Abstract
Alternatively activated (M2) macrophages play critical roles in diverse chronic diseases, including parasite infections, cancer, and allergic responses. However, little is known about the acquisition and maintenance of their phenotype. We report that M2-macrophage marker genes are epigenetically regulated by reciprocal changes in histone H3 lysine-4 (H3K4) and histone H3 lysine-27 (H3K27) methylation; and the latter methylation marks are removed by the H3K27 demethylase Jumonji domain containing 3 (Jmjd3). We found that continuous interleukin-4 (IL-4) treatment leads to decreased H3K27 methylation, at the promoter of M2 marker genes, and a concomitant increase in Jmjd3 expression. Furthermore, we demonstrate that IL-4–dependent Jmjd3 expression is mediated by STAT6, a major transcription factor of IL-4–mediated signaling. After IL-4 stimulation, activated STAT6 is increased and binds to consensus sites at the Jmjd3 promoter. Increased Jmjd3 contributes to the decrease of H3K27 dimethylation and trimethylation (H3K27me2/3) marks as well as the transcriptional activation of specific M2 marker genes. The decrease in H3K27me2/3 and increase in Jmjd3 recruitment were confirmed by in vivo studies using a Schistosoma mansoni egg–challenged mouse model, a well-studied system known to support an M2 phenotype. Collectively, these data indicate that chromatin remodeling is mechanistically important in the acquisition of the M2-macrophage phenotype.
Introduction
Chronic immune responses are often distinguished by a characteristic cytokine profile, as exemplified by the dominance of either T helper type 1 (Th1) cytokines, such as interferon-γ (IFN-γ) or T helper type 2 (Th2) cytokines, such as interleukin-4 (IL-4) and IL-13.1 This skewed cytokine environment is important in dictating the type of immune response required to efficiently target antigens or pathogens, including the activation and polarization of specific macrophage subsets. Depending on the phenotype, macrophages can be designated as either classically activated (M1) or alternatively activated (M2) macrophages; each type plays a specialized role in response to environmental signals, such as cytokines and microbial products in inflamed tissue.2,3 Because M2-macrophage activation is mediated by IL-4 and/or IL-13 (Th2 cytokines), these macrophages are normally associated with immune responses that possess a Th2-skewed cytokine environment, as observed in parasite infections and allergic inflammation.4,5 In addition, M2-macrophages are also involved in tissue repair and remodeling,6 insulin resistance,7 atherosclerosis,8 and tumor progression.9,10
Both M1- and M2-macrophages can be characterized by a set of marker genes, which are significantly induced by the dominance of either Th1 or Th2 cytokines. For example, IL-4 (a Th2 cytokine) is a potent inducing factor for the expression of Ym1, found in inflammatory zone-1 (FIZZ1), and Arginase 1 by M2-macrophages.2,11-13 On the other hand, M1-macrophages are induced by the Th1 cytokine, IFN-γ, either alone or with a microbial trigger. The prototypic marker of M1 activation is the generation of nitric oxide by inducible nitric oxide synthase (iNOS).2
It is well established that IL-4 and IL-13 can activate the JAK-STAT6 signaling pathway. This results in STAT6 translocation into the nucleus where it binds to the promoter region of target genes and regulates the expression of specific genes,14 including those involved in the differentiation of naive T-cell precursors into Th2 cells15-17 and characteristic genes expressed by M2-macrophages.13,18,19 Although it is well documented that M2-macrophages contribute to the pathogenesis of various diseases, little is known about the mechanisms underlying the acquisition and maintenance of the M2 phenotype.
Chromatin remodeling, via histone modifications, is one of the key epigenetic mechanisms known to regulate normal embryonic development,20 cancer,21 and the evolution of specific acquired immune responses.22 Of particular importance is histone methylation, which plays a pivotal role in the maintenance of both active and suppressed states of gene expression, depending on the sites of methylation.23,24 The methylation of histone H3 at lysine-4, -36, and -79 (H3K4, H3K36, and H3K79) is implicated in activation of transcription, whereas methylation of histone H3 at lysine-9 and -27, and histone H4 at lysine-20 (H3K9, H3K27, and H4K20) is correlated with repression of transcription.
Historically, lysine methylation has been thought to be stable, as early studies indicated a low turnover rate for the methyl group on lysine.24 However, accumulating evidence has revealed that lysine methylation is often reversible and can be removed by site-specific demethylases, including amine oxidase LSD1 and members of the Jumonji C (JmjC) domain protein family.25 Jumonji domain containing 3 (Jmjd3) and ubiquitously transcribed tetratricopeptide repeat gene, X chromosome, both members of the JmjC protein family, were recently shown to be specific demethylases of H3K27me2/3.26-29 These demethylases were first studied during animal embryogenesis where ubiquitously transcribed tetratricopeptide repeat gene, X chromosome was shown to be involved in zebrafish posterior development,26 whereas the Jmjd3 homologue was shown to regulate Caenorhabditis elegans gonadal development.27 Furthermore, Jmjd3 is induced by external inflammatory stimuli, including lipopolysaccharide (LPS).28
In the present study, we demonstrate that characteristic M2 marker genes, which phenotypically define these immune cells, are epigenetically regulated. A novel aspect of this epigenetic regulation is the result of the STAT6-dependent induction of the H3K27 demethylase Jmjd3. As a downstream target of STAT6, the level of Jmjd3 was clearly increased by IL-4 and was reduced after withdrawing IL-4 from the medium. Importantly, the levels of Jmjd3 are correlated with changes in H3K27 methylation at the promoters of M2-macrophage marker genes. Furthermore, in vivo studies suggest that the increased recruitment of Jmjd3 can be associated with a decrease in H3K27 methylation in M2-macrophages recovered from Schistosoma mansoni egg–challenged mice, a well-studied in vivo system known to skew macrophages to an M2 phenotype.30-32
Methods
Mice
Female wild-type (WT) BALB/c mice, WT C57BL/6 mice, WT Swiss-Webster mice, and Stat4−/− and Stat6−/− mice on BALB/c background (6-10 weeks old) were purchased from The Jackson Laboratory. S mansoni–infected Swiss-Webster mice were provided by Dr Fred Lewis (Biomedical Research Laboratories). All mice, including female Myd88−/− and Trif−/− C57BL/6 mice, were housed in the University Laboratory Animal Medicine Facility at the University of Michigan Medical School. All animal experiments were approved by the Animal Use Committee at the University of Michigan.
Reagents
Antibody to Ym1 was from StemCell Technologies. Antibodies to Arginase 1 and STAT6 for immunoblotting were from BD Biosciences. Antibody to Jmjd3 for immunoblotting was from Abgent. Antibodies to phospho-STAT6, STAT1, and histone H3 were from Cell Signaling Technology. Antibody to STAT6 for chromatin immunoprecipitation (ChIP) assay was from Santa Cruz Biotechnology. Antibodies to acetyl histone H3, H3K27me2, H3K27me3, and H3K9me3 were from Millipore. Antibodies to Gapdh and H3K4me3 were from Abcam. LPS from Escherichia coli (O55:B5) was from Sigma-Aldrich. Mouse cytosine-phosphate-guanosine (CpG) DNA was from Cell Sciences. Recombinant mouse IL-4, IL-13, IL-10, CCL17, and IFN-γ were from R&D Systems.
Cell isolation and culture
Bone marrow cells were collected by flushing femurs and tibias of mice with RPMI 1640. Bone marrow–derived macrophages (BMDMs) were generated as previously described.33 On day 6, the cells were replated. After resting overnight, cells were incubated with or without IL-4 (10 ng/mL) for the indicated times. Medium was changed every 2 days, and cells were cultured without IL-4 from day 2 (IL-4 medium group) or restimulated with IL-4 (10 ng/mL) every 2 days (IL-4 continuous group). BMDCs were generated in the presence of granulocyte-macrophage colony-stimulating factor as previously described.34 Peritoneal exudate cells were harvested by phosphate-buffered saline peritoneal lavage and were incubated in RPMI medium for 2 hours at 37°C. More than 94% of adherent cells were confirmed as peritoneal macrophages by cytospin analysis. Unless stated, the concentration of IL-4, IL-13, IL-10, and CCL17 was 10 ng/mL, and the concentrations of LPS, IFN-γ, and CpG DNA were 100 ng/mL, 100 U/mL, and 1 μM, respectively.
S mansoni egg–challenged mouse model
Live S mansoni eggs were purified from the livers of S mansoni–infected Swiss-Webster mice as previously described.35 Mice were intraperitoneally injected with 1.7% sodium chloride solution alone (sham control group) or 5000 S mansoni eggs in 1.7% sodium chloride solution (Schisto group; ie, Schisto ip in naive mice group). For some experiments, the S mansoni–infected mice were also intraperitoneally injected with 5000 S mansoni eggs in 1.7% sodium chloride solution (Schisto ip in Schisto mice group). After 7 days, cells from peritoneal exudates were harvested.
Quantitative RT-PCR
RNA was isolated from BMDMs, BMDCs, and peritoneal macrophages using TRIzol (Invitrogen) according to the manufacturer's instructions. Total RNA (2 μg) was reverse-transcribed to yield cDNA as previously described.33 Quantitative real-time PCR (RT-PCR) analysis was performed by 7500 Real-Time PCR System (Applied Biosystems). Gene expression assays for Ym1 (Chi3l3), Arginase 1 (Arg1), and Jarid1d were purchased from Applied Biosystems. Gapdh (Applied Biosystems) was used as a loading control. The entire list of primers can be found in supplemental Figure 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Generation of FIZZ1- and Jmjd3-specific antibodies
The FIZZ1-specific antibody for immunoblotting and Jmjd3-specific antibody for ChIP assay were prepared by multiple-site immunization of New Zealand white rabbits with recombinant mouse FIZZ1 (R&D Systems) and mouse Jmjd3 (raised against antigen aa 1326-1642) in CFA, and boosted with FIZZ1 and Jmjd3 in IFA, respectively, as in previously described procedures from our laboratory.36 Polyclonal antibodies were titered by direct enzyme-linked immunosorbent assay against FIZZ1 or Jmjd3, and titers were 107 and 105, respectively.
Immunoblotting
Cells were lysed in lysis buffer (Cell Signaling Technology), briefly sonicated, kept on ice for 30 minutes, and centrifuged at 15 000g for 15 minutes. The supernatant was collected and stored at −80°C until use. Total protein concentration of the samples was measured by bicinchroninic acid protein assay (Pierce Chemical). Equal amounts (15-30 μg) of cell lysates were fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (Nupage; Invitrogen). Then the proteins were transferred onto nitrocellulose membrane (Invitrogen). After the overnight incubation with appropriate primary antibody, the membrane was counterstained with horseradish peroxidase-conjugated rabbit or mouse IgG antibody and visualized with enhanced chemiluminescence detection reagents (GE Healthcare).
ChIP assay
ChIP assay was carried out using a ChIP assay kit (Millipore), as previously described.37 The list of ChIP primers can be found in supplemental Figure 2.
Short-interfering RNA assay
A total of 1.5 × 106 BMDMs were transfected with 2 μg of a mixture of Jmjd3-specific (QIAGEN or Dharmacon) or nontargeting control siRNAs (QIAGEN, or Dharmacon, respectively), using mouse macrophage nucleofector kit (Lonza Walkersville) according to the manufacturer's instructions and plated in a 6-well plate. After 24 hours, cells were used for the experiment.
Statistical analysis
Statistical analysis was performed with the unpaired Student t test or analysis of variance followed by the Tukey multiple comparison test. A P value the less than .05 was used to indicate statistical significance.
Results
Induction and maintenance of M2-macrophage marker genes by IL-4
To examine the role of IL-4 on the induction and maintenance of the M2 phenotype in macrophages, BMDMs were stimulated with IL-4 as indicated (Figure 1). The mRNAs of Ym1 (Chi3l3), FIZZ1 (Retnla), and Arginase 1 (Arg1), genes characteristically expressed by M2-macrophages, were up-regulated 2 days after IL-4 incubation (Figure 1A). Importantly, expression of these genes requires the continuous presence of IL-4. Withdrawing IL-4 after day 2 led to a gradual decrease in both mRNA (Figure 1A) and protein levels (Figure 1B) for all 3 M2 marker genes. To examine the role of IL-4 on the expression of the M1 marker genes, we measured the expression of the M1 marker iNOS (Nos2).2 In contrast to M2 marker genes, the expression of Nos2 remained very low in the presence of continuous exposure to IL-4 but was up-regulated on removal of IL-4 from the culture media (IL-4 medium group; Figure 1C).
Critical roles of IL-4 on induction and maintenance of alternative activation in BMDMs. BMDMs were incubated with IL-4 for 2 days; then the cells were incubated with (IL-4 continuous group) or without (IL-4 medium group) IL-4 for the indicated time. (A) The mRNA levels of Ym1 (Chi3l3), FIZZ1 (Retnla), and Arginase 1 (Arg1) were analyzed by quantitative RT-PCR. The data shown are “fold induction” relative to that in untreated cells. (B) Expressions of Ym1, FIZZ1, and Arginase 1 (Arg1) were measured by immunoblotting. Gapdh was used as a loading control. (C) The mRNA levels of iNOS (Nos2) were analyzed by quantitative RT-PCR. The data shown are “fold induction” relative to that in untreated cells. #P < .05 compared with the level on day 0. (D) Expression of phosphorylated (pY)-STAT6 was measured by immunoblotting. Gapdh was used as a loading control. Data are representative of 3 independent experiments (A-D) and are expressed as mean ± SEM (A,C). *P < .05, **P < .01, ***P < .001, compared with IL-4 medium group.
Critical roles of IL-4 on induction and maintenance of alternative activation in BMDMs. BMDMs were incubated with IL-4 for 2 days; then the cells were incubated with (IL-4 continuous group) or without (IL-4 medium group) IL-4 for the indicated time. (A) The mRNA levels of Ym1 (Chi3l3), FIZZ1 (Retnla), and Arginase 1 (Arg1) were analyzed by quantitative RT-PCR. The data shown are “fold induction” relative to that in untreated cells. (B) Expressions of Ym1, FIZZ1, and Arginase 1 (Arg1) were measured by immunoblotting. Gapdh was used as a loading control. (C) The mRNA levels of iNOS (Nos2) were analyzed by quantitative RT-PCR. The data shown are “fold induction” relative to that in untreated cells. #P < .05 compared with the level on day 0. (D) Expression of phosphorylated (pY)-STAT6 was measured by immunoblotting. Gapdh was used as a loading control. Data are representative of 3 independent experiments (A-D) and are expressed as mean ± SEM (A,C). *P < .05, **P < .01, ***P < .001, compared with IL-4 medium group.
Because STAT6 is a major transcription factor for IL-4–mediated signaling responses,14 the expression of phosphorylated STAT6 was measured in IL-4–stimulated macrophages. We found that phosphorylated STAT6 was expressed at 6 hours after IL-4 stimulation and was higher at days 4 and 6 in cells with continuous IL-4 exposure (IL-4 continuous group) compared with IL-4 medium group (Figure 1D).
Epigenetic gene regulation in M2-macrophages
Because there were dramatic changes in gene expression after prolonged IL-4 treatment, we next tested whether there are changes in the chromatin modification status at M2-macrophage marker genes. ChIP assay was used to test the levels of H3K4 methylation, a mark for transcription activation, and H3K27 methylation, a mark for transcription repression. We found that H3K4me3 was significantly up-regulated, whereas H3K27me2/3 was significantly decreased at the promoters of the M2 marker genes (ie, Chi3l3, Retnla, and Arg1) after IL-4 treatment (Figure 2A). These epigenetic changes are also observed at the promoter region of mannose receptor, a marker of alternative activation for both human and mouse macrophages (data not shown).2 In contrast, the H3K27me3 level at the promoter region of Nos2 (an M1 marker gene) was not changed by IL-4 treatment (supplemental Figure 3). The changes of histone modifications at the promoter of Chi3l3, Retnla, and Arg1 were specific because there was no global change of H3K4me3 and H3K27me2/3 after IL-4 stimulation (supplemental Figure 4).
Dynamic epigenetic gene regulations in IL-4–induced M2-macrophages. (A) BMDMs were incubated with or without IL-4 for 2 days. ChIP assay was performed on the promoter regions of Chi3l3, Retnla, and Arg1 using indicated antibodies. (B-C) BMDMs were incubated with IL-4 for 2 days and then were cultured with (IL-4 continuous group) or without (IL-4 medium group) IL-4 for an additional 2 days. ChIP assay was performed on the promoter regions of M2 marker genes using antibodies for H3K4me3 (B) and H3K27me3 (C). Untreated cells were used as a baseline control (medium-medium group). Data are representative of 3 to 5 independent experiments and are expressed as mean ± SEM. *P < .05; **P < .01.
Dynamic epigenetic gene regulations in IL-4–induced M2-macrophages. (A) BMDMs were incubated with or without IL-4 for 2 days. ChIP assay was performed on the promoter regions of Chi3l3, Retnla, and Arg1 using indicated antibodies. (B-C) BMDMs were incubated with IL-4 for 2 days and then were cultured with (IL-4 continuous group) or without (IL-4 medium group) IL-4 for an additional 2 days. ChIP assay was performed on the promoter regions of M2 marker genes using antibodies for H3K4me3 (B) and H3K27me3 (C). Untreated cells were used as a baseline control (medium-medium group). Data are representative of 3 to 5 independent experiments and are expressed as mean ± SEM. *P < .05; **P < .01.
Furthermore, the level of H3K4me3, once increased after initial IL-4 treatment, did not respond to the IL-4 withdraw, as it remained the same between the IL-4 continuous and the IL-4 medium groups (Figure 2B). Interestingly, there was a very significant change in the H3K27me3 level on IL-4 withdrawal. H3K27me3 was significantly increased in the IL-4 medium group, restoring its level to that before IL-4 treatment (Figure 2C). These results indicate that H3K27 methylation on M2 marker genes can be dynamically modified depending on exposure to IL-4.
Induction of Jmjd3 in IL-4–induced M2-macrophages
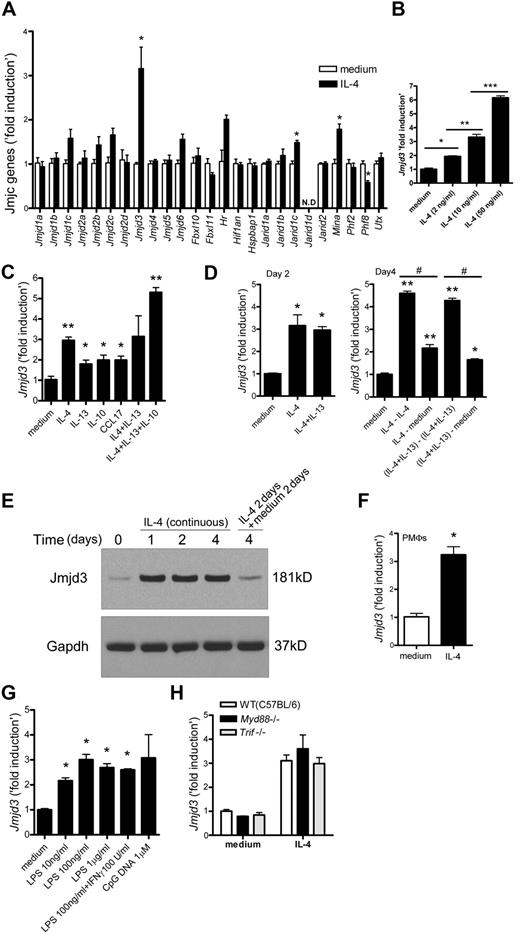
As the H3K27 methylation at the promoters of M2 marker genes was dynamically regulated in IL-4–induced M2-macrophages, we predicted that a specific histone demethylase might contribute to this process. The largest known group of histone demethylase enzymes is the JmjC family proteins.25 To examine the role of JmjC family proteins in IL-4–induced M2-macrophages, BMDMs were stimulated with IL-4 for 6 hours and the expression of 25 JmjC genes was analyzed. Of the JmjC family, Jmjd3, a recently discovered H3K27me2/3-specific demethylase, was clearly induced by IL-4 (Figure 3A). As shown in Figure 3B, the response of Jmjd3 to IL-4 was dose-dependent.
Jmjd3 induction in IL-4–treated macrophages. (A) BMDMs were incubated with IL-4 for 6 hours. The mRNA levels of JmjC family proteins were measured by quantitative RT-PCR. N.D. indicates not detected. (B-D) Jmjd3 mRNA levels were measured by quantitative RT-PCR after the following treatments. BMDMs were incubated with IL-4 (B) and indicated cytokines (C) for 6 hours. BMDMs were incubated with indicated cytokines for 2 days (D left). On day 2, cells were washed with phosphate-buffered saline and incubated with or without fresh IL-4, or with or without a cocktail of IL-4 and IL-13 for an additional 2 days as indicated (D right). #P < .05. (E) BMDMs were incubated with IL-4 as indicated. Jmjd3 expression was measured by immunoblotting. Gapdh was used as a loading control. Representative data were shown from 3 independent experiments. (F-H) Jmjd3 mRNA levels were measured by quantitative RT-PCR after the following treatments. Peritoneal macrophages (PMΦs) were incubated with IL-4 for 6 hours (F). BMDMs were incubated with LPS, a cocktail of LPS and IFN-γ, or CpG DNA (in indicated concentration) for 6 hours (G). BMDMs from WT (C57BL/6), Myd88−/−, and Trif−/− mice were incubated with or without IL-4 for 6 hours (H). All data of quantitative RT-PCR shown are “fold induction” relative to that in untreated WT cells. Data are representative of 3 independent experiments and are expressed as mean ± SEM. *P < .05; **P < .01; ***P < .001.
Jmjd3 induction in IL-4–treated macrophages. (A) BMDMs were incubated with IL-4 for 6 hours. The mRNA levels of JmjC family proteins were measured by quantitative RT-PCR. N.D. indicates not detected. (B-D) Jmjd3 mRNA levels were measured by quantitative RT-PCR after the following treatments. BMDMs were incubated with IL-4 (B) and indicated cytokines (C) for 6 hours. BMDMs were incubated with indicated cytokines for 2 days (D left). On day 2, cells were washed with phosphate-buffered saline and incubated with or without fresh IL-4, or with or without a cocktail of IL-4 and IL-13 for an additional 2 days as indicated (D right). #P < .05. (E) BMDMs were incubated with IL-4 as indicated. Jmjd3 expression was measured by immunoblotting. Gapdh was used as a loading control. Representative data were shown from 3 independent experiments. (F-H) Jmjd3 mRNA levels were measured by quantitative RT-PCR after the following treatments. Peritoneal macrophages (PMΦs) were incubated with IL-4 for 6 hours (F). BMDMs were incubated with LPS, a cocktail of LPS and IFN-γ, or CpG DNA (in indicated concentration) for 6 hours (G). BMDMs from WT (C57BL/6), Myd88−/−, and Trif−/− mice were incubated with or without IL-4 for 6 hours (H). All data of quantitative RT-PCR shown are “fold induction” relative to that in untreated WT cells. Data are representative of 3 independent experiments and are expressed as mean ± SEM. *P < .05; **P < .01; ***P < .001.
In addition to IL-4, other cytokines, including IL-13, IL-10, and the chemokine CCL17, have been shown to support the skewing of macrophages to the M2 phenotype2,38,39 ; we next examined the effects of these cytokines on Jmjd3 expression. Jmjd3 was up-regulated by each of these cytokines alone, with IL-4 causing the highest levels of Jmjd3 induction (Figure 3C). Interestingly, the cocktail of IL-4, IL-13, and IL-10 induced an even higher Jmjd3 response (Figure 3C). We further examined the ability of either IL-4 or IL-4 plus IL-13 challenged macrophages to induce Jmjd3 gene expression after 2 and 4 days in culture (Figure 3D). Jmjd3 was expressed by BMDMs continuously treated with IL-4 or IL-4 plus IL-13; however, the up-regulation of Jmjd3 was significantly reduced in BMDMs stimulated with IL-4 or with IL-4 plus IL-13 for 2 days followed by incubation with medium alone for 2 days (Figure 3D). We next examined the expression of Jmjd3 protein by immunoblotting and found an increase at 1 day after IL-4 stimulation, which was sustained in the presence of IL-4 at day 4. In contrast, Jmjd3 level was clearly decreased after terminating IL-4 stimulation (Figure 3E). To examine whether IL-4 can induce Jmjd3 expression in other macrophage populations, we examined the IL-4–induced Jmjd3 level in peritoneal macrophages. Jmjd3 induction was also observed in IL-4–stimulated primary peritoneal macrophage cultures in addition to BMDMs (Figure 3F).
Previous investigations demonstrated that both LPS and CpG DNA can induce Jmjd3 in macrophages and the LPS-induced expression of Jmjd3 was MyD88-dependent.28 We confirmed that both LPS and CpG DNA can induce Jmjd3 expression in primary BMDM cultures similar to IL-4 (Figure 3G), whereas IFN-γ alone did not induce Jmjd3 up-regulation (supplemental Figure 5A left panel). We next examined whether IL-4–dependent Jmjd3 expression relies on MyD88 or TRIF, a major signaling molecule of MyD88-independent pathway. To this end, BMDMs from WT, Myd88−/−, or Trif−/− mice were stimulated with IL-4 and the Jmjd3 levels were measured. As shown in Figure 3H, Jmjd3 levels were comparable among WT, Myd88−/−, and Trif−/− BMDMs. In contrast to LPS, IL-4 up-regulated Jmjd3 in a MyD88- or TRIF-independent manner.
Jmjd3 expression depends on STAT6 in IL-4–induced M2-macrophages
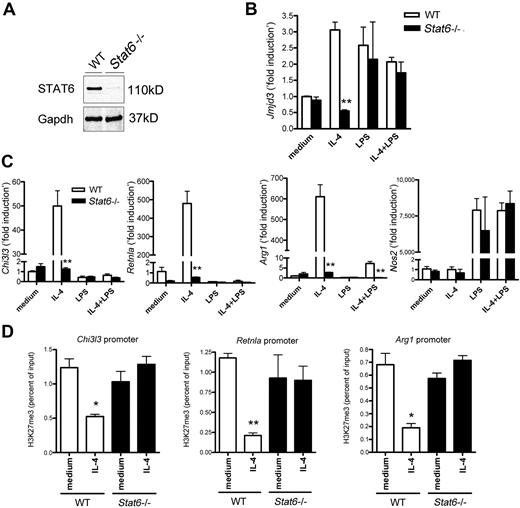
Because STAT6 is a major transcription factor for IL-4–mediated signaling response, we examined the role of STAT6 on the expression of the following genes: Jmjd3, Nos2 (an M1 marker gene), Chi3l3, Retnla, and Arg1 (M2 marker genes). BMDMs from WT and Stat6−/− mice were stimulated with IL-4, LPS, or a cocktail of IL-4 and LPS. STAT6 expression was not detected in Stat6−/− BMDMs (Figure 4A). The up-regulation of Jmjd3 was abolished in IL-4–treated Stat6−/− BMDMs, whereas the level of Jmjd3 was comparable between LPS-treated macrophages from WT and Stat6−/− mice (Figure 4B). This result suggests that IL-4–dependent up-regulation of Jmjd3 is specifically mediated by STAT6. As expected, we confirmed that the up-regulation of M2 marker genes was significantly suppressed in IL-4–treated Stat6−/− BMDMs (Figure 4C).
STAT6-dependent Jmjd3 induction in IL-4–induced M2-macrophages. (A) Immunoblotting for STAT6 in BMDMs from WT and Stat6−/− mice. Gapdh was used as a loading control. (B-C) The expression of Jmjd3 (B), Chi3l3, Retnla, Arg1, and Nos2 (C) in WT and Stat6−/− BMDMs was measured by quantitative RT-PCR after the indicated treatments. The data shown are “fold induction” relative to that in untreated WT cells. (D) Anti-H3K27me3 ChIP assay was performed on the promoter regions of M2 marker genes in WT and Stat6−/− BMDMs after IL-4 stimulation. Data are representative of 3 independent experiments and are expressed as mean ± SEM. *P < .05, **P < .01, compared with WT mice group (B-C) or unstimulated (medium alone) group (D).
STAT6-dependent Jmjd3 induction in IL-4–induced M2-macrophages. (A) Immunoblotting for STAT6 in BMDMs from WT and Stat6−/− mice. Gapdh was used as a loading control. (B-C) The expression of Jmjd3 (B), Chi3l3, Retnla, Arg1, and Nos2 (C) in WT and Stat6−/− BMDMs was measured by quantitative RT-PCR after the indicated treatments. The data shown are “fold induction” relative to that in untreated WT cells. (D) Anti-H3K27me3 ChIP assay was performed on the promoter regions of M2 marker genes in WT and Stat6−/− BMDMs after IL-4 stimulation. Data are representative of 3 independent experiments and are expressed as mean ± SEM. *P < .05, **P < .01, compared with WT mice group (B-C) or unstimulated (medium alone) group (D).
As STAT4 is a major transcription factor for Th1 immunity and its deficiency often results in enhancement of Th2-skewed immune response, such as parasite infections and allergic responses,40 we hypothesized that macrophages recovered from Stat4−/− mice may enhance the M2 phenotype. However, the gene levels of Jmjd3 and M2 marker genes were comparable between WT and Stat4−/− BMDMs when cells were treated with IL-4 (supplemental Figure 6).
Because we showed that IL-4–induced expression of Jmjd3 and M2 marker genes is STAT6-dependent, we next examined H3K27me3 levels at the promoters of M2 marker genes in untreated or IL-4–treated BMDMs from WT and Stat6−/− mice. Whereas IL-4 stimulation resulted in a significant decrease in H3K27me3 levels at all 3 promoters examined in WT macrophages, there was no decrease in the H3K27me3 levels in Stat6−/− macrophages (Figure 4D).
Direct regulation of Jmjd3 by STAT6 in M2-macrophages
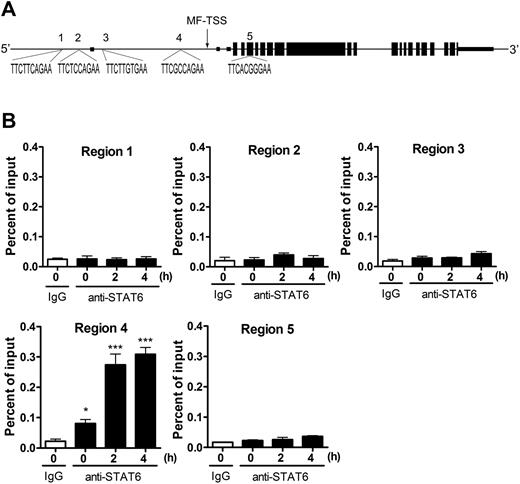
To test whether Jmjd3 is a direct downstream target of STAT6, we examined the promoter of Jmjd3. The macrophage transcriptional start site (MF-TSS) of the Jmjd3 gene has previously been reported, as shown in Figure 5A.28 It is known that several IL-4–dependent genes contain the consensus STAT6-binding site TTCnnnnGAA, where n represents any nucleotide.41 We found 5 STAT6 consensus sequences at Jmjd3 gene loci: 4 of them are within 6500 bp upstream of the MF-TSS (regions 1-4) and 1 is within 2500 bp downstream of MF-TSS (region 5; Figure 5A). To test whether STAT6 binds to any of these consensus sites after IL-4 stimulation, BMDMs were incubated with or without IL-4 for 2 and 4 hours. Recruitment of STAT6 to these regions of Jmjd3 gene was examined by ChIP assay. We found STAT6 binding only to region 4, which is immediately upstream of MF-TSS of Jmjd3 gene and was significantly increased after IL-4 stimulation (Figure 5B). This result strongly argues that Jmjd3 is a direct downstream target of STAT6.
Direct control of Jmjd3 induction by STAT6. (A) The structure of mouse Jmjd3 gene (NM_001017426). Small boxes represent untranslated regions; larger boxes, coding regions. The putative STAT6-binding sites (regions 1-5) and their sequences are shown. MF-TSS indicates macrophages-transcriptional start site.28 (B) Anti-STAT6 ChIP assay was performed with primers surrounding STAT6-binding site–containing regions of Jmjd3 gene in BMDMs after IL-4 stimulation. Data are representative of 3 independent experiments and are expressed as mean ± SEM. *P < .05, ***P < .001, compared with IgG control.
Direct control of Jmjd3 induction by STAT6. (A) The structure of mouse Jmjd3 gene (NM_001017426). Small boxes represent untranslated regions; larger boxes, coding regions. The putative STAT6-binding sites (regions 1-5) and their sequences are shown. MF-TSS indicates macrophages-transcriptional start site.28 (B) Anti-STAT6 ChIP assay was performed with primers surrounding STAT6-binding site–containing regions of Jmjd3 gene in BMDMs after IL-4 stimulation. Data are representative of 3 independent experiments and are expressed as mean ± SEM. *P < .05, ***P < .001, compared with IgG control.
Jmjd3 plays important roles in regulating the expression of M2-macrophage maker genes
Given the direct and specific response of Jmjd3 expression to IL-4–mediated STAT6 signal transduction pathway, we hypothesized that Jmjd3 plays important roles in down-regulating the level of H3K27me2/3 at M2 marker gene promoters and control M2 marker gene expression. To examine this, we first assessed the Jmjd3 recruitment to the promoter regions of M2 marker genes by ChIP assay. We found that the Jmjd3 recruitment at M2 marker genes was significantly increased in IL-4–induced M2-macrophages compared with untreated macrophages (Figure 6A), whereas Jmjd3 recruitment at Nos2 gene was not significantly increased (data not shown). The specificity of the Jmjd3 antibody was confirmed by ChIP assay in Jmjd3-knocked down BMDMs. The signal for Jmjd3 binding to selected M2 marker genes was greatly reduced in the knocked down cells (supplemental Figure 7). Next, we examined the role of Jmjd3 on H3K27me2/3 levels by knocking down the Jmjd3 gene. We confirmed that the Jmjd3 mRNA and protein levels were reduced in Jmjd3 siRNA-treated macrophages (Figure 6B-C). We then compared H3K27me2/3 levels between Jmjd3 siRNA- and control siRNA-treated macrophages. Knocking down the Jmjd3 rescued most of the H3K27me3 levels at all 3 M2 marker genes in IL-4–treated BMDMs (Figure 6D). Jmjd3 knockdown also partially rescued the H3K27me2 levels at the promoter regions of Chi3l3 and Retnla, with the exception at the Arg1 promoter region (Figure 6D). The site may be less sensitive to changes in Jmjd3 level or may involve other demethylase activities. Furthermore, the expression levels of M2 marker genes were significantly lower in Jmjd3 siRNA-treated macrophages compared with control siRNA-treated macrophages, whereas Nos2 levels were comparable between control and Jmjd3 siRNA-treated macrophages (Figure 6E). These results suggest that Jmjd3 contributes to maintaining M2 marker genes in a transcriptionally active state (Figure 6F).
M2 marker genes are targets of Jmjd3. (A) Anti-Jmjd3 ChIP assay was performed on the indicated M2 marker promoter regions in BMDMs 48 hours after IL-4 stimulation. (B-E) BMDMs were transfected with Jmjd3 siRNA or control siRNA. The cells were then incubated with IL-4 for 6 hours (B,E) or 24 hours (C), and the expression of indicated genes was measured by quantitative RT-PCR (B,E) or, in the case of Jmjd3, also by immunoblotting (C). Gapdh was used as a loading control. The data shown in panels B and E are “fold induction” relative to that in unstimulated control siRNA-treated cells. Jmjd3 siRNA- or control siRNA-transfected BMDMs were incubated with IL-4 for 48 hours (D). Anti-H3K27me2/3 ChIP assay was performed on the promoter regions of M2 marker genes (D). (F) The schema shows that STAT6-dependent Jmjd3 induction decreases H3K27me2/3 level on M2 marker promoter regions and helps to maintain M2 marker genes in a transcriptionally active state. Data of ChIP assay and quantitative RT-PCR are representative of 3 independent experiments and are expressed as mean ± SEM. *P < .05, **P < .01 compared with unstimulated (medium alone) group (A) or control siRNA group (B,E). #P < .05 compared with control siRNA–IL-4 group (D).
M2 marker genes are targets of Jmjd3. (A) Anti-Jmjd3 ChIP assay was performed on the indicated M2 marker promoter regions in BMDMs 48 hours after IL-4 stimulation. (B-E) BMDMs were transfected with Jmjd3 siRNA or control siRNA. The cells were then incubated with IL-4 for 6 hours (B,E) or 24 hours (C), and the expression of indicated genes was measured by quantitative RT-PCR (B,E) or, in the case of Jmjd3, also by immunoblotting (C). Gapdh was used as a loading control. The data shown in panels B and E are “fold induction” relative to that in unstimulated control siRNA-treated cells. Jmjd3 siRNA- or control siRNA-transfected BMDMs were incubated with IL-4 for 48 hours (D). Anti-H3K27me2/3 ChIP assay was performed on the promoter regions of M2 marker genes (D). (F) The schema shows that STAT6-dependent Jmjd3 induction decreases H3K27me2/3 level on M2 marker promoter regions and helps to maintain M2 marker genes in a transcriptionally active state. Data of ChIP assay and quantitative RT-PCR are representative of 3 independent experiments and are expressed as mean ± SEM. *P < .05, **P < .01 compared with unstimulated (medium alone) group (A) or control siRNA group (B,E). #P < .05 compared with control siRNA–IL-4 group (D).
Epigenetic regulation in macrophages recovered from S mansoni egg–challenged mice
Because we showed that IL-4–induced M2 marker genes are epigenetically regulated in M2-macrophages in vitro, we examined whether M2-macrophages from S mansoni egg–challenged mice, an established in vivo model for the generation of M2-macrophages, are epigenetically regulated. We first confirmed that peritoneal macrophages from S mansoni egg–intraperitoneally challenged mice have an M2 phenotype (Figure 7A) and the H3K27me2/3 levels at specific M2 marker gene promoters were significantly reduced in macrophages from S mansoni egg–challenged mice, which are consistent with our in vitro results (Figures 2A, 7B). We also examined the global Jmjd3 levels in macrophages recovered from control (sham) and S mansoni egg–challenged mice, which are either naive mice (Schisto ip in naive mice group) or mice that had been previously infected with S mansoni (Schisto ip in Schisto mice group). Jmjd3 levels were significantly higher in macrophages recovered from S mansoni egg–challenged mice (either Schisto ip in naive mice group or Schisto ip in Schisto mice group) compared with nonchallenged control (sham) mice. Interestingly, prior exposure to S mansoni led to a strong skewing to the M2 phenotype (supplemental Figure 8) and enhanced levels of Jmjd3 after the S mansoni egg challenge (Schisto ipy in Schisto mice group; Figure 7C). Finally, we found that Jmjd3 recruitment to M2 marker gene promoters in macrophages from S mansoni egg–challenged mice was significantly increased compared with sham controls (Figure 7D).
Epigenetic gene regulation of M2-macrophages from S mansoni egg–challenged mice in vivo. Naive mice were intraperitoneally injected with 1.7% sodium chloride solution alone (sham group) or with 5000 S mansoni eggs in 1.7% sodium chloride solution (Schisto group; ie, Schisto ip in naive mice group) for 7 days. (A) The peritoneal macrophages from Schisto and the sham control group were incubated with or without IL-4 for 6 hours. The expression of Chi3l3, Retnla, Arg1, and Nos2 was measured by quantitative RT-PCR. The data shown are “fold induction” relative to that in untreated cells from the sham group. (B) ChIP assay was performed to determine H3K27 methylation status on the promoter regions of Chi3l3, Retnla, and Arg1 in macrophages from both Schisto and sham groups. (C) Naive mice were intraperitoneally injected with 5000 S mansoni eggs (Schisto ip in naive group; ie, Schisto group). In addition, S mansoni–infected mice were intraperitoneally injected with 5000 S mansoni eggs (Schisto ipy in Schisto group). Jmjd3 mRNA levels in peritoneal macrophages from indicated group were measured by quantitative RT-PCR. The data shown are “fold induction” relative to that in cells from the sham group. (D) Anti-Jmjd3 ChIP assay was performed on M2 marker promoter regions in macrophages from Schisto and control groups. Data are represented as mean ± SEM of 8 to 10 mice per group. *P < .05, **P < .01, ***P < .001, compared with sham group.
Epigenetic gene regulation of M2-macrophages from S mansoni egg–challenged mice in vivo. Naive mice were intraperitoneally injected with 1.7% sodium chloride solution alone (sham group) or with 5000 S mansoni eggs in 1.7% sodium chloride solution (Schisto group; ie, Schisto ip in naive mice group) for 7 days. (A) The peritoneal macrophages from Schisto and the sham control group were incubated with or without IL-4 for 6 hours. The expression of Chi3l3, Retnla, Arg1, and Nos2 was measured by quantitative RT-PCR. The data shown are “fold induction” relative to that in untreated cells from the sham group. (B) ChIP assay was performed to determine H3K27 methylation status on the promoter regions of Chi3l3, Retnla, and Arg1 in macrophages from both Schisto and sham groups. (C) Naive mice were intraperitoneally injected with 5000 S mansoni eggs (Schisto ip in naive group; ie, Schisto group). In addition, S mansoni–infected mice were intraperitoneally injected with 5000 S mansoni eggs (Schisto ipy in Schisto group). Jmjd3 mRNA levels in peritoneal macrophages from indicated group were measured by quantitative RT-PCR. The data shown are “fold induction” relative to that in cells from the sham group. (D) Anti-Jmjd3 ChIP assay was performed on M2 marker promoter regions in macrophages from Schisto and control groups. Data are represented as mean ± SEM of 8 to 10 mice per group. *P < .05, **P < .01, ***P < .001, compared with sham group.
Discussion
A successful immune response is based on the host's ability to respond to a foreign challenge with a programmed repertoire of functions, which provides broad responses to innumerable antigens. This dynamic process is dependent on immune cells undergoing activation, differentiation, and trafficking events, which are dictated by mechanisms that either activate or silence the expression of various genes, resulting in a tailored response to a particular challenge. Whereas the development of T-cell and macrophage subsets is a good example of cell systems that are defined by genes they express during specific host responses, the molecular mechanisms that regulate these expression patterns are ill understood.
Recent investigations have brought some clarity to the mechanisms that control, in part, the cytokine phenotype that defines a Th1 or Th2 lymphocyte subset.42 In these studies, epigenetic regulation via chromatin remodeling was an important event that controlled the expression pattern of specific lymphocyte genes. Macrophage subsets are also classified by a set of proteins they express, as they in turn respond to the cytokine phenotype generated by specific T-cell subsets. M1-macrophages, as identified by their characteristic phenotype, are induced by IFN-γ derived from Th1 cells, and M2-macrophages, as identified by their characteristic M2 marker proteins, are induced by IL-4 and/or IL-13 derived from Th2 cells. In this study, we demonstrate that M2-macrophages are epigenetically regulated, and an H3K27 demethylase, Jmjd3, provides a supportive molecular activity that dictates the M2 expression profile.
Epigenetic modifications of histone tails are known to affect chromatin structure and consequently gene expression programs. Among them, methylations of histone tails at lysine residues are often correlated with transcriptional activation or repression. For instance, histone H3K4 methylation is linked to active transcription, whereas histone H3K27 methylation is linked to repressive transcription.23,25 Historically, histone methylation was considered as a static modification.24 However, recent discovery of histone lysine demethylases indicated that histone methylation can be dynamically regulated.25,43
Jmjd3 was recently identified as an H3K27me2/3-specific demethylase, and it was shown to be an important regulator of Hox genes.26,28,29 Because H3K27me2/3 was decreased on M2 marker gene promoter regions in IL-4–stimulated macrophages in parallel with up-regulation of M2 marker genes, we hypothesized that Jmjd3 participates in these processes. We demonstrated that Jmjd3 was specifically induced by IL-4 and recruited to the promoter regions of M2 marker genes. Because STAT6 is a pivotal transcription factor for IL-4–mediated signaling,14 we further hypothesized that STAT6 participates in these IL-4–induced Jmjd3 activities. Our studies of Stat6−/− macrophages documented that IL-4–induced Jmjd3 induction is STAT6-dependent. Furthermore, STAT6 directly bound to the Jmjd3 promoter region, confirming that Jmjd3 is a direct downstream target of STAT6.
We also found that IL-4 caused a decrease in H3K27me2/3 levels on the promoter regions of M2 marker genes that was partially restored by knocking down Jmjd3, suggesting that Jmjd3 is actively involved in the dynamic regulation of H3K27me2/3. In contrast to H3 K27 methylation, H3K4 methylation is induced by IL-4 treatment; however, it remains high after IL-4 withdrawal. Given the correlation of H3K27 methylation with the expression of M2 marker gene, it is probable that M2 marker genes are directly regulated by the dynamic interactions between H3K27 methyltransferase and demethylase instead of the antagonism between H3K4 methylation and H3K27 methylation that was widely used for gene expression control. The incomplete recovery of H3K27me2/3 levels in these studies may be the result of unknown histone demethylases. Considering that histone lysine methylation is mediated by approximately 50 histone methyltransferases,44 a comparable number of histone demethylases may also exist.25
Our findings of impaired up-regulation of M2 marker genes in IL-4–treated Jmjd3 knocked down macrophages as well as the direct regulation of Jmjd3 expression by STAT6 suggest that Jmjd3 is an important downstream effector of the IL-4–mediated STAT6 signaling pathway for the efficient induction of M2 marker genes. Jmjd3 knockdown led to significant, but not complete, reduction of specific M2 marker genes. This could be the result of the possibility that other functional pathways are involved in the regulation of specific M2 marker genes.
Jmjd3 has also been shown to be up-regulated by LPS in macrophages.28 In the previous study, the macrophage cell line (Raw264.7 cells) appeared to be more susceptible to LPS-induced Jmjd3 expression than primary macrophage cultures.28 Consistently, we found that Jmjd3 was induced to similar levels by either LPS or IL-4 in our primary macrophages. The fact that LPS is a potent activating agent for macrophages with an M1 phenotype may seem at odds with our finding that Jmjd3 appears to be important for the M2-macrophage phenotype. However, up-regulation of global Jmjd3 expression by LPS does not necessarily mean increased recruitment of Jmjd3 to specific gene promoters. Indeed, Jmjd3 recruitment to M2 marker gene promoters was not increased in LPS-stimulated macrophages (supplemental Figure 9).
Our studies suggest that M2-macrophages from S mansoni egg–challenged mice are subject to chromatin regulation in vivo. There is an increase of Jmjd3 recruitment and a corresponding reduction of H3K27me2/3 at the promoters of M2 marker genes. These results led us to speculate that M2-macrophages are probably epigenetically regulated through chromatin modifications in a clinical setting, and these concepts are worthy for therapeutic interventions. Interestingly, a significant amount of information suggests an important role for M2-macrophages in cancer9,10 and diabetic patients,7,45 whose disease phenotype may be controlled by dynamic chromatin alterations. Jmjd3 has been reported to be expressed in prostate cancer tissues, and the expression profiles paralleled the disease severity.46 Tumor-associated macrophages, which generally assume an M2 phenotype,9,10 could also be dependent on an protein profile influenced by the expression of Jmjd3.
In conclusion, we demonstrate that M2-macrophage marker genes are epigenetically regulated by reciprocal changes in histone H3 lysine-4 (H3K4) and histone H3 lysine-27 (H3K27) methylation. A novel aspect of this epigenetic regulation is that STAT6-dependent induction of H3K27 demethylase Jmjd3 contributes to decreasing H3K27 methylation on the promoter region of M2 marker genes and subsequent maintenance of M2 marker genes in a transcriptionally active state. Further, our in vivo studies suggest that the increased recruitment of Jmjd3 can be associated with a decrease in H3K27 methylation in macrophages obtained from S mansoni egg–challenged mice. Our studies highlight the mechanisms that underlie the acquisition and maintenance of the M2-macrophage phenotype, a phenotype that may be important in the development of a variety of chronic diseases.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Pamela Lincoln, Holly Evanoff, and Aaron Berlin for technical assistance; Robin Kunkel for her artistic work; Judith Connett for critical reading of the manuscript; and Colin Duckett for providing advice.
This work was supported by the National Institutes of Health (HL31327, HL89216, HL31963, and HL092845; S.L.K.), the Kanae Foundation for the Promotion of Medical Science in Japan (M.I.), and the Uehara Memorial Foundation (M.I.).
National Institutes of Health
Authorship
Contribution: Y.D., C.M.H., N.W.L., and S.L.K. designed the experiments; M.I. performed the experiments and collected and analyzed data; H.W., C.A.S.C., T.L., A.L.C., R.M.A., W.F.C., K.A.C., and X.L. contributed to some of the experiments; M.I., Y.D., C.M.H., N.W.L., and S.L.K. contributed to manuscript preparation; and S.L.K. supervised the project.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Steven L. Kunkel, Department of Pathology, University of Michigan Medical School, Rm 4071, Biomedical Sciences Research Building, 109 Zina Pitcher Pl, Ann Arbor, MI 48109-2200; e-mail: slkunkel@umich.edu.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal