockdown cells compared with control B-ALL cells. Thus, an intense apoptotic stimulus such as chemotherapy is not required to observe effects of Mer inhibition on cancer cell survival. Furthermore, the tumor microenvironment, which is known to play critical roles in tumorigenesis, is not recapitulated in vitro. It is likely that the tumor microenvironment presents multiple apoptotic stimuli in vivo such as hypoxic growth conditions. Our results suggest that clinical application of a Mer inhibitor would inhibit leukemogenesis and increase the efficacy of standard chemotherapy, permitting dose reduction and decreasing associated toxicity.
The 5 cytotoxic drugs evaluated in the metabolic assay act by different mechanisms, suggesting that Mer inhibition increases the efficacy of these agents via common downstream pathways. Previous reports demonstrated that Mer prevents apoptosis by stimulating canonical cell survival pathways.21,23 The Erk 1/2, Akt, and mTOR protein kinases are well defined regulators of cell proliferation, survival, and growth which have frequently been associated with malignant transformation. Here, we describe Mer-dependent modification of these signaling pathways in human leukemia cells. Although previous studies have identified Erk 1/2 and Akt as downstream targets of Mer, this study represents the first report of a link between Mer and mTOR signaling.
Erk 1/2 has a well-defined role in protecting cancer cells from apoptosis. In a study of 131 adult ALL patients, activation of Erk 1/2 was associated with poor prognostic factors and a lower probability of achieving complete response to treatment.42 In this study, inhibition of Mer expression prevented chemotherapy-induced activation of Erk 1/2 indicating that aberrant expression of Mer enables B-ALL cells to activate the Erk 1/2 survival pathway in response to cytotoxic agents. Targeted inhibition of Mer may therefore prevent Erk 1/2 activation resulting in a more favorable response to cytotoxic agents and improved prognosis.
Previous studies have demonstrated that stimulation of Mer results in activation of Akt in murine leukemia cells and macrophages.23,43 Based on these findings, one might expect that inhibition of Mer would lead to reduced Akt activation in human B-ALL cells. However, in these studies phosphorylation of Akt (S473) was up-regulated in Mer knockdown cells. Phosphorylation of S473 is required for complete activation of Akt,44 and increased phosphorylation of this residue has been observed in cancer cells after treatment with mTORC1 inhibitors such as rapamycin and RAD001.34,35,45,46 Further investigation revealed that mTORC1 regulates an Akt negative feedback loop.35,45 A commonly reported indicator of mTORC1 activation is phosphorylation of mTOR (S2448).47,48 Concomitant with the observed increase in Akt phosphorylation, we report decreased phosphorylation of mTOR (S2448) in response to reduced expression of Mer in human B-ALL cells suggesting that Mer regulates mTOR activity in this context. The observed reduction in mTOR S2448 phosphorylation did not correlate with changes in the phosphorylation state of S6 ribosomal protein or 4EBP1 (data not shown). Reduced phosphorylation of these mTORC1 effector proteins is typically coincident with augmentation of Akt S473 phosphorylation in rapamycin-treated cells.34,35,46 Thus, Mer inhibition and rapamycin treatment induce distinct biochemical changes, suggesting alteration of mTORC1 signaling via different mechanisms. Similar to our results, transient disruption of the mTORC1 complex in human lung cancer cells resulted in increased phosphorylation of Akt S473 without a detectable effect on S6K1 phosphorylation confirming that these 2 events are not always concurrent.49 Our results clearly indicate a role for Mer in mTOR-mediated feedback inhibition of Akt. Mer may also play a role in regulation of mTOR activity. The proposed connection between Mer and mTOR is supported by a previous study which identified S6K1 as a downstream target of a chimeric Mer receptor exogenously expressed in NIH3T3 fibroblasts.50 Furthermore, S6K1 was shown to be required for Gas6-mediated proliferation and survival of serum starved NIH3T3 cells,51 which express the Mer-related tyrosine kinases Axl and Tyro3. Further investigation of this novel link between Mer and mTOR is necessary.
The mechanism(s) of aberrant Mer expression in B-ALL and other cancers remains unknown. Given that E2A and PBX1 are both transcription factors, we considered the possibility that the E2A-PBX1 fusion protein drives expression of Mer in the t(1;19) subset of B-ALL. However, expression of E2A-PBX1 under control of an inducible promoter in an ALL cell line does not result in expression of Mer (K.K.S. and D.K.G., unpublished data, May 2006). This result is consistent with our finding that Mer is expressed in both E2A-PBX1+ and E2A-PBX1− human B-ALL cell lines. Aberrant oncogene expression can occur via other mechanisms including increased gene copy number, dysregulation of DNA methylation, or abnormal microRNA expression. Expression of the Mer-related RTK Axl was inversely correlated with promoter methylation in AML cells.33 In metastatic breast cancer, overexpression of Mer was correlated with loss of microRNA-335.52 It will be interesting to see whether these or other mechanisms contribute to ectopic expression of Mer in B-ALL and additional cancers.
In conclusion, we have provided the first evidence that Mer is ectopically expressed at the protein level in pediatric B-ALL. Inhibition of Mer expression reduces pro-survival signaling, dramatically increases the sensitivity of leukemia cells to cytotoxic agents, and significantly delays the onset of disease in a mouse model of leukemia. Furthermore, we provide the first evidence of an interaction between the Mer and mTOR signaling pathways. Our results demonstrate that Mer is a novel biologic target for the treatment of pediatric leukemia. Although current cure rates for pediatric leukemia approach 80%, survivors remain at significant risk of late effects. Our results suggest that new agents that target Mer may directly inhibit leukemogenesis and interact synergistically with standard chemotherapeutic agents, permitting dose reduction and decreased toxicity. Furthermore, Mer inhibitors may be most beneficial for patients who currently have a poor prognosis, namely those with chemoresistant or recurrent disease. While the focus of this study was pediatric leukemia, the widespread aberrant expression of Mer in human cancers suggests that novel agents which target Mer may also prove useful for treatment of adult leukemia, other hematologic malignancies, and solid tumors. Active investigation of novel Mer inhibitors is warranted, as these compounds would make excellent candidates for clinical investigations.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Stephen Hunger and the COG (grants CA98543 and CA114766) for providing E2A-PBX1+ leukemia patient samples; Dr Mark Gregory for technical assistance generating the shMer knockdown lines; and the University of Colorado Cancer Center Core Facilities (Flow Cytometry, Real time PCR, Tissue Culture, and DNA Sequencing) for expert technical assistance. We also thank Drs. James DeGregori, Gail Eckhardt, Lia Gore, Stephen Hunger, and Kelly Maloney for thoughtful discussions and critical reading of the manuscript.
This research was supported in part by grants from the Gabrielle's Angel Foundation for Cancer Research (#030, D.K.G.), the National Heart, Lung, and Blood Institute (F32HL096416, L.B.), and the Brent Eley Foundation (A.K.K.). D.K.G. is the Damon Runyon–Novartis Clinical Investigator supported in part by the Damon Runyon Cancer Research Foundation (CI-39-07).
Authorship
Contributions: R.M.A.L., D.D., L.B., K.K.S, K.M.J., and A.K.K. designed and performed experiments and analyzed data; X.L. analyzed data; and R.M.A.L., D.D., and D.K.G. wrote the manuscript.
The current address for Kelly K. Sawczyn is All Children's Hospital, 880 6th St South, Suite 140, St Petersburg, FL 33701.
Conflict-of-interest disclosure: R.M.A.L., D.D., L.B., A.K.K., and D.K.G. have submitted a patent application related to the work described in the present study. The remaining authors declare no competing financial interests.
Correspondence: Douglas K. Graham, University of Colorado Denver School of Medicine, Department of Pediatrics, P18-4401, Mail Stop 8302, PO Box 6511, Aurora, CO 80045; e-mail: doug.graham@ucdenver.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal