Abstract
The gene(s) responsible for natural killer (NK)–cell lymphoma/leukemia have not been identified. In the present study, we found that in NK-cell lymphoma lines (n = 10) and specimens of primary lymphoma (n = 10), levels of miR-21 and miR-155 expression were inversely related and were significantly greater than those found in normal natural killer (CD3−CD56+) cells (n = 8). To determine the functions of these microRNAs in lymphomagenesis, we examined the effects of antisense oligonucleotides (ASOs) targeting miR-21 (ASO-21) and/or miR-155 (ASO-155) in NK-cell lymphoma lines overexpressing one or both of these miRNAs. Conversely, cells showing little endogenous expression of miR-21 or miR-155 were transduced by the use of lentiviral vectors, leading to their overexpression. Reducing expression of miR-21 or miR-155 led to up-regulation of phosphatase and tensin homologue (PTEN), programmed cell death 4 (PDCD4), or Src homology-2 domain-containing inositol 5-phosphatase 1 (SHIP1). ASO-21– and ASO-155–treated cell lines all showed down-regulation of phosphorylated AKTser473. Moreover, transduction with either miR-21 or miR-155 led to down-regulation of PTEN and PDCD4 or SHIP1 with up-regulation of phosphorylated AKTser473. Collectively, these results provide important new insight into the pathogenesis of NK-cell lymphoma/leukemia and suggest targeting miR-21 and/or miR-155 may represent a useful approach to treating NK-cell lymphoma/leukemia.
Introduction
Natural killer (NK)–cell lymphomas/leukemias are characterized groups of highly aggressive lymphoid malignancies, which are composed of “extranodal NK/T-cell lymphoma, nasal type” and “aggressive NK-cell leukemia.”1 Notably, these 2 subtypes show many similarities in their morphologic features, immunophenotypes, and genotypes and are invariably associated with Epstein-Barr virus (EBV), which suggests they may share the same genetic alterations. To assign a classification, the World Health Organization classification uses cytogenetic and molecular features to characterize lymphoma subtypes.1 For example, it is known that various genomic translocations and genetic alterations, including BCL2, CCDN1, and c-MYC, occur in B-cell lymphomas. These disease-specific genetic translocations characterize lymphoma subtypes, such as follicular lymphoma characterized by BCL2 rearrangement, mantle-cell lymphoma characterized by CCDN1 rearrangement, and Burkitt lymphoma characterized by c-MYC rearrangement. However, although the World Health Organization classification recognizes NK-cell lymphomas/leukemias as distinct clinicopathologic entities, disease-specific translocations and the gene(s) affected in the 2 subtypes have not yet been identified. It was previously reported that a 6q deletion occurs in approximately 10% to 20% of NK-cell lymphomas/leukemias2-7 ; however, this loss may not be disease specific because it has been observed in a variety of cancers, including solid tumors and hematologic malignancies. It is currently unclear whether the loss is a primary or progression-associated event.
It was recently discovered that some microRNAs (miRNAs) are oncogenic in B-cell lymphomas. For example, aberrant overexpression of 2 miRNAs, miR-17-92 and miR-155, is closely associated with B-cell lymphomagenesis.8 With respect to miR-17-92, we recently demonstrated that the polycistron can down-regulate CDKN1A/p21 in B-cell lymphomagenesis and promote cell-cycle regulation.8 Furthermore, it is unlikely that aberrant expression of miRNAs is restricted to B-cell lymphomas, and it may occur in other lymphoma subtypes, including T/NK-cell lymphomas. In the present study, therefore, we used Northern and quantitative polymerase chain reaction (PCR) analyses to screen for and quantitatively assess miRNA expression in NK-cell lymphomas/leukemias and found that miR-21 and miR-155 were overexpressed in NK-cell lymphoma/leukemia. Moreover, the effects of antisense oligonucleotides (ASOs) revealed that miR-21 and miR-155 act as oncomiRNAs, promoting NK-cell lymphomagenesis through dysregulation of AKT signaling.
Methods
Cell lines
The following 10 NK-cell lymphoma/leukemia cell lines commonly show CD2+, sCD3−, CD3ϵ+, CD5−, CD19−, CD56+, TCRα/β −, and TCRγ/δ − phenotypes: NKL, KHYG1, YT, NK92, HANK1, KAI3, SNK1, SNK6, DERL7, and MOTN1.9-18 The culture medium was RPMI1640 supplemented with 10% fetal calf serum or 5% human serum with 200 U (NKL, KHYG1, YT, KAI3, NK92, HANK1, DERL7, and MOTN1) or 700 U (SNK1 and SNK6) of IL-2. Rat-1 was derived from rat fibroblasts that were obtained from the RIKEN Bioresource Center and cultured in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum.
Primary NK-cell lymphoma/leukemia samples and normal sCD3− and CD56+ cells
Ten specimens of primary NK-cell lymphoma/leukemia, including 7 “extranodal NK/T cell lymphoma, nasal type” and 3 “aggressive NK-cell leukemia,” were collected from 10 patients (9 men and 1 woman, ranging in age from 44 to 82 years; Table 1). Samples were obtained from 10 patients with an Institutional Review Board of Akita University–approved protocol. Informed consent was obtained from all patients according to the Declaration of Helsinki before collection of the specimens. In addition, none of the patients had a history of malignant lymphoma/leukemia. All the samples were obtained from tumors at the time of diagnosis before any treatment was administered. All samples showed CD2 and CD56 positivity. Eight cases were also positive for CD3ϵ but negative for sCD3. All cases showed either EBER-ISH positive or cytotoxic molecules (TIA-1 or Granzyme B) positive or both. Normal sCD3−CD56+ cells also were collected from 8 healthy donors (3 women and 5 men; 3 of these 8 also were used as control samples in Northern blot analyses) by the use of a magnetic cell-sorting system (Miltenyi Biotec) with CD3–fluorescein isothiocyanate (FITC), CD19-FITC, and CD56-FITC antibodies and anti-FITC MicroBeads. Southern blot analysis was performed for 4 of 10 cases by the use of TCR-Cβ-1 probe.
Summary of primary NK-cell lymphoma/leukemia cases
| Patient no. . | Sex . | Age . | Diagnosis . | Origin . | EBER-ISH . | sCD3 . | CD2 . | CD5 . | CD20 . | CD56 . | TCRre . | Cytotoxic molecules . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NKpt1 | M | 84 | Extranodal NK/T-cell lymphoma, nasal type | Orbit | + | − | + | − | − | + | ND | TIA-1+, granzyme B+ |
| NKpt2 | M | 57 | Extranodal NK/T-cell lymphoma, nasal type | Skin | − | + | + | − | − | + | − | TIA-1+, granzyme B+ |
| NKpt3 | M | 58 | Aggressive NK-cell leukemia | Peripheral blood | + | − | + | − | − | + | ND | TIA-1−, granzyme B− |
| NKpt4 | M | 68 | Extranodal NK/T-cell lymphoma, nasal type | Nasal cavity | + | + | + | − | − | + | ND | TIA-1+, granzyme B+ |
| NKpt5 | M | 44 | Extranodal NK/T-cell lymphoma, nasal type | Nasal cavity | + | − | + | − | − | + | − | TIA-1+, granzyme B+ |
| NKpt6 | M | 53 | Extranodal NK/T-cell lymphoma, nasal type | Nasal cavity | + | − | + | − | − | + | − | TIA-1+, granzyme B− |
| NKpt7 | F | 68 | Extranodal NK/T-cell lymphoma, nasal type | Nasal cavity | ND | − | + | − | − | + | ND | TIA-1+, granzyme B+ |
| NKpt8 | M | 60 | Aggressive NK-cell leukemia | Bone marrow | + | − | + | − | − | + | − | ND |
| NKpt9 | M | 75 | Extranodal NK/T-cell lymphoma, nasal type | Nasal cavity | + | − | + | − | − | + | ND | TIA-1+, granzyme B− |
| NKpt10 | M | 56 | Aggressive NK-cell leukemia | Bone marrow | + | − | + | − | − | + | ND | TIA-1+, granzyme B+ |
| Patient no. . | Sex . | Age . | Diagnosis . | Origin . | EBER-ISH . | sCD3 . | CD2 . | CD5 . | CD20 . | CD56 . | TCRre . | Cytotoxic molecules . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NKpt1 | M | 84 | Extranodal NK/T-cell lymphoma, nasal type | Orbit | + | − | + | − | − | + | ND | TIA-1+, granzyme B+ |
| NKpt2 | M | 57 | Extranodal NK/T-cell lymphoma, nasal type | Skin | − | + | + | − | − | + | − | TIA-1+, granzyme B+ |
| NKpt3 | M | 58 | Aggressive NK-cell leukemia | Peripheral blood | + | − | + | − | − | + | ND | TIA-1−, granzyme B− |
| NKpt4 | M | 68 | Extranodal NK/T-cell lymphoma, nasal type | Nasal cavity | + | + | + | − | − | + | ND | TIA-1+, granzyme B+ |
| NKpt5 | M | 44 | Extranodal NK/T-cell lymphoma, nasal type | Nasal cavity | + | − | + | − | − | + | − | TIA-1+, granzyme B+ |
| NKpt6 | M | 53 | Extranodal NK/T-cell lymphoma, nasal type | Nasal cavity | + | − | + | − | − | + | − | TIA-1+, granzyme B− |
| NKpt7 | F | 68 | Extranodal NK/T-cell lymphoma, nasal type | Nasal cavity | ND | − | + | − | − | + | ND | TIA-1+, granzyme B+ |
| NKpt8 | M | 60 | Aggressive NK-cell leukemia | Bone marrow | + | − | + | − | − | + | − | ND |
| NKpt9 | M | 75 | Extranodal NK/T-cell lymphoma, nasal type | Nasal cavity | + | − | + | − | − | + | ND | TIA-1+, granzyme B− |
| NKpt10 | M | 56 | Aggressive NK-cell leukemia | Bone marrow | + | − | + | − | − | + | ND | TIA-1+, granzyme B+ |
EBER-ISH indicates EBER in situ hybridization; ND, not determined; NK, natural killer; TCRre, T-cell receptor rearrangement; +, positive stain; and −, negative stain.
Northern blot analysis
Northern blotting for mature microRNAs was performed as described elsewhere.8 In brief, total RNA was extracted from the cell lines by the use of the acid-phenol precipitation method, after which 2-μg normal control samples or 5-μg cell line samples were separated on 15% denaturing polyacrylamide gels. The blots for all 4 membranes hybridized by γ32-dATP (1.85 MBq per 10 pmol probe; for 5S tRNA, 1 pmol)–antisense oligo for microRNAs were exposed at the same conditions: 8 hours at −80°C with BioMax films (Kodak).
Western blot analysis
Western analyses were performed as described elsewhere.8 Antibodies against phosphatase and tensin homologue (PTEN; A2B1) and Src homology-2 domain-containing inositol 5-phosphatase 1 (SHIP1; P1C1) were purchased from Santa Cruz Biotechnology. Antibodies against programmed cell death protein 4 (PDCD4), phospho-NFκB (phospho-p65ser536; 93H1), phosphor-AKT ser473 (pAKT), total AKT, Apaf-1, RECK (D8C7), p21WAF1/CIP1 (p21), and p27KIP1 (p27) were all purchased from Cell Signaling Technology (Cell Cycle Regulation Sampler Kit). Anti-Bim (AAP-330) was from Stressgen Bioreagents. Anti-Bcl2 (BCL-2-100) was from Sigma. Anti-p53 (DO-7) was from Dako Cytomation.
Real-time quantitative PCR for miRNAs (Taqman PCR)
A Recover All Total Nucleic Acid Isolation kit (Applied Biosystems) was used to extract total RNA from primary specimens embedded in paraffin. Quantitative stem-loop reverse transcription (RT) was then performed by the use of a Taqman microRNA RT kit (Applied Biosystems), after which quantitative PCR for mature miRNAs was performed by the use of Taqman microRNA assays (Applied Biosystems). All reactions were run in duplicate. Mean cycle threshold (Ct) values for all miRNAs were quantified by the use of sequence detection system software (SDS, version 2.1; Applied Biosystems). The miRNA expression was normalized to U47 mRNA expression, yielding a −ΔCt value. The −ΔΔCt value was then calculated by subtracting the −ΔCt value for a standard normal sCD3−CD56+ sample from the respective −ΔCt values from the cancer cells. The −ΔΔCt values were then imported into Microsoft Excel, and P values were calculated by the use of a Student t test. Quantitative RT-PCR was performed by the use of a Universal probe library (Roche Diagnostics) and Light Cycler 480 probe master (Roche Diagnostics) with the Taqman method.
Construction of plasmids and transduction
Lentiviral vectors for the delivery of microRNA were designed and produced by use of the reagents and protocols included in the BLOCK-iT Lentiviral Pol II miR RNA interference expression system (Invitrogen). The premicro-RNA (miR-21 and miR-155) and its reverse complement were annealed and ligated into the pcDNA6.2-GW/EmGFP-miR vector, which contains the full premicro-RNA 5′- and 3W′-flanking regions, as well as the cocistronic Emerald GFP (EmGFP) gene. After sequence verification, the EmGFP-premicro-RNA cassette was transferred to the pLenti6/V5 expression construct by the use of BP/LR recombination reactions. Three micrograms of the resulting pLenti6/EmGFP-premicro-RNA vector, together with 9 μg of ViraPower Packaging mix, was transfected by the use of Lipofectamine 2000 into 293FT producer cells. After overnight culture, medium was exchanged to remove transfection reagents. The following day, virus stocks were harvested. Viral supernatants were harvested 48 to 72 hours after transfection. Cells stably expressing miR-21 or miR-155 were sorted for green fluorescent protein (GFP) expression by the use of a Dako Cytomation MoFlo, after which individual clones were isolated. In Rat-1 cells, the pre miR-155 was cloned into the appropriate cloning site of pMXspuro, after which Rat-1 cells were stably transfected. Detailed transfection methods was described elsewhere.8
ASO assay
ASO and their respective scrambled control oligonucleotides were synthesized as hybrid deoxyribonucleotide molecules linked between the 2′-O and 4′-C-methylene bridge (locked nucleic acid) modification of G and C residues (Greiner). The ASOs used were as follows: AS-miR-21 (ASO-21), 5′-TCAACATCAGTCTGATAAGCTA-3′ and AS-miR-155 (ASO-155), 5′-TCCCCTATACACGATTAGCATTAA-3′. The SCOs used were SC-miR-21 (SCO-21), 5′-TAACGTCACTTCGACTGAACTGCT-3′ and SC-miR-155 (SCO-155), 5′-ATCTCATACTACACTTGAACACT-3′. To assess their effects, cells were plated to a density of 105 cells/well in 24-well dishes and then transfected with oligonucleotides (20 nmol/L) by the use of Lipofectamine 2000 (day 1). Two days later (day 3), the cells were harvested and resuspended in a fresh medium. Then cells were transfected again with oligonucleotides (20 nmol/L) by the use of Lipofectamine 2000 by day 5 and analyzed.
Cell-cycle analysis
For cell-cycle analysis, the cells were suspended in a mixture containing 0.2 mL of 0.9% NaCl and 3 mL of 70% EtOH, after which the nuclei were stained with propidium iodide (Sigma). The cellular DNA content was then measured with a FACSCalibur flow cytometer running the CELL Quest program (BD Biosciences).
Apoptosis analysis
For apoptosis analysis, an annexin V–PE apoptosis detection kit (BD Biosciences) was used to assess the incidence of apoptosis among GFP-positive cells. In addition, an annexin V–FITC apoptosis detection kit (Sigma) was used to assess the incidence of apoptosis among GFP-negative cells. Cells were exposed by 50 μmol/L etoposide for 2 hours (KAI3 cells) or 100 μmol/L etoposide for 4 hours (NKL, YT, and MOTN1 cells), after which the assays were performed.
Luciferase reporter assay
The pGL3 control vector (Promega) encoding firefly luciferase was modified such that the INPP5D/SHIP1 3′UTR was inserted into the XbaI site immediately downstream from the stop codon. Rat-1 cells expressing miR-155 were cultured to 80% to 90% confluence in 24-well plates and then transfected with 0.8 μg of the firefly luciferase reporter vector and 0.16 μg of pRL-TK control vector (Promega) encoding Renilla luciferase with Lipofectamine 2000 in a final volume of 1.0 mL. Thirty hours later, firefly and Renilla luciferase activities were measured consecutively in Dual-luciferase assays (Promega). Two independent experiments were conducted in triplicate. In addition, the following sequences were inserted into the PGL3 control vector: wild-type INPP5D/SHIP1 3′UTR, 5′-CACCAGTTTAAAACGGTGTGTGTTCGGAGGGGTGAAAGCATTAAGAAGCCCAGTGCCCTCCTGGAGTGAG-3′ (the underlined sequence is the conserved target of miR-155) and mutated INPP5D/SHIP1 3′ UTR, 5′- CACCAGTTTAAAACGGTGTGTGTTCGGAGGGGTGAAtGCATTAtGAAGCCCAGTGCCCTCCTGGAGTGAG-3′ (mutations are indicated by lowercase characters).
Results
Detection of aberrant overexpression of microRNAs in NK-cell lymphoma/leukemia lines
To detect aberrant overexpression of miRNAs in NK-cell lymphomas/leukemias, we initially performed a Northern analysis with 66 probe sets (supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article) with 12 lymphoma/leukemia cell lines, which included 6 NK-cell, 2 T-cell, and 4 B-cell lymphomas/leukemias. The probe sets for the miRNAs were selected based on recent reports on cancer-associated miRNAs. We found that miR-21, miR-23a, miR-29 family (miR-29a-c), and miR-155 were all more highly expressed in NK-cell lymphoma cell lines than in B-cell lymphoma cell lines. miR-20, miR-26a, miR-92, miR-103, and miR-181 were also ubiquitously overexpressed in the examined samples.
Because our interest was to investigate NK-cell lymphoma–specific overexpression of miRNA, we further examined expression of miR-21, miR-23a, miR29 family (miR-29a-c), and miR-155 in 10 NK-cell lymphoma cell lines. We defined “overexpression” as a level of miRNA expression that was more than 2-fold greater than the average in corresponding normal cells. The expression levels of these 4 miRNAs in 2 T-cell lines, 6 B-cell lymphoma/leukemia lines, and 3 normal sCD3−CD56+ samples (control) are compared in Figure 1A-B. Expression of miR-21 and miR-155 in KAI3 cells was 4.7- to 8.8-fold greater than the average expression level observed in normal control cells (averages: 0.17 for miR-21; 0.36 for miR-155; Figure 1B). Expression of miR-21 in KAI3, YT, NK92, NKL, KHYG1, HANK1, SNK1, SNK6, and DERL7 was 3.1- to 15.7-fold greater than in normal control cells. miR-155 expression in KAI3, KHYG1, HANK1, SNK1, SNK6, and MOTN1 was 2.6- to 10.2-fold greater than in normal control cells (Figure 1A-B). Expression of miR-21 and miR-155 in these cell lines were overexpressed than normal control cells. By contrast, levels of miR-21 in MOTN1 cells and levels of miR-155 in YT, NK92, NKL, and DERL7 cell were the same or less than 2-fold lower than in control cells. Expression of miR-23a in KAI3 cells was similar to that in the normal control samples, whereas levels of miR-29a were slightly greater in the KAI3 cells than in the control samples.
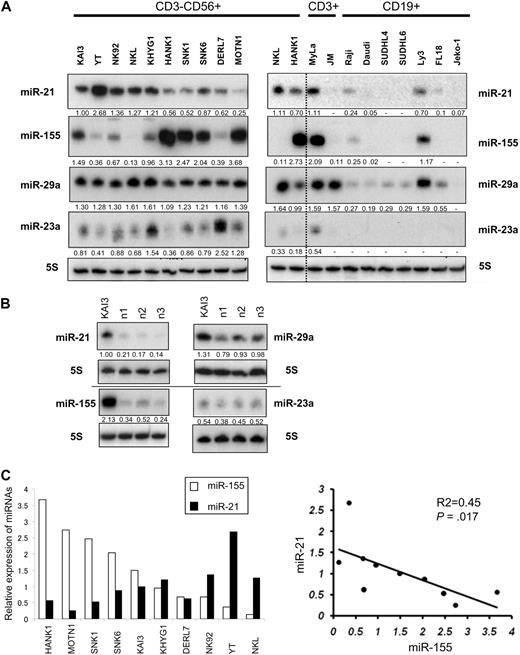
Northern analysis of miR-21, miR-23a, miR-29a, and miR-155 in NK-cell lymphoma/leukemia lines. (A) Northern analysis of miR-21, miR-23a, miR-29a, and miR-155 in 10 NK-cell (CD3−CD19−CD56+; left panel), 2 T-cell (CD3+CD19−CD56−; right panel), and 7 B-cell (CD3−CD19+CD56−; right panel) lymphoma/leukemia lines. NKL and HANK1 are shown in both panels in duplicate to compare miRNA expressions. Fold changes in miRNA were determined by densitometry and are shown below the gels after normalization to the level of miR-21 in KAI3 cells, which was assigned a value of 1.00. The blots for all 4 membranes hybridized by γ32-dATP-antisense oligo for microRNAs were exposed at the same condition (see “Methods”). (B) Expression of miR-21, miR-23a, miR-29a, and miR-155 measured in 3 samples of normal cells (n1, n2, and n3) expressing sCD3−CD56+. The level of miR-21 expression in KAI3 cells was assigned a value of 1.00. (C) Correlation between miR-21 and miR-155 expression. In the left panel, □ and ■ depict relative expression values for miR-155 and miR-21 expression, respectively. In the right panel, y-axis, expression values for miR-21 expression; x-axis, expression values for miR-155 expression. The P value was calculated by simple regression analysis.
Northern analysis of miR-21, miR-23a, miR-29a, and miR-155 in NK-cell lymphoma/leukemia lines. (A) Northern analysis of miR-21, miR-23a, miR-29a, and miR-155 in 10 NK-cell (CD3−CD19−CD56+; left panel), 2 T-cell (CD3+CD19−CD56−; right panel), and 7 B-cell (CD3−CD19+CD56−; right panel) lymphoma/leukemia lines. NKL and HANK1 are shown in both panels in duplicate to compare miRNA expressions. Fold changes in miRNA were determined by densitometry and are shown below the gels after normalization to the level of miR-21 in KAI3 cells, which was assigned a value of 1.00. The blots for all 4 membranes hybridized by γ32-dATP-antisense oligo for microRNAs were exposed at the same condition (see “Methods”). (B) Expression of miR-21, miR-23a, miR-29a, and miR-155 measured in 3 samples of normal cells (n1, n2, and n3) expressing sCD3−CD56+. The level of miR-21 expression in KAI3 cells was assigned a value of 1.00. (C) Correlation between miR-21 and miR-155 expression. In the left panel, □ and ■ depict relative expression values for miR-155 and miR-21 expression, respectively. In the right panel, y-axis, expression values for miR-21 expression; x-axis, expression values for miR-155 expression. The P value was calculated by simple regression analysis.
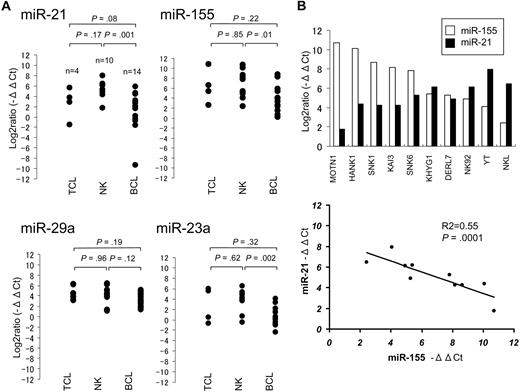
Comparison of the miRNA expression in NK-cell lymphoma/leukemia lines revealed a significant inverse relation between the expression of miR-21 and miR-155 (P = .017; Figure 1C), but no significant relations were detected for any other miRNA pairs (eg, miR-21 vs miR-23a; data not shown). We next performed quantitative real-time PCR (Taqman PCR) by the use of 28 lymphoma cell lines, including 10 NK-cell, 4 T-cell, and 14 B-cell lymphoma/leukemia lines, as well as 8 normal control samples. These cell lines and control cells include all samples presented in Figure 1. Taqman PCR showed significantly greater expression of these miRNAs in the NK-cell and T-cell lymphoma/leukemia lines than in the B-cell lymphoma lines (Figure 2A), but there was no significant difference in their expression between NK-cell and T-cell lymphomas/leukemias. Moreover, Taqman PCR also showed an inverse correlation between the expression of miR-21 and miR-155 in the NK-cell lymphoma/leukemia cell lines (Figure 2B).
Comparison of miRNA expression in T-, B-, and NK-cell lymphoma/leukemia lines with quantitative RQ-PCR (Taqman PCR). (A) Taqman PCR analysis of miR-21, miR-23a, miR-29a, and miR-155 expression in 4 T-cell, 10 NK-cell, and 14 B-cell lymphoma/leukemia lines. The significance of differences between T- and NK-cell, between NK- and B-cell, and between T- and B-cell lymphoma/leukemia lines were assessed by use of the Student t test. The following B- and T-cell lymphoma/leukemia lines were used for this study. SUDHL4, SUDHL6, OCI-Ly3, and OCI-Ly7 cells were derived from diffuse large B-cell lymphoma; Raji, Daudi, Ramous, Namalwa, and D.G-75 cells were derived from Burkitt lymphoma; SP-49 and Jeko-1 cells were derived from mantle cell lymphoma; FL-18 cells were derived from follicular lymphoma; Karpass 1718 and SLVL cells were derived from splenic virus lymphoma; ATN1 cells were derived from adult T-cell leukemia; JM and MOLT4 cells were derived from T-cell leukemia; and MyLa cells were derived from peripheral T-cell lymphoma unspecified. (B) Correlation between expression miR-21 and miR-155 in 10 NK-cell lymphoma/leukemia lines. In the top panel, □ and ■ bars depict −ΔΔCt values for miR-155 and miR-21 expression, respectively. In the bottom panel: y-axis, −ΔΔCt values for miR-21 expression; x-axis, −ΔΔCt values for miR-155 expression are shown. The P value was calculated by simple regression analysis.
Comparison of miRNA expression in T-, B-, and NK-cell lymphoma/leukemia lines with quantitative RQ-PCR (Taqman PCR). (A) Taqman PCR analysis of miR-21, miR-23a, miR-29a, and miR-155 expression in 4 T-cell, 10 NK-cell, and 14 B-cell lymphoma/leukemia lines. The significance of differences between T- and NK-cell, between NK- and B-cell, and between T- and B-cell lymphoma/leukemia lines were assessed by use of the Student t test. The following B- and T-cell lymphoma/leukemia lines were used for this study. SUDHL4, SUDHL6, OCI-Ly3, and OCI-Ly7 cells were derived from diffuse large B-cell lymphoma; Raji, Daudi, Ramous, Namalwa, and D.G-75 cells were derived from Burkitt lymphoma; SP-49 and Jeko-1 cells were derived from mantle cell lymphoma; FL-18 cells were derived from follicular lymphoma; Karpass 1718 and SLVL cells were derived from splenic virus lymphoma; ATN1 cells were derived from adult T-cell leukemia; JM and MOLT4 cells were derived from T-cell leukemia; and MyLa cells were derived from peripheral T-cell lymphoma unspecified. (B) Correlation between expression miR-21 and miR-155 in 10 NK-cell lymphoma/leukemia lines. In the top panel, □ and ■ bars depict −ΔΔCt values for miR-155 and miR-21 expression, respectively. In the bottom panel: y-axis, −ΔΔCt values for miR-21 expression; x-axis, −ΔΔCt values for miR-155 expression are shown. The P value was calculated by simple regression analysis.
Overexpression of microRNAs in primary NK-cell lymphoma/leukemia cases
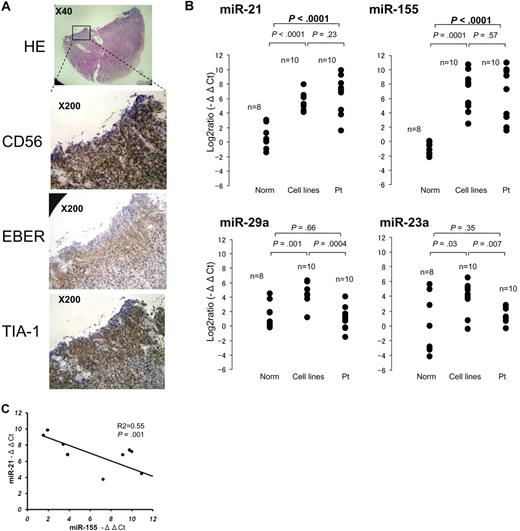
To examine the expression of candidate microRNAs in primary extranodal NK-cell lymphoma (Table 1), we extracted total RNA from paraffin-embedded tissues, from CD56+ tumor cells (Figure 3A) collected by microscopic dissection. Aggressive NK-cell leukemia samples were obtained from peripheral blood or bone marrow. Total RNA from normal sCD3−CD56+ cells were obtained from peripheral blood samples collected from 8 healthy donors. Comparison of patient samples to normal sCD3−CD56+ cells revealed no significant differences in the expression of miR-23a and miR-29a (Figure 3B). We therefore excluded these miRNAs from further investigation. By contrast, miR-21 and miR-155 were expressed at significantly (P < .001) greater levels in cancer cells than normal sCD3−CD56+ cells (Figure 3B), and expression levels of miR-21 and miR-155 were also inversely related in primary specimens, as was observed in cell lines (P = .001; Figure 3C).
Quantitative PCR analysis of miRNA levels in specimens of primary NK-cell lymphoma/leukemia. (A) Histopathologic features of a typical specimen of primary extranodal NK/T-cell lymphoma, nasal type (NKpt5). Hematoxylin and eosin–stained (×40), CD56-stained, EBER-ISH–stained, and TIA-1-stained (×200) cells are shown. (B) Quantitative PCR analysis of mature miR-21, miR-23a, miR-29a, and miR-155 in normal sCD3−CD56+ cells (n = 8), NK-cell lymphoma/leukemia lines (n = 10), and specimens of primary NK-cell lymphoma/leukemia (n = 10). Y-axis: −ΔΔCt values for miRNA expression. The significance of differences between groups was evaluated by the use of the Student t test. (C) Correlation between miR-21 and miR-155 expression in 10 primary lymphoma/leukemia specimens. y-axis, −ΔΔCt values of miR-21 expression; x-axis, −ΔΔCt values miR-155 expression. The P value was calculated by simple regression analysis.
Quantitative PCR analysis of miRNA levels in specimens of primary NK-cell lymphoma/leukemia. (A) Histopathologic features of a typical specimen of primary extranodal NK/T-cell lymphoma, nasal type (NKpt5). Hematoxylin and eosin–stained (×40), CD56-stained, EBER-ISH–stained, and TIA-1-stained (×200) cells are shown. (B) Quantitative PCR analysis of mature miR-21, miR-23a, miR-29a, and miR-155 in normal sCD3−CD56+ cells (n = 8), NK-cell lymphoma/leukemia lines (n = 10), and specimens of primary NK-cell lymphoma/leukemia (n = 10). Y-axis: −ΔΔCt values for miRNA expression. The significance of differences between groups was evaluated by the use of the Student t test. (C) Correlation between miR-21 and miR-155 expression in 10 primary lymphoma/leukemia specimens. y-axis, −ΔΔCt values of miR-21 expression; x-axis, −ΔΔCt values miR-155 expression. The P value was calculated by simple regression analysis.
miR-21 induces dysregulation of PTEN/AKT signaling in NK-cell lymphoma/leukemia
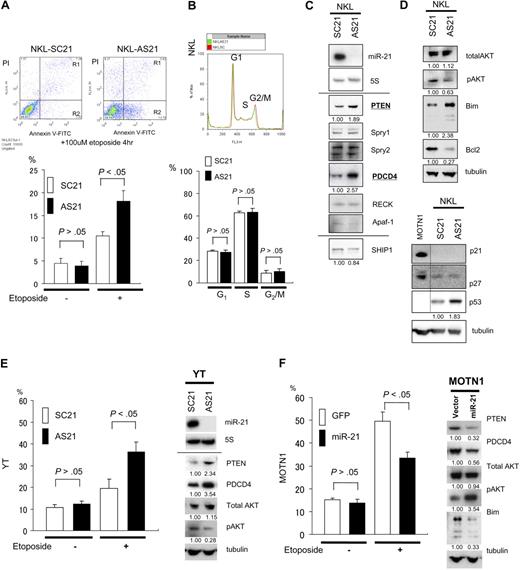
miR-21 and miR-155 are known to be oncogenic in a variety of cancers, including hematologic malignancies. To determine whether these microRNAs are associated with NK-cell lymphomagenesis, we examined the effects of antisense oligonucleotides targeted to miR-21 (ASO-21) and miR-155 (ASO-155) in NK-cell lymphoma/leukemia lines, which constitutively overexpress miR-21 but show very little expression of miR-155. Northern analysis revealed no expression of miR-21 in NKL cells treated with ASO-21 compared with cells treated with scrambled control oligonucleotide (SCO-21). To investigate effects of ASO assay, we performed apoptosis and cell assays. We found that ASO-21 increased the incidence of apoptosis among NKL cells exposed to 100 μmol/L etoposide for 4 hours (Figure 4A). When we performed a cell-cycle analysis of ASO-21–treated NKL cells, we detected no significant differences in the respective cell-cycle stages between SCO-21–treated and ASO-21–treated cells (Figure 4B). Because Apaf-1, RECK, PDCD4, Spry1, Spry2, and PTEN are all candidate targets of miR-21 in tumorigenesis,19-23 we next conducted Western analyses with cells treated with SCO-21 and ASO-21 (Figure 4C). We found that among these candidate targets, PTEN and PDCD4 were up-regulated in ASO-21–treated cells compared with SCO-21–treated cells. Moreover, we found that levels of activated AKT (pAKT), a downstream target in the PTEN/AKT signaling pathway, were reduced, whereas those of the proapoptotic protein Bim, a downstream target of pAKT via FoxO3, were increased.24,25 Western analysis of p21 levels in SCO-21– and ASO-21–treated NKL cells revealed no expression of p21 but a slight increase in p53 levels in ASO-21–treated cells. There was no difference in the levels of p27 expression between SCO- and ASO-treated cells. To confirm the function of miR-21, we examined the effects ASO-21 in YT cells, which overexpress miR-21 with little expression of miR-155. As with NKL cells, cell- cycle analysis showed no difference between SCO-21–treated and ASO-21–treated YT cells (data not shown). By contrast, ASO-21 enhanced apoptotic activity in YT cells after exposure to 100 μmol/L etoposide (Figure 4E). YT cells treated with ASO-21 also showed up-regulation of PTEN and down-regulation of pAKT, as was seen in NKL cells (Figure 4E). These results suggest that miR-21 contributes to NK-cell lymphomagenesis by reducing apoptotic activity but not cell-cycle progression.
Targets of miR-21 and the function in NK-cell lymphoma/leukemia. (A) Apoptosis among NKL cells. ASO-21– and SCO-21–treated cells were exposed to 100 μmol/L etoposide for 4 hours. Top panels show flow cytometric analysis of ASO-21–treated and SCO-21–treated NKL cells: y-axis, cells stained by propidium iodide (PI); x-axis, cells stained by annexin V–FITC. Apoptotic cells are in Regions 1 (R1) and 2 (R2). Apoptotic (R1 + R2) cells of ASO-21–treated cells are showing significantly greater percentages than those cells treated with SCO-21. Bottom panel shows the percentages of apoptotic cells (R1 + R2). □ and ■ depict percent apoptosis (R1 + R2) among NKL cells treated with SCO-21 and ASO-21, respectively. Symbols and bars indicate means and SDs of triplicate samples. (B) Cell-cycle analysis for NKL cells treated with ASO-21 or SCO-21. In the top panel, red lines show the cell-cycle pattern of cells treated with SCO-21, whereas green lines show the cell-cycle pattern of those treated with ASO-21. G1, S, and G2/M phases are shown as described. Average (n = 3) percentages among SCO-treated NKL cells: G1, 29.3%; S, 61.3%; and G2/M, 9.4%. Average (n = 3) percentages among ASO-treated NKL cells: G1, 25.3%; S, 66.5%; and G2/M, 8.2%. (C) Western analysis for direct candidate targets of miR-21 of NKL cell line treated with ASO-21 and SCO-21. Northern blot analysis of miR-21 in NKL cells treated with ASO-21 and SCO-21 are also shown. Fold changes in proteins expression are shown below the gels and normalized to the level in NKL SCO-21 treated cells, which are assigned a value of 1.00. (D) Western analysis for candidate downstream targets of miR-21 of NKL cell line treated with ASO-21 and SCO-21. Fold changes in proteins expression are shown below the gels and normalized to the level in NKL SCO-21–treated cells, which are assigned a value of 1.00. Bottom panel shows an immunoblot analysis of p21, p27, and p53 in NKL cells treated with ASO-21 or SCO-21. (E) Incidence of apoptosis among YT cells. In the left panel, cells treated with ASO-21 or SCO-21 were exposed to 100 μmol/L etoposide for 4 hours. □ and ■ depict percent apoptosis among YT cells treated with SCO-21 and ASO-21, respectively. Symbols and bars indicate means and SDs of triplicate samples. Apoptotic cells of ASO-21–treated cells show significantly greater percentages than those cells treated with SCO-21. In the right panel, Western analysis of miR-21 targets in YT cells treated with ASO-21 or SCO-21. Fold changes in protein expression are shown below the gels and normalized to the level in SCO-21–treated cells, which was assigned a value of 1.00. (F) Incidence of apoptosis among transduced MOTN1 and control cells. In the left panel, MOTN1 cells transduced with miR-21 plus GFP or GFP alone (control) were exposed to 100 μmol/L etoposide for 4 hours. □ and ■ depict percent apoptosis among GFP-transduced (control) and miR-21–transduced cells, respectively. Symbols and bars indicate means and SDs of triplicate samples. Apoptotic cells of miR-21 transduced cells show significantly lower percentages than those of control cells. In the right panel, Western analysis of PTEN, pAKT, total AKT, and Bim expression in MOTN1 cells. Fold changes in protein expression are shown below the gels and are normalized to the level in GFP-transduced control cells, which was assigned a value of 1.00.
Targets of miR-21 and the function in NK-cell lymphoma/leukemia. (A) Apoptosis among NKL cells. ASO-21– and SCO-21–treated cells were exposed to 100 μmol/L etoposide for 4 hours. Top panels show flow cytometric analysis of ASO-21–treated and SCO-21–treated NKL cells: y-axis, cells stained by propidium iodide (PI); x-axis, cells stained by annexin V–FITC. Apoptotic cells are in Regions 1 (R1) and 2 (R2). Apoptotic (R1 + R2) cells of ASO-21–treated cells are showing significantly greater percentages than those cells treated with SCO-21. Bottom panel shows the percentages of apoptotic cells (R1 + R2). □ and ■ depict percent apoptosis (R1 + R2) among NKL cells treated with SCO-21 and ASO-21, respectively. Symbols and bars indicate means and SDs of triplicate samples. (B) Cell-cycle analysis for NKL cells treated with ASO-21 or SCO-21. In the top panel, red lines show the cell-cycle pattern of cells treated with SCO-21, whereas green lines show the cell-cycle pattern of those treated with ASO-21. G1, S, and G2/M phases are shown as described. Average (n = 3) percentages among SCO-treated NKL cells: G1, 29.3%; S, 61.3%; and G2/M, 9.4%. Average (n = 3) percentages among ASO-treated NKL cells: G1, 25.3%; S, 66.5%; and G2/M, 8.2%. (C) Western analysis for direct candidate targets of miR-21 of NKL cell line treated with ASO-21 and SCO-21. Northern blot analysis of miR-21 in NKL cells treated with ASO-21 and SCO-21 are also shown. Fold changes in proteins expression are shown below the gels and normalized to the level in NKL SCO-21 treated cells, which are assigned a value of 1.00. (D) Western analysis for candidate downstream targets of miR-21 of NKL cell line treated with ASO-21 and SCO-21. Fold changes in proteins expression are shown below the gels and normalized to the level in NKL SCO-21–treated cells, which are assigned a value of 1.00. Bottom panel shows an immunoblot analysis of p21, p27, and p53 in NKL cells treated with ASO-21 or SCO-21. (E) Incidence of apoptosis among YT cells. In the left panel, cells treated with ASO-21 or SCO-21 were exposed to 100 μmol/L etoposide for 4 hours. □ and ■ depict percent apoptosis among YT cells treated with SCO-21 and ASO-21, respectively. Symbols and bars indicate means and SDs of triplicate samples. Apoptotic cells of ASO-21–treated cells show significantly greater percentages than those cells treated with SCO-21. In the right panel, Western analysis of miR-21 targets in YT cells treated with ASO-21 or SCO-21. Fold changes in protein expression are shown below the gels and normalized to the level in SCO-21–treated cells, which was assigned a value of 1.00. (F) Incidence of apoptosis among transduced MOTN1 and control cells. In the left panel, MOTN1 cells transduced with miR-21 plus GFP or GFP alone (control) were exposed to 100 μmol/L etoposide for 4 hours. □ and ■ depict percent apoptosis among GFP-transduced (control) and miR-21–transduced cells, respectively. Symbols and bars indicate means and SDs of triplicate samples. Apoptotic cells of miR-21 transduced cells show significantly lower percentages than those of control cells. In the right panel, Western analysis of PTEN, pAKT, total AKT, and Bim expression in MOTN1 cells. Fold changes in protein expression are shown below the gels and are normalized to the level in GFP-transduced control cells, which was assigned a value of 1.00.
Because MOTN1 cells showed less expression of miR-21 than the other NK-cell lymphoma cell lines, we transduced these cells by using a lentiviral vector encoding both miR-21 and GFP, which yielded greater than 95% GFP positivity. Subsequent Taqman PCR analysis showed that miR-21 expression was 2.6-fold greater in the transduced MOTN1 cells than in control cells. As with the other cells tested, we detected no difference in the respective cell-cycle stages between control cells and miR-21 transductants (data not shown). However, apoptotic activity was diminished in MOTN1 cells transduced with miR-21 (Figure 4F), whereas Western analysis showed that levels of PTEN and PDCD4 were reduced and those of pAKT were increased in the transductant cells (Figure 4F). Total AKT levels were unaffected by miR-21. Collectively, these findings suggest miR-21 acts as an oncomiRNA through dysregulation of the PTEN/AKT signaling pathway, resulting in diminished apoptotic activity.
miR-155 induces dysregulation of SHIP1/AKT signaling in NK-cell lymphomas/leukemias
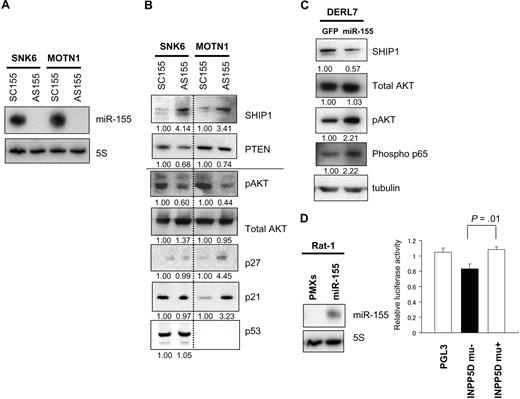
We next used ASO-155 to examine the effects of reducing miR-155 expression in SNK6 and MOTN1 cells, both of which overexpress miR-155 (Figure 5A). By using Target Scan software,21 we determined that one likely target of miR-155 is SHIP1, an inositol 5-phosphatase expressed in hemopoietic cells. By hydrolyzing the 5-phosphates from PtdIns(3,4,5)P(3) and Ins(1,3,4,5)P,(4), SHIP1 negatively regulates signaling in the phosphoinositide 3-kinase (PI3K)–AKT pathway. Consequently, inhibition of SHIP1expression by miR-155 would likely increase activity in the AKT pathway.
miR-155 induces dysregulation of SHIP1 signaling to AKT in NK-cell lymphoma/leukemia lines. (A) Northern analysis of miR-155 in SNK6 and MOTN1 cells treated with ASO-155 or SCO-155. Controls (5S tRNA) are shown below the miR-155 blots. (B) Western analysis of PTEN, pAKT, total AKT, p21, p27, and p53 expression in MOTN1 and SNK6 cells treated with ASO-155 or SCO-155. Fold changes in protein expressions are shown below the gels and are normalized to the levels in SCO-treated SNK6 or MOTN1 cells, respectively, which were assigned a value of 1.00. (C) Western analysis of SHIP1, total AKT, pAKT, and phospho-NFκB (Phospho p65) in DERL7 cells transduced with miR-155 plus GFP or GFP alone (control). Fold changes in protein expression are shown below the gels and are normalized to the level in the GFP-transduced control cells, which was assigned a value of 1.00. (D) Luciferase reporter assay of INPP5D/SHIP expression in Rat-1 cells transfected with miR-155 (PMXs-miR-155-puro) or empty vector (PMXs-puro). Blots showing MiR-155 expression are beside the bars. The significance of differences between wild-type pGL3-INPP5D 3′UTR and pGL3-INPP5D was evaluated with the Student t test.
miR-155 induces dysregulation of SHIP1 signaling to AKT in NK-cell lymphoma/leukemia lines. (A) Northern analysis of miR-155 in SNK6 and MOTN1 cells treated with ASO-155 or SCO-155. Controls (5S tRNA) are shown below the miR-155 blots. (B) Western analysis of PTEN, pAKT, total AKT, p21, p27, and p53 expression in MOTN1 and SNK6 cells treated with ASO-155 or SCO-155. Fold changes in protein expressions are shown below the gels and are normalized to the levels in SCO-treated SNK6 or MOTN1 cells, respectively, which were assigned a value of 1.00. (C) Western analysis of SHIP1, total AKT, pAKT, and phospho-NFκB (Phospho p65) in DERL7 cells transduced with miR-155 plus GFP or GFP alone (control). Fold changes in protein expression are shown below the gels and are normalized to the level in the GFP-transduced control cells, which was assigned a value of 1.00. (D) Luciferase reporter assay of INPP5D/SHIP expression in Rat-1 cells transfected with miR-155 (PMXs-miR-155-puro) or empty vector (PMXs-puro). Blots showing MiR-155 expression are beside the bars. The significance of differences between wild-type pGL3-INPP5D 3′UTR and pGL3-INPP5D was evaluated with the Student t test.
Western analysis revealed that treating SNK6 and MOTN1 with ASO-155 increased expression of SHIP1 (Figure 5B) and reduced levels of pAKT in both cell lines. Interestingly, MOTN1 cells treated with ASO-155 showed greater levels of p21 and p27 than SCO-treated cells. ASO-155–treated SNK6 cells also showed greater levels of SHIP1 and lower levels of pAKT, but unlike MOTN1 cells they did not show up-regulation of p21. This finding may reflect the fact that, in MOTN1 cells, p21 expression is regulated by SHIP1/AKT signaling and is unaffected by p53, whereas in SNK6 cells p21 expression can be regulated by wild-type p53. It thus appears that continuous overexpression of miR-155 leads to increased production of pAKT via down-regulation of SHIP1. It is noteworthy that transduction of MOTN1 cells with miR-21 leads to further up-regulation of pAKT (Figure 4F) in the context of continuous overexpression of miR-155. This finding suggests that forced expression of miR-21 further enhances pAKT expression via down-regulation of PTEN.
Levels of miR-155 expression were lower in DERL7 cells than in the other NK-cell lymphoma lines tested (Figure 1A) but were increased 3.1-fold by transduction with miR-155 using a lentiviral vector. This increase in miR-155 expression led to down-regulation of SHIP1 and up-regulation of pAKT (Figure 5C). Moreover, immunoblotting of downstream targets of AKT, including Bim, p21, p27, and phosphorylated NF-κB (phospho-p65), revealed that levels of phospho-p65 were increased in the miR-155 transductants, which suggests NF-κB activation via pAKT may also contribute to lymphomagenesis.
To determine whether expression of INPP5D/SHIP, which encodes SHIP1, is directly regulated by miR-155, we performed luciferase reporter assays by using Rat-1 fibroblasts stably expressing miR-155. Rat-1 fibroblast stably expressing miR-155 was established by retrovirus transduction with PMXs-premiR-155-puromycine vector. We found that when the cells were transfected with a reporter construct harboring wild-type INPP5D/SHIP, luciferase activity was significantly repressed compared with cells transfected with a construct harboring a INPP5D/SHIP mutant (Figure 5D). These findings, combined with the results obtained with the use of ASO-155 in NK-cell lymphoma cells, suggest that INPP5D/SHIP1 is a direct downstream target of the miR-155.
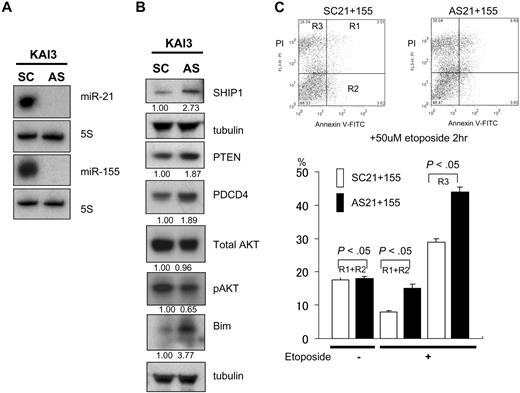
Antisense oligonucleotide assay of miR-21 and miR-155 for a KAI3, which is overexpressing the microRNAs, showed dysregulation of PTEN/AKT and SHIP1/AKT signaling
KAI3 cells overexpress both miR-21 and miR-155 (Figures 1,Figure 2–3). When we treated these cells with ASO-21 or ASO-155, we detected no change in the expression of pAKT. However, when we treated the cells with a combination of ASO-21 and ASO-155, which reduced expression of both miR-21 and miR-155 (Figure 6A), levels of PTEN and SHIP1 were increased (Figure 6B), and those of pAKT were reduced. The incidences of apoptotic and necrotic cells among ASO-21/155–treated cells were greater than among SCO-treated cells, which suggests that reducing expression of miR-21 and miR-155 enhances apoptotic activity in KAI3 cells (Figure 6C). In addition, up-regulation PDCD4 was also detected (Figure 6B). Thus miR-21 and miR-155 may act in concert to enhance expression of pAKT in KAI3 cells. Moreover, the observation that MOTN1 cells transfected with miR-21 also showed enhanced activation of pAKT signaling suggests miR-21 and miR-155 also act together to enhance expression in the pAKT signaling pathway.
Effects of ASO-21 and ASO-155 in KAI3 cells. (A) Northern analysis of miR-21 and miR-155 in KAI3 cells treated with both ASO-21 and ASO-155 and in cells treated with SCO-21 and SCO-155. (B) Western analysis of PTEN and SHIP1 and their downstream targets. Fold changes in protein levels are shown below the gels and are normalized to the level in SCO-treated cells, which was assigned a value of 1.00. (C) Incidence of apoptosis among KAI3 cells. Cells treated with both ASOs or SCOs were exposed to 50 μmol/L etoposide for 2 hours. (Top panels) Flow cytometric analysis is shown. y-axis: number of cells stained with PI; x-axis: cells stained with annexin V–FITC. Apoptotic cells are in R1 and R2; necrotic cells are in R3. (Bottom panel) □ and ■ indicate percentage of apoptosis (R1 + R2) or necrosis (R3) among SCO-21/155–treated and ASO-21/155–treated cells, respectively. Symbols and bars indicate means and SDs of triplicate samples. Both apoptotic (R1 + R2) and necrotic (R3) cells of ASO-21/155–treated cells show significantly greater percentages than those cells treated with SCO-21/155.
Effects of ASO-21 and ASO-155 in KAI3 cells. (A) Northern analysis of miR-21 and miR-155 in KAI3 cells treated with both ASO-21 and ASO-155 and in cells treated with SCO-21 and SCO-155. (B) Western analysis of PTEN and SHIP1 and their downstream targets. Fold changes in protein levels are shown below the gels and are normalized to the level in SCO-treated cells, which was assigned a value of 1.00. (C) Incidence of apoptosis among KAI3 cells. Cells treated with both ASOs or SCOs were exposed to 50 μmol/L etoposide for 2 hours. (Top panels) Flow cytometric analysis is shown. y-axis: number of cells stained with PI; x-axis: cells stained with annexin V–FITC. Apoptotic cells are in R1 and R2; necrotic cells are in R3. (Bottom panel) □ and ■ indicate percentage of apoptosis (R1 + R2) or necrosis (R3) among SCO-21/155–treated and ASO-21/155–treated cells, respectively. Symbols and bars indicate means and SDs of triplicate samples. Both apoptotic (R1 + R2) and necrotic (R3) cells of ASO-21/155–treated cells show significantly greater percentages than those cells treated with SCO-21/155.
Our results demonstrate that overexpression of miR-21 and/or miR-155 play an essential role in NK-cell lymphomagenesis through activation of AKT signaling, which diminishes apoptosis (Bim down-regulation) and promotes cell-cycle progression (p21 and p27 down-regulation) and nuclear factorκB activation (phospho-p65 up-regulation). Among these, the proapoptotic protein Bim is the most likely the target of miR-21 during NK-cell lymphomagenesis because Bim disruption was repeatedly observed in our experiments. By contrast, miR-155 may control a variety of downstream targets via AKT disruption.
Discussion
Our data demonstrated that nearly all NK-cell lymphoma/leukemia lines, as well as specimens of primary lymphoma, overexpress miR-21, miR-155, or both. Interestingly, these 2 miRNAs were irreversibly expressed in both NK-cell lymphoma cell lines and patient samples. This finding suggests that miR-21 and miR-155, the functions of which are complementary, may contribute to lymphomagenesis through disruption of the same targets. Indeed, we found that these 2 miRNAs acted via PTEN and SHIP1, respectively, to regulate AKT, a serine-threonine kinase involved in various cellular functions, including the regulation of cell survival and proliferation. However, because our results were obtained from a limited numbers of samples, confirmation with the use of a larger number of test samples will be needed before we can conclude that the expression of these miRNAs in NK-cell lymphomas is actually irreversible. But irrespective of whether the expression patterns of these miRNAs are irreversible, the overexpression of either or both could play a key role in NK-cell lymphomagenesis.
miR-21 has been identified as the best hit in several medium-scale profiling experiments designed to detect miRNAs dysregulated in tumors, including cancers of the breast, colon, lung, pancreas, prostate, and stomach.26 Recent studies indicate that miR-21 is also up-regulated in lymphomas/leukemias, including chronic lymphocytic leukemia, aggressive diffuse large B-cell lymphoma, both de novo and transformed cases, and follicular center lymphoma and Hodgkin lymphoma, whereas miR-155 is overexpressed in Burkitt lymphoma, chronic lymphocytic leukemia, and Hodgkin lymphoma.27-32 However, there have been no studies investigating the involvement of miRNA in NK-cell lymphomas/leukemias; consequently, the roles played by miRNAs in the pathogenesis of these diseases remained largely unknown. In the present study, however, we found that miR-21 and miR-155 are both overexpressed in NK-cell lymphomas/leukemias. Furthermore, the expression levels of these miRNAs are significantly higher in NK-cell than in B-cell lymphomas/leukemias including EBV-positive B-cell lymphoma. How are these microRNAs overexpressed in NK-cell lymphomas?
In B-cell lymphoma, overexpression of miR-17-92 is caused by genomic amplification of the 13q31 locus.8 miR-21 is located at 17q23, whereas miR-155 is at 21q21. Despite efforts by several groups2-7 to detect specific genomic alterations in NK-cell lymphomas, no genomic amplifications or translocations of the 17q23 or 21q21 locus have been reported, which suggests genomic structural alterations are not involved in the up-regulation of these miRNAs in cancer.
One of possible alternative mechanism is infection of normal NK cells by EBV. Because nearly all NK-cell lymphomas/leukemias and endemic Burkitt lymphoma show infection by EBV, it is reasonable to suspect that the overexpression of these miRNAs is related to the virus. Consistent with that idea, recent reports suggest that EBV infection can lead directly to up-regulation of miR-21 and miR-155. For instance, Mrázek et al33 reported that miR-21 is strongly up-regulated in EBV-infected human B lymphocytes, whereas Rahadiani et al34 showed that EBV infection induces up-regulation of miR-155 in Burkitt lymphoma cell lines. Moreover, several groups35-38 have shown that EBV-encoded LMP1 can induce transactivation of miR-155, which supports our idea that up-regulation of miR-21 and/or miR-155 by EBV infection is not limited to Burkitt lymphoma but also occurs in NK-cell lymphomas/leukemias. Up-regulation of these miRNAs via EBV infection could be the first-hit genetic alterations during NK-cell lymphomagenesis.
miRNAs are also up-regulated by oncogenic activation of transcription factors. For instance, expression of both miR-21 and miR-155 can be regulated by transcription factors such as AP-1.39,40 Indeed, oncogenic transformation is frequently associated with enhancement of endogenous AP-1 activity mediated through various signal transduction pathways, which suggests AP-1–induced expression of miRNAs may also contribute to NK-cell lymphomagenesis.
In the present study, we found that miR-21 down-regulates the tumor suppressors PTEN and PDCD4. Meng et al20 were the first to report that miR-21 regulates expression of PTEN and downstream PI3K signaling in human cholangiocarcinoma and hepatocellular carcinoma cells, and a corresponding modulation of PTEN protein expression by miR-21 was subsequently detected in a colon cancer cell line and during vascular neointimal lesion formation.41,42 However, it remains unclear whether miR-21 regulates PTEN directly because the miR-21 binding site on the PTEN mRNA has not yet been characterized. Whether direct or not, regulation of PTEN by miR-21 appears to be cell type specific. miR-21 also down-regulated the tumor suppressor PDCD4, which is consistent with earlier findings that miR-21 directly down-regulates PDCD4 in breast, brain, and colorectal cancers.43,44 PDCD4 inhibits PMA-induced neoplastic transformation, tumor promotion and progression, and intravasation,45-47 which suggests its down-regulation by miR-21 could contribute to the aggressive clinicopathologic features of NK-cell lymphoma/leukemia, although we did investigate the mechanism by which PDCD4 functions in lymphomagenesis in the present study.
We found that miR-155 directly down-regulates SHIP1, which appears to regulate both p21 and p27 via pAKT in MOTN1 cells, suggesting SHIP1 can affect cell-cycle progression via the AKT-signaling pathway. Horn et al48 showed that forced expression of SHIP1 in Jurkat cells, a T-cell leukemia cell line, induces G1-S arrest by repressing p27, and Freeburn et al49 showed that SHIP1 can influence the constitutive levels of PI(3,4,5)P(3) and the activity of downstream PI3K effectors, including AKT, in T lymphocytes. These results strongly support our finding that SHIP1 down-regulates p21 and p27 via the AKT pathway. Moreover, it was very recently reported that endogenous expression of SHIP1 is repressed in hematopoietic cells overexpressing miR-155 in vitro and in vivo, which leads to increased activation of AKT.50 This, too, is consistent with our findings.
In summary, we have shown that dysregulation of miRNAs is a key feature of the pathogenesis of NK-cell lymphoma/leukemia. Overexpression of miR-21 and/or miR-155, which might be up-regulated by EBV infection, affects AKT signaling via their respective repression of PTEN and SHIP1. These findings provide new insight into the pathogenesis of NK-cell lymphoma/leukemia and suggest targeting miR-21 and/or miR-155 may represent a useful approach to treating NK-cell lymphoma/leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We wish to express our appreciation to A. Kobayashi, B. Kataho, and C. Kikuchi (Akita University) for their outstanding technical assistance; and Drs M. Saitoh, M. Kume, and S. Takahashi (Hiraka General Hospital) and H. Nanjo (Akita University) for histologic and/or clinical diagnosis of lymphoma. pMX vector was kindly provided by Dr T. Kitamura (Tokyo University).
This work was supported in by the Grant-in-Aid Scientific Research (C) from the Japan Society for the Promotion of Science (to H.T.) and by the Grant Global Center of Excellence Program (G-COE) of the Ministry of Education, Science, Technology, Sports, and Culture of Japan (to K.S.).
Authorship
Contribution: Y.Y. performed experiments, analyzed data, and constructed the tables; A.W.,Y.-M. G., K.I., and J.Y. performed the experiments; N.T., H.S., Y.K., N.S., R.I., N.T., and K.S. analyzed data; and H.T. designed and performed experiments, analyzed data, wrote the paper, and organized the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hiroyuki Tagawa, MD, PhD, Department of Hematology, Nephrology, and Rheumatology, Akita University Graduate School of Medicine, 1-1-1 Hondo, Akita 0108543, Japan; e-mail: htagawa0279jp@yahoo.co.jp.
References
Author notes
*Y.Y. and H.T. share first authorship.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal