Abstract
Active gene promoters are associated with covalent histone modifications, such as hyperacetylation, which can modulate chromatin structure and stabilize binding of transcription factors that recognize these modifications. At the β-globin locus and several other loci, however, histone hyperacetylation extends beyond the promoter, over tens of kilobases; we term such patterns of histone modifications “hyperacetylated domains.” Little is known of either the mechanism by which these domains form or their function. Here, we show that domain formation within the murine β-globin locus occurs before either high-level gene expression or erythroid commitment. Analysis of β-globin alleles harboring deletions of promoters or the locus control region demonstrates that these sequences are not required for domain formation, suggesting the existence of additional regulatory sequences within the locus. Deletion of embryonic globin gene promoters, however, resulted in the formation of a hyperacetylated domain over these genes in definitive erythroid cells, where they are otherwise inactive. Finally, sequences within β-globin domains exhibit hyperacetylation in a context-dependent manner, and domains are maintained when transcriptional elongation is inhibited. These data narrow the range of possible mechanisms by which hyperacetylated domains form.
Introduction
A crucial feature of gene activation is the interaction between transcription factors and chromatin. All eukaryotic genomic DNA, with limited exceptions, is packaged with core histones to form chromatin. The fundamental subunit of chromatin is the nucleosome, consisting of approximately 147 bp of DNA wrapped in approximately 1.75 turns about an octamer of core histones; a variable length of linker DNA extends between nucleosomes and can in turn be partially sequestered by interactions with core histone amino-terminal “tail” regions and/or linker histones, such as histone H1. The resulting structure, when observed on low-salt spreads by electron microscopy, has been termed “beads on a string” based on its appearance.1,2 At physiologically relevant salt concentrations, however, this structure spontaneously condenses, first to a helical array of nucleosomes termed the 30-nm fiber, then through additional levels of higher-order structure, which are not well understood.3 Nevertheless, this packaging renders the eukaryotic DNA relatively inaccessible to transcription factors.4-6
The transcriptional machinery possesses mechanisms for modulating chromatin structure. One of these is covalent modification of core histones, including acetylation, methylation, phosphorylation, ubiquitylation, SUMOylation, and ADP ribosylation, in short, the gamut of modifications known to occur on cellular proteins. Different modifications can lead to different functional consequences. Histone acetylation is associated with transcriptional activation; indeed, core histones proximal to active gene promoters are universally hyperacetylated.7,8 Histone methylation can be associated with activation or repression, depending on the exact residue modified; methylation of lysine 4 of histone H3 is, like hyperacetylation, associated with active promoters, whereas methylation of lysine 9 of histone H3 localizes with repressive heterochromatin.9,10 Different histone modification patterns can stabilize or destabilize the interaction of transcription factors and thus mediate functional effects.
The transcriptional regulation of the β-globin genes has been the subject of intense study for several decades. The mammalian β-globin genes are confined to a single cluster and are expressed exclusively in erythroid cells. High-level β-globin gene expression requires sequences located 5′ of the genes and termed the locus control region (LCR).11 Expression from within the cluster is developmentally regulated. Initially during embryogenesis, immature primitive erythroblasts derived from the yolk sac enter the bloodstream as nucleated cells and differentiate as a semisynchronous cohort.12 Subsequently, definitive erythrocytes, having matured and enucleated within the fetal liver or postnatal marrow, enter the circulation. Corresponding to these changes, mammals undergo a switch in globin gene expression on the transition from primitive to definitive erythropoiesis, and some, including humans, have fetal-specific β-globin genes as well.13-15
Active β-globin gene promoters in erythroid cells are, as expected, associated with nucleosomes that exhibit hyperacetylation and dimethylation at histone H3K4. Within this locus, however, these modifications are not confined to promoter-proximal regions; each of the active genes resides within blocks of 11 to 12 kb or more within which all sequences are associated with hyperacetylated histones.15,16 We term these regions “hyperacetylated domains,” although additional modifications, such as H3K4 dimethylation, comap with histone acetylation.17
Genome-wide studies of histone modifications have demonstrated that hyperacetylated domains are not a general phenomenon.18,19 To date, all known examples of hyperacetylated domains occur over highly expressed, tissue-specific genes, most of which are also known to be controlled by distal enhancers.17,20 Within transgenic human growth hormone loci, for example, mutation of a pituitary enhancer approximately 15 kb 5′ of the genes eliminates transcription and the hyperacetylated domain.21 In another study, histone modifications at the λ5/VpreB1 locus, active in pre-B lymphocytes, were monitored in cell lines modeling different stages of differentiation. When the genes were maximally active in pre-B cells, they were also located within a hyperacetylated domain. Interestingly, histone hyperacetylation in embryonic stem (ES) cells was limited to a regulatory element located between the 2 genes; in cell lines modeling later stages of differentiation, the region associated with hyperacetylation appeared to spread outward from this site.22
Other than this correlation with tissue-specific gene activation by distal regulatory elements, our understanding of the formation and function of hyperacetylated domains is limited. We have therefore further examined the hyperacetylated domains that form within the murine β-globin locus in erythroid cells. Our results indicate that domain formation precedes both high-level β-globin expression and commitment to the erythroid lineage, suggesting that they are not a consequence of transcription but instead precede it. Deletions of the β-globin LCR or of individual β-globin gene promoters do not eliminate the domains, implying that DNA sequence requirements for domain formation reside elsewhere. On the other hand, we show that the embryonic β-globin gene promoters are required for suppression of domain formation over the embryonic genes in definitive erythroid cells, where these genes are normally silenced. We also demonstrate that histone hyperacetylation over at least some regions of the locus is not an intrinsic function of DNA sequences within these regions but depends on the presence of other sequences or on relative position within the locus. Coupled with the observation that inhibition of RNA polymerase II elongation has no effect on maintenance of histone hyperacetylation, our work narrows the range of potential models for domain formation and provides support for a mechanism involving spreading of the histone modification signature from specific sequences.
Methods
Cell culture
Cell lines were maintained at 37°C in a 5% CO2-humidified atmosphere. MEL745a cells were cultured in Dulbecco modified Eagle medium containing 10% fetal bovine serum, 1% Glutamax (Invitrogen), and 1% penicillin/streptomycin. For induction, 2% dimethyl sulfoxide (Sigma-Aldrich) was added to the culture medium. Factor-dependent cell Paterson mixed potential cells (FDCP-Mix) were cultured in Iscove modified Dulbecco medium with 20% horse serum (Invitrogen), 1% Glutamax, 1% penicillin/streptomycin, and 10 ng/mL murine IL-3 (PeproTech).
ES cell and embryoid body culture and differentiation
Murine ES cells were differentiated into embryoid bodies (EBs) as previously described.23 At day 2, 4, 6, and 8 of differentiation, EBs were dissociated with trypsin (Invitrogen) to give single-cell suspensions. These cells were harvested for RNA, chromatin immunoprecipitation assay (ChIP), cell counts, and benzidine staining. Details of these protocols are listed in “Benzidine staining of EBs,” “Quantitative RT-PCR,” and “ChIP.”
Collection of embryonic peripheral blood and fetal liver
Mice were mated overnight and the vaginal plug verified the next morning indicating E0.3. At E12.5 and E14.5, pregnant mice were killed by cervical dislocation for collection of embryonic blood and fetal liver.12,15 Animal experiments were approved by the University Committee on Animal Resources at the University of Rochester.
Benzidine staining of EBs
To identify heme-containing cells, trypsinized EBs were stained with benzidine dihydrochloride (Sigma-Aldrich) as described24 and counted on a hemocytometer (VWR International).
Quantitative RT-PCR
RNA was isolated using TRIzol (Invitrogen) according to manufacturer recommendations. A total of 1 μg RNA was used to synthesize cDNA using the iScript cDNA synthesis kit (Bio-Rad). Reverse transcription (RT) was also performed without enzyme as a negative control. Primers used amplified ϵy-, βh1-, and combined β1- and β2-globin cDNAs. QuantiTect Primers (QIAGEN) for 18S mRNA were used as an internal control. cDNA was amplified using iQ SYBR green supermix (Bio-Rad) and detected using the MyiQ Single Color Real Time PCR detection system and iCycler (Bio-Rad).
ChIP
Cells were formaldehyde crosslinked, sonicated and immunoprecipitated, and DNA isolated as previously described.16,25 Cells were washed with phosphate-buffered saline and crosslinked at room temperature in 10 mL volume containing 1% formaldehyde. After addition of 0.5 mL of 2.5 M glycine and a phosphate-buffered saline wash, cells were resuspended in 3 mL and sonicated for a total of 80 seconds using a Misonix Sonicator 3000 at 36 to 39 W output. An “input” fraction was then removed to serve as a control. Antibodies used for immunoprecipitation were (1) normal rabbit IgG (Upstate-Millipore) as a control; (2) antihistone H3 acetylated at K9 and/or K16 (Upstate-Millipore); (3) antihistone H4 acetylated at K5, K8, K12, and/or K16 (Upstate-Millipore); (4) antihistone H3 dimethylated at K4 (Upstate-Millipore and Abcam); (5) antihistone H3 trimethylated at K4 (Upstate-Millipore); and (6) anti-RNA Pol II (Santa Cruz Biotechnology). Analysis was performed using quantitative polymerase chain reaction (PCR) in “Quantitative RT-PCR.” Primers were designed to amplify regions within the murine β-globin locus, and regions within loci that are inactive in erythroid cells as controls, including amylase, protamine, and T-cell receptor β-chain. Primers and calculation of enrichments were described previously.15 Error bars represent the SEM from a minimum of 3 independent ChIP experiments. Some enrichment and SEM bars were truncated to demonstrate the significance of lower enrichment levels. These exact numerical values are listed in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Results
Murine β-globin hyperacetylated domain formation precedes high-level expression and erythroid commitment
Hyperacetylated domains, each of which is at least 10 kb in extent, encompass the expressed β1- and β2-globin genes in definitive erythroid cells from adult mice.16 In primitive erythroid cells, where both the adult genes (at low levels) and the embryonic ϵy- and βh1-globin genes are expressed, the individual β1- and β2-globin domains are present, whereas the ϵy- and βh1-globin genes reside within a distinct hyperacetylated domain of more than 19 kb.15 In these studies, histone modifications were assayed using spleens from anemic adult mice or circulating primitive erythroblasts, both of which consist primarily of late-stage erythroid cells expressing high levels of β-globins.
To assay for hyperacetylated domain formation before high-level β-globin gene expression, we used murine erythroleukemia (MEL) cells. Under normal conditions of cell culture, MEL cells express low levels of the adult β1- and β2-globin genes. When cultured in media containing 2% dimethyl sulfoxide (DMSO), however, MEL cells terminally differentiate, ceasing to proliferate and becoming visibly smaller and red with hemoglobin, reflecting an increase of 75- to 150-fold in adult globin mRNA levels (Figure 1A-B).
Histone modifications across the β-globin locus in MEL745a and FDCP-Mix cells. The murine β-globin locus is depicted to scale as a line at the top, with the 4 developmentally regulated β-globin genes labeled over black arrows, and exons represented by black bars on the line. Unlabeled, thicker black bars on the line represent β-globin pseudogenes. HS2 indicates hypersensitive site 2 of the locus control region (LCR). Black bars below the line depict the locations of PCR amplimers used in the ChIP assay, which is shown in turn in bar graph format for each of the antibodies used for IP. (A) Quantitative RT-PCR of combined β1- and β2-globin mRNA expression levels relative to 18s mRNA in FDCP-Mix, and in both uninduced (Un-Ind.) MEL745a and MEL745a induced with 2% DMSO for 72 hours (Ind.). (B) Fold induction of β1- and β2-globin mRNA expression levels in MEL745a on culture in 2% DMSO for 72 hours, relative to uninduced MEL745a. (C) Bar graph depicting H3Ac and (D) H3K4 dimethyl enrichment levels, relative to inactive gene loci (“ChIP”), in both uninduced (Un-ind.) and induced (Ind.) MEL745a. The positions of each bar correspond to the positions of the corresponding amplimers shown to scale at the top. (E) H3K4 trimethyl enrichment levels, relative to inactive gene loci, for uninduced and induced MEL745a. (F) H3Ac and H3K4 dimethyl enrichment levels for FDCP-Mix.
Histone modifications across the β-globin locus in MEL745a and FDCP-Mix cells. The murine β-globin locus is depicted to scale as a line at the top, with the 4 developmentally regulated β-globin genes labeled over black arrows, and exons represented by black bars on the line. Unlabeled, thicker black bars on the line represent β-globin pseudogenes. HS2 indicates hypersensitive site 2 of the locus control region (LCR). Black bars below the line depict the locations of PCR amplimers used in the ChIP assay, which is shown in turn in bar graph format for each of the antibodies used for IP. (A) Quantitative RT-PCR of combined β1- and β2-globin mRNA expression levels relative to 18s mRNA in FDCP-Mix, and in both uninduced (Un-Ind.) MEL745a and MEL745a induced with 2% DMSO for 72 hours (Ind.). (B) Fold induction of β1- and β2-globin mRNA expression levels in MEL745a on culture in 2% DMSO for 72 hours, relative to uninduced MEL745a. (C) Bar graph depicting H3Ac and (D) H3K4 dimethyl enrichment levels, relative to inactive gene loci (“ChIP”), in both uninduced (Un-ind.) and induced (Ind.) MEL745a. The positions of each bar correspond to the positions of the corresponding amplimers shown to scale at the top. (E) H3K4 trimethyl enrichment levels, relative to inactive gene loci, for uninduced and induced MEL745a. (F) H3Ac and H3K4 dimethyl enrichment levels for FDCP-Mix.
We analyzed histone acetylation and H3K4 dimethylation within the β-globin locus in MEL cells by ChIP before and after differentiation in 2% DMSO (Figure 1C-D). Under both conditions, the pattern of histone modifications across the region containing the adult globin genes was similar to that observed in definitive erythroid cells from fetal liver, with domains of hyperacetylation and H3K4 dimethylation extending across approximately 10 kb encompassing either gene. Notably, with the exceptions of the gene promoters, we observed no significant change in histone modifications on induction of terminal differentiation, despite the increase in expression of these genes.
Lysines can be monomethylated, dimethylated, or trimethylated; and for histone H3K4, these methylation states occur with different patterns across the genome and active gene loci.9,10 Previously, we have examined solely H3K4 dimethylation, so we determined the pattern of H3K4 trimethylation across the β-globin locus in MEL cells. Unlike the dimethyl H3K4 pattern, we find significant levels of H3K4 trimethylation only within the transcribed regions of the β1- and β2-globin genes, and then only in DMSO-induced MEL cells (Figure 1E). Uninduced MEL cells, expressing low levels of β-globin, show minimal H3K4 trimethylation even within the genes. Probes located at the promoters or downstream of the genes, or anywhere else in the locus, do not exhibit this modification. This indicates that hyperacetylated domains are associated specifically with dimethyl H3K4.
In addition to MEL cells, we have examined histone modifications in FDCP-Mix, a murine bone marrow-derived cell line that is commonly used as a model for multipotential myeloid progenitors.26 These cells are not committed to differentiation along a particular pathway, and can be induced to differentiate into a variety of myeloid cell types, including erythroid; β-globin expression in these cells is extremely low (Figure 1A). In these cells, under nondifferentiating conditions, we again observe hyperacetylated domains over the regions containing the adult β-globin genes (Figure 1F) that do not differ significantly from those observed in primary erythroid cells or MEL cells (Figure 1E). Taken together, these observations suggest that domain formation is independent of high-level β-globin gene expression and indeed precedes commitment to the erythroid lineage.
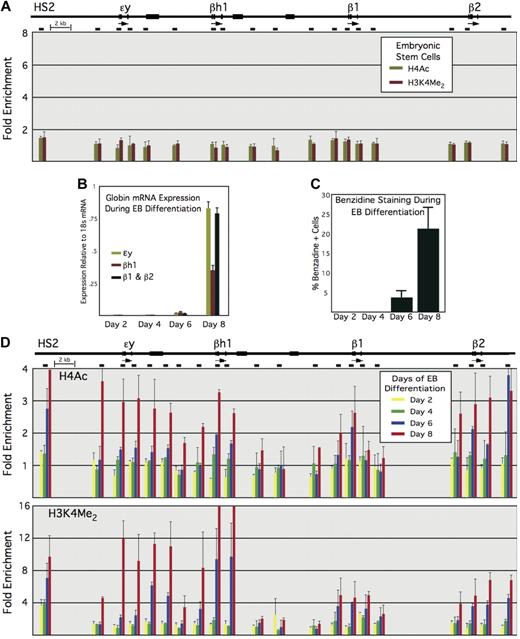
In murine ES cells, we do not observe appreciable enrichments for histone hyperacetylation or H3K4 dimethylation anywhere within the β-globin locus (Figure 2A), indicating that the hyperacetylated domains that form over the LCR and the active β-globin genes are established at a specific stage of development and/or differentiation. To examine this in more detail, we performed in vitro differentiation of murine ES cells by culturing them in suspension, which results in the formation of EBs.23 Over time in suspension culture, hematopoietic cells begin to appear in differentiating EBs, with benzidine-positive (heme-containing) cells and β-globin mRNA first evident at day 6, albeit at very low levels, and then prevalent by day 8, with nearly 25% of all cells staining benzidine-positive (Figure 2B-C). We similarly find that histone hyperacetylation and H3K4 dimethylation are first evident within the β-globin locus at day 6, when β-globin expression is first detectable (Figure 2D). The data are roughly consistent with results from the MEL and FDCP-Mix cell lines, in that histone hyperacetylation, and by extension domain formation, appear to precede full β-globin expression.
Histone modifications across the β-globin locus during ES cell differentiation. The murine β-globin locus and locations of amplimers used for quantitative PCR are shown at the top, as in Figure 1. (A) Enrichments for H4Ac and H3K4 dimethyl within the β-globin locus in undifferentiated ES cells. (B) Globin mRNA expression during days 2 to 8 of EB differentiation, normalized to 18s mRNA. (C) Benzidine staining during days 2 to 8 of EB differentiation. (D) Enrichments for H4Ac (top) and H3K4 dimethyl (bottom) in EBs differentiated for 2, 4, 6, and 8 days.
Histone modifications across the β-globin locus during ES cell differentiation. The murine β-globin locus and locations of amplimers used for quantitative PCR are shown at the top, as in Figure 1. (A) Enrichments for H4Ac and H3K4 dimethyl within the β-globin locus in undifferentiated ES cells. (B) Globin mRNA expression during days 2 to 8 of EB differentiation, normalized to 18s mRNA. (C) Benzidine staining during days 2 to 8 of EB differentiation. (D) Enrichments for H4Ac (top) and H3K4 dimethyl (bottom) in EBs differentiated for 2, 4, 6, and 8 days.
Domain formation does not require the β-globin LCR
The β-globin LCR is a broadly distributed (25-30 kb) set of regulatory sequences located upstream of the gene cluster. Deletion of this region from the endogenous mouse β-globin locus results in a 25- to 100-fold reduction in β-globin gene expression.11 The LCR is not continuous with the hyperacetylated domains encompassing the adult β-globin genes but does appear to be continuous with the hyperacetylated domain encompassing the embryonic β-globin genes in primitive erythroblasts. Prior studies have shown that deletion of the LCR has no effect on histone modifications at the active β-globin gene promoters,25 but the effect of the LCR deletion on the formation of hyperacetylated domains has not previously been determined.
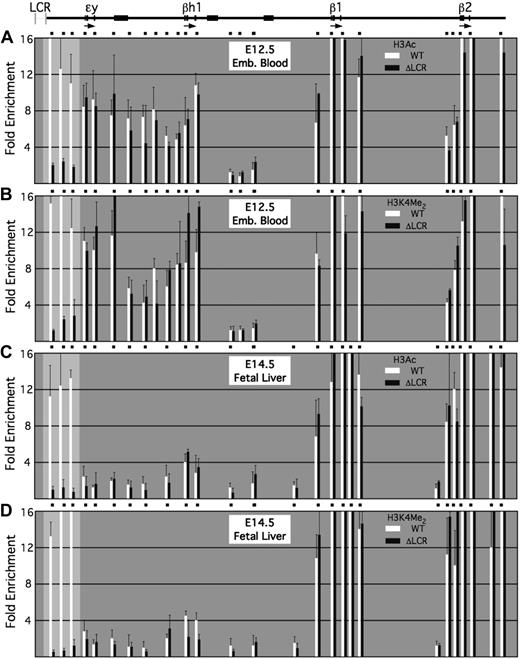
We examined histone modifications across the β-globin locus in mice homozygous for the LCR deletion; this deletion is lethal in mice but is rescued by a transgenic human β-globin locus.11,27 ChIP assays were performed using E12.5 embryonic blood (Figure 3A) and E14.5 fetal liver (Figure 3B), representing primitive and definitive erythroid cells, respectively. On β-globin alleles lacking the LCR, histone hyperacetylation and H3K4 dimethylation occur in domains encompassing both the embryonic ϵy- and βh1-globin genes and the adult β1- and β2-globin genes in primitive erythroid cells. In definitive erythroid cells, only the domains encompassing the β1- and β2-globin genes are present.
Histone modifications within the β-globin locus in the absence of the LCR. ChIP assays were performed using anti-H3Ac and anti-H3K4 dimethyl antibodies in primary cells isolated from wild-type (WT) mice and mice homozygous for deletion of the endogenous β-globin LCR (ΔLCR). The β-globin locus and positions of PCR amplimers are shown to scale at the top. Enrichments in E12.5 embryonic blood for (A) H3Ac and (B) H3K4 dimethyl and in E14.5 fetal liver for (C) H3Ac and (D) H3K4 dimethyl are shown in bar graph format beneath. The more lightly shaded portion of the graph represents the region in which enrichments for the indicated histone modifications were significantly lower than in erythroid cells from wild-type mice.
Histone modifications within the β-globin locus in the absence of the LCR. ChIP assays were performed using anti-H3Ac and anti-H3K4 dimethyl antibodies in primary cells isolated from wild-type (WT) mice and mice homozygous for deletion of the endogenous β-globin LCR (ΔLCR). The β-globin locus and positions of PCR amplimers are shown to scale at the top. Enrichments in E12.5 embryonic blood for (A) H3Ac and (B) H3K4 dimethyl and in E14.5 fetal liver for (C) H3Ac and (D) H3K4 dimethyl are shown in bar graph format beneath. The more lightly shaded portion of the graph represents the region in which enrichments for the indicated histone modifications were significantly lower than in erythroid cells from wild-type mice.
These patterns of histone modification are similar to those observed at the wild-type β-globin locus (Figure 3), suggesting that the LCR is not required for domain formation over any of the β-globin genes. Notably, however, probes that are normally interposed between the LCR and the ϵy-globin gene are not hyperacetylated in the absence of the LCR in either primitive or definitive erythroid cells (Figure 3). This region exhibits hyperacetylation and H3K4 dimethylation at both developmental stages in the wild-type allele, suggesting that histone modifications associated with these sequences are LCR-dependent. Furthermore, sequences located between the LCR and the ϵy-globin gene are apparently not part of the hyperacetylated domain encompassing the embryonic β-globin genes in primitive erythroblasts, and are part of a distinct domain that includes the LCR.
Domain formation does not require the embryonic β-globin gene promoters
Based partly on the absence of a role for the LCR, we hypothesize that DNA sequences required for the formation of hyperacetylated domains will reside within or at the boundaries of the domains themselves. The β-globin gene promoters are the most obvious candidate sequences. As such, we examined histone modifications across β-globin loci harboring deletions of the embryonic β-globin gene promoters.
Targeted deletion of the ϵy- and βh1-globin gene promoters, alone and in combination, has been performed previously28,29 (Figure 4A). In agreement with these studies, we find that each promoter deletion eliminates expression of the corresponding β-globin gene, but expression of the other globin genes is either not affected (ϵy in the βh1 promoter deletion, βh1 in the ϵy promoter deletion), or slightly affected (β1/β2 expression in the ϵy/βh1 double promoter deletion) (Figure 4B).
Histone modifications within the β-globin locus in primitive erythroblasts from mice harboring deletions of the ϵy- and/or βh1-globin gene promoters. Histone modification patterns and mRNA expression levels were analyzed in E12.5 peripheral blood from mice homozygous for the deletion of either one (Δϵy Pr. and Δβh1 Pr.) or both (Δϵy/Δβh1 Pr.) of the embryonic globin gene promoters. (A) Schematic of the β-globin locus showing a closer view of the regions being analyzed and the location of each deletion. (B) Globin expression levels using quantitative RT-PCR. Values are standardized to 18S mRNA levels and are presented as the fold change with respect to the levels seen in wild-type mice. (C) Enrichments for H3Ac (top) and H3K4 dimethyl (bottom) at the indicated locations within the β-globin locus in E12.5 blood from the indicated embryonic promoter deletion mice.
Histone modifications within the β-globin locus in primitive erythroblasts from mice harboring deletions of the ϵy- and/or βh1-globin gene promoters. Histone modification patterns and mRNA expression levels were analyzed in E12.5 peripheral blood from mice homozygous for the deletion of either one (Δϵy Pr. and Δβh1 Pr.) or both (Δϵy/Δβh1 Pr.) of the embryonic globin gene promoters. (A) Schematic of the β-globin locus showing a closer view of the regions being analyzed and the location of each deletion. (B) Globin expression levels using quantitative RT-PCR. Values are standardized to 18S mRNA levels and are presented as the fold change with respect to the levels seen in wild-type mice. (C) Enrichments for H3Ac (top) and H3K4 dimethyl (bottom) at the indicated locations within the β-globin locus in E12.5 blood from the indicated embryonic promoter deletion mice.
We performed ChIP analysis of these β-globin alleles using E12.5 circulating primitive erythroblasts to evaluate the hyperacetylated domain encompassing the embryonic β-globin genes. The pattern of histone modifications is identical to the unaltered locus in both single promoter deletions as well as the double deletion (Figure 4C-D); all probes from approximately 2 kb 5′ of the ϵy-globin promoter to approximately 2 kb 3′ of the βh1-globin gene exhibit histone hyperacetylation and H3K4 dimethylation. These results suggest that neither the embryonic β-globin gene promoters nor transcription of either gene is necessary for domain formation.
Embryonic β-globin gene promoters are required to suppress domain formation in definitive erythroid cells
In definitive erythroid cells, the embryonic β-globin genes are transcriptionally inactive, and the hyperacetylated domain that encompasses these genes in primitive erythroblasts is absent. Incubation with histone deacetylase inhibitors, however, results in an increase in histone H4 acetylation levels over the embryonic genes, implying that deacetylases are actively recruited to the region.30 Because the promoters are known to be sufficient for embryonic β-globin gene silencing in definitive erythroid cells, we examined histone modifications across the β-globin locus in fetal liver from mice harboring deletions of the embryonic gene promoters.
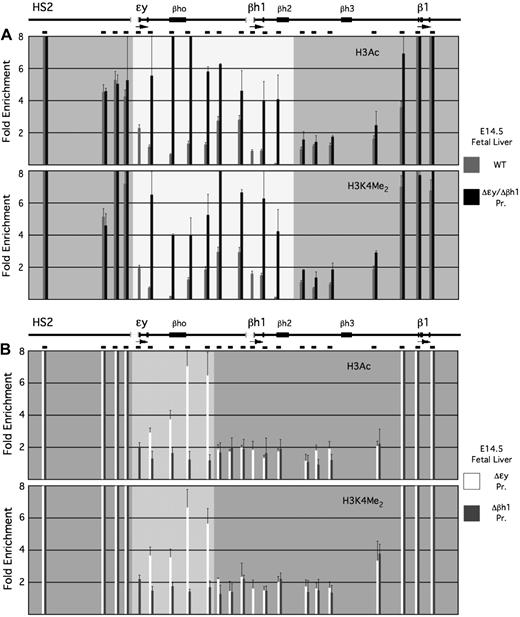
Deletion of both embryonic gene promoters results in the establishment of the hyperacetylated domain, normally specific to primitive erythroblasts, in definitive erythroid cells derived from fetal liver (Figure 5A). Histone hyperacetylation and H3K4 dimethylation are increased throughout the embryonic domain, and the effect is specific to this region because significant changes in histone modifications are not observed elsewhere in the locus. Thus, although the promoters are not required for formation of the embryonic domain in primitive erythroid cells, they are required for suppression of domain formation in definitive erythroid cells.
Histone modifications within the β-globin locus in definitive erythroid cells from mice harboring deletions of the ϵy- and/or βh1-globin gene promoters. (A) Enrichments for H3Ac (top) and H3K4 dimethyl (bottom) at the indicated locations within the β-globin locus in E14.5 fetal liver from wild-type and Δϵy/Δβh1 Pr. mice. (B) Enrichments for H3Ac (top) and H3K4 dimethyl (bottom) at the indicated locations within the β-globin locus in E14.5 fetal liver from Δϵy Pr. and Δβh1 Pr. mice. The locus is shown to scale above the bar graphs as before, with the promoter deletions indicated by gray brackets. The more lightly shaded portions of the graphs in panels A and B represent regions in which a significant difference in histone modification levels between erythroid cells derived from wild-type and the promoter deletion mice is evident.
Histone modifications within the β-globin locus in definitive erythroid cells from mice harboring deletions of the ϵy- and/or βh1-globin gene promoters. (A) Enrichments for H3Ac (top) and H3K4 dimethyl (bottom) at the indicated locations within the β-globin locus in E14.5 fetal liver from wild-type and Δϵy/Δβh1 Pr. mice. (B) Enrichments for H3Ac (top) and H3K4 dimethyl (bottom) at the indicated locations within the β-globin locus in E14.5 fetal liver from Δϵy Pr. and Δβh1 Pr. mice. The locus is shown to scale above the bar graphs as before, with the promoter deletions indicated by gray brackets. The more lightly shaded portions of the graphs in panels A and B represent regions in which a significant difference in histone modification levels between erythroid cells derived from wild-type and the promoter deletion mice is evident.
Deletions of individual embryonic β-globin promoters exhibit distinct phenotypes in definitive erythroid cells (Figure 5B). We observe no significant changes in histone hyperacetylation or H3K4 dimethylation on the allele lacking the βh1-globin promoter, indicating that the ϵy-globin promoter is sufficient to suppress domain formation. Deletion of the ϵy-globin promoter alone, however, does not have the same effect as deletion of both promoters; on this allele, significant enrichments for histone hyperacetylation and H3K4 methylation extend from the ϵy-globin gene to 7 to 8 kb 3′ of the deletion, and no further. This partial restoration of the domain suggests that the βh1-globin promoter suppresses domain formation in definitive erythroid cells, even though its deletion has no effect by itself.
The mouse β-globin allele on which the embryonic β-globin promoter deletions were performed contains an additional gene, βh0, downstream of ϵy-globin. In primitive erythroblasts, βh0 is up-regulated on deletion of the ϵy-, but not the βh1-globin, promoter, which probably reflects transcriptional interference from the ϵy-globin gene.29 In definitive erythroid cells, however, we cannot detect βh0 expression from the allele lacking the ϵy- and βh1-globin gene promoters (data not shown), indicating that domain formation is insufficient for βh0 expression and that βh0 transcription is still suppressed by other mechanisms. This also demonstrates that domain formation is not a consequence of transcription of any of the embryonic β-globin genes.
An intergenic sequence exhibits histone modifications in a position-dependent fashion
In considering mechanisms by which a hyperacetylated domain might form, it is possible that all sequences within a domain comprise regulatory regions that recruit histone-modifying complexes; we term this mechanism “mass action.” Histone hyperacetylation and H3K4 dimethylation associated with sequences located between the LCR and the ϵy-globin gene, however, require the presence of the LCR (Figure 3), arguing against this model.
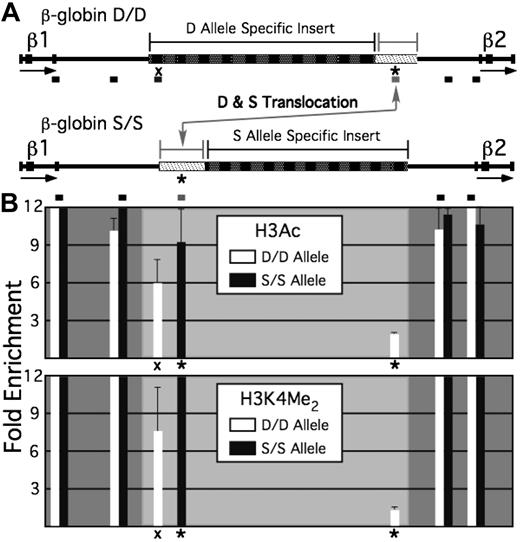
In addition, we tested the “mass action” model by comparison of the naturally occurring D and S alleles of the β-globin locus in mice. Restriction site polymorphisms between these alleles have been used previously to distinguish between transcripts originating from D versus S β-globin loci.11 Such polymorphisms occur within noncoding regions as well, and repetitive sequences (eg, LINE and SINE) occur with different patterns. In particular, a 6-kb insertion is present in the D allele between the β1- and β2-globin genes; this insertion is not present in the S allele, which instead harbors a different insertion between the β1- and β2-globin genes of 7 kb. The insertion sites on the 2 alleles are not identical, resulting in a different location, relative to the 2 adult β-globin genes, for a region of approximately 1.4 kb of unique sequence; this region is located closer to the β2-globin gene on the D allele and closer to the β1-globin gene on the S allele (Figure 6A).
Histone modification state of a unique sequence with a different positioning within 2 alleles of the β-globin locus. (A) Schematic of the region of the β-globin locus between the β1- and β2-globin genes for the D and S alleles. The block of unique sequence that occupies different relative positions within each allele is indicated by the thicker bar with a light fill, whereas the large insertions of repetitive DNA specific for each allele are indicated by thicker bars with dark fill. Positions of PCR amplimers used in analysis of ChIP assays are shown by the short bars beneath the representation of each allele. * represents the amplimer located within the unique sequence; “X,” an amplimer within a unique region that is found in the D but not the S allele. (B) Bar graphs showing enrichment levels for H3Ac (top) and H3K4 dimethyl (bottom), relative to inactive gene loci. Bars within the dark-shaded regions represent portions of the locus that are common to both the D and S alleles; lightly shaded regions, portions of the locus unique to the D or S alleles. * represents enrichments observed for the translocated unique sequence common to D and S; “X,” enrichment observed for the sequence unique to D.
Histone modification state of a unique sequence with a different positioning within 2 alleles of the β-globin locus. (A) Schematic of the region of the β-globin locus between the β1- and β2-globin genes for the D and S alleles. The block of unique sequence that occupies different relative positions within each allele is indicated by the thicker bar with a light fill, whereas the large insertions of repetitive DNA specific for each allele are indicated by thicker bars with dark fill. Positions of PCR amplimers used in analysis of ChIP assays are shown by the short bars beneath the representation of each allele. * represents the amplimer located within the unique sequence; “X,” an amplimer within a unique region that is found in the D but not the S allele. (B) Bar graphs showing enrichment levels for H3Ac (top) and H3K4 dimethyl (bottom), relative to inactive gene loci. Bars within the dark-shaded regions represent portions of the locus that are common to both the D and S alleles; lightly shaded regions, portions of the locus unique to the D or S alleles. * represents enrichments observed for the translocated unique sequence common to D and S; “X,” enrichment observed for the sequence unique to D.
ChIP analysis of the D and S alleles shows that chromatin within this region is differentially modified (Figure 6B). On the D allele, this sequence is located approximately 2 kb 5′ of the β2-globin gene promoter and approximately 10 kb 3′ of the β1-globin gene. In this position, it does not exhibit histone hyperacetylation or H3K4 dimethylation; it is apparently too far 3′ of the β1-globin domain and outside the 5′ boundary of the β2-globin domain. On the S allele, this sequence is located 3 to 4.4 kb 3′ of the β1-globin gene and 8.2 to 9.6 kb 5′ of the β2-globin gene. In this position, it exhibits histone hyperacetylation and H3K4 dimethylation as part of the β1-globin hyperacetylated domain. These data suggest that histone hyperacetylation in erythroid cells is not an intrinsic function of this sequence but is position-dependent and directed by neighboring sequences.
Hyperacetylated domains are maintained in the absence of elongation by RNA polymerase II
If the modification state associated with some sequences within the domain is not intrinsic to those sequences, it follows that it is a consequence of regulatory phenomena that originate elsewhere and establish a chromatin modification pattern that then spreads to include these sequences. Among potential “spreading” models for domain formation, an attractive possibility involves elongation by RNA polymerase II. This model originates from 2 observations: (1) at the human β-globin locus, and elsewhere, transcription is not limited to the genes but occurs throughout intergenic regions as well, although the function of such transcription is unknown31-34 ; (2) histone acetyltransferases and methyltransferases interact with elongating RNA polymerase II.35,36 Thus, hyperacetylated domains could form via transcriptional elongation within the β-globin locus. Previously, however, it has been demonstrated that incubation of MEL cells in dichloro-1-beta-D-ribofuranosyl benzimidazole (DRB), an inhibitor of RNA polymerase II elongation, has no effect on histone modifications over the β1-globin gene in MEL cells.37
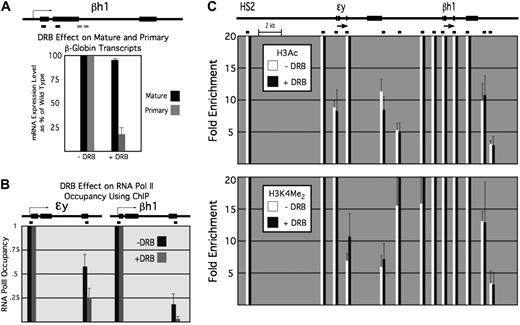
To extend this observation, we isolated E12.5 mouse embryonic blood, which consists almost exclusively of primitive erythroid cells, and incubated them in DRB. Under these conditions, primary βh1-globin transcript levels decreased drastically, whereas mature (spliced) transcript levels were unaffected, as expected given the stability of mature β-globin mRNA (Figure 7A). Occupancy of the β-globin promoters by RNA polymerase II was unaffected, as demonstrated by ChIP, but polymerase levels were reduced within the ϵy- and βh1-globin genes, as expected in the absence of elongation (Figure 7B).
Domain formation after inhibition of RNA Pol II elongation by DRB. (A) DRB effect on mature and primary mRNA transcripts of the embryonic βh1 globin gene. (B) ChIP results for RNA Pol II at the promoters and the 3′ region of ϵy and βh1 in untreated (■) and DRB-treated ( ) samples. (C) H3Ac (top) and H3K4 dimethyl (bottom) ChIP results for E12.5 peripheral blood across the region harboring the embryonic β-globin genes in untreated (□) and DRB-treated (■) cells.
) samples. (C) H3Ac (top) and H3K4 dimethyl (bottom) ChIP results for E12.5 peripheral blood across the region harboring the embryonic β-globin genes in untreated (□) and DRB-treated (■) cells.
Domain formation after inhibition of RNA Pol II elongation by DRB. (A) DRB effect on mature and primary mRNA transcripts of the embryonic βh1 globin gene. (B) ChIP results for RNA Pol II at the promoters and the 3′ region of ϵy and βh1 in untreated (■) and DRB-treated ( ) samples. (C) H3Ac (top) and H3K4 dimethyl (bottom) ChIP results for E12.5 peripheral blood across the region harboring the embryonic β-globin genes in untreated (□) and DRB-treated (■) cells.
) samples. (C) H3Ac (top) and H3K4 dimethyl (bottom) ChIP results for E12.5 peripheral blood across the region harboring the embryonic β-globin genes in untreated (□) and DRB-treated (■) cells.
Under these conditions, we observed no significant change in histone hyperacetylation or H3K4 methylation across the embryonic domain, compared with cells in the absence of DRB (Figure 7C). Given the dynamic nature of histone acetylation, this suggests that maintenance of the hyperacetylated domain does not require elongation by RNA polymerase II, and thus that the histone modification pattern characteristic of the hyperacetylated domain spreads by a distinct mechanism.
Discussion
Based on genome-wide studies of histone modifications in mammalian cells, hyperacetylated domains seem to occur over a small proportion of active gene loci, at least in the cell types examined so far.18,19 As a general rule, histone hyperacetylation and H3K4 dimethylation occur at active gene promoters, with modification levels returning to background within a few hundred base pairs to either side, suggesting that only the most proximal nucleosomes are modified.18,19,35 Among published examples of hyperacetylated domains, all are associated with tissue-specific genes under the control of distal regulatory elements, such as enhancers. Perhaps the most striking example of this specificity is provided by the mammalian α-globin gene cluster. This locus is embedded within a region containing several genes that are constitutively expressed in a wide range of tissues. Despite this arrangement, a hyperacetylated domain forms only in erythroid cells, encompassing the α-globin cluster and regulatory elements located within the introns of a neighboring gene.38
The models provided by the MEL and FDCP-Mix cell lines suggest that the hyperacetylated domains within the β-globin locus in erythroid cells are established well before high-level transcription occurs, and even before commitment to the erythroid lineage. This argues against a mechanism in which hyperacetylated domains are a consequence of high-level transcription during terminal differentiation, although it does not rule out a role for transcription per se because the β-globin genes are transcribed at low levels in these cell lines. In addition, the timing of domain formation remains to be determined in primary erythroid cells, which could differ from the immortalized or transformed lines we use here. Nevertheless, coupled with the fact that domain formation is limited to a minority of gene loci, the available evidence suggests that hyperacetylated domains represent a specific and regulated feature of the activation of certain genes.
Our data provide a basis for differentiating between mechanisms for domain formation. The modification state of nucleosomes located 5′ of the ϵy-globin gene promoter is LCR-dependent; we also find that a sequence located between the β1- and β2-globin genes exhibits different histone modification states on different alleles, within which it resides in different relative locations. Thus, the modifications exhibited by histones at specific sequences are dependent on their context. These results suggest that histone modifications associated with these sequences are a downstream consequence of regulatory phenomena that originate elsewhere.
It has been speculated that nongenic/intergenic transcription could account for the spread of histone modifications, but we demonstrate that the domains are unaffected by inhibition of RNA polymerase II elongation with DRB.31,32,34 Our DRB incubation times (15 minutes in Figure 7; up to 4 hours, data not shown) are sufficient to demonstrate that maintenance of histone hyperacetylation does not require RNA polymerase elongation because this modification is highly dynamic; a typical acetylation event lasts approximately 5 to 15 minutes.39-41 Hyperacetylated domains are also associated with histone H3K4 dimethylation, however, which is more stable and indeed may not be removed in the absence of cell division.9 It is therefore possible that this modification depends on RNA polymerase II elongation, although then the maintenance of histone hyperacetylation would require this or another stable modification, either for active recruitment of histone acetyltransferases or to inhibit histone deacetylases.
We find that the embryonic β-globin gene promoters are not required for formation of the hyperacetylated domain encompassing these genes, suggesting that sequence determinants of domain function reside elsewhere. This result also argues against a role for genic transcription in domain formation, although we cannot rule out a role for transcription per se because the βh0-globin gene is transcribed on the allele harboring the ϵy- and βh1-globin promoter deletions.
Interestingly, we find that a hyperacetylated domain forms over this region in definitive erythroid cells on the allele lacking the ϵy- and βh1-globin gene promoters; on this allele, the βh0 promoter is still intact, but we are unable to detect any transcription. Taken together, the data suggest that the domain is governed by a regulatory sequence distinct from the promoters. This activity is suppressed by the embryonic promoters in definitive erythroid cells; but in the absence of these sequences, it is still functional. The identity of this sequence is not known, but we note that on deletion of the ϵy-globin gene promoter, the domain is partially restored, extending from the gene to a region 7 to 8 kb 3′ of the deletion, where it abruptly ends. This region therefore represents a candidate for a cis-acting sequence element involved in domain formation.
The establishment of a hyperacetylated domain over the embryonic β-globin genes in definitive erythroid cells in the absence of the gene promoters has ramifications for the mechanism by which these genes are developmentally silenced. Previously, it has been shown that incubation of erythroid cells from E14.5 fetal liver with histone deacetylase inhibitors results in an increase in histone acetylation levels at the ϵy-globin promoter, suggesting that histone deacetylase activity is recruited there.42 Our results indicate that the embryonic β-globin gene promoters are required for recruitment throughout the domain. Given that we do not observe any transcription originating from the intact βh0-globin gene promoter, domain formation appears to be insufficient to achieve embryonic β-globin gene activation in definitive erythroblasts. The lack of detectable transcripts, however, suggests that domain formation is a prerequisite to gene activation, not a consequence.
Our results, along with previous studies, suggest that hyperacetylated domains represent a regulated feature of the activation of specific gene loci. Notably, the best-supported model for formation of heterochromatic domains involves the nucleation of heterochromatin at specific sequences, which then spreads bidirectionally to encompass neighboring sequences as well. As yet, however, we cannot conclude that hyperacetylated domains are formed by a completely analogous mechanism. It remains to be determined what cis-acting regulatory sequences govern domain formation and maintenance, and the function they provide in gene activation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Institutes of Health (R01 DK070687), a Burroughs Wellcome Career Development Award and a Beckman Foundation Young Investigator award (M.B.), and the National Institutes of Health T32 training grant (G.F.).
National Institutes of Health
Authorship
Contribution: G.F. and M.B. designed research, analyzed data, and wrote the paper; G.F. and C.d.V. performed research; and M.A.B., M.G., S.F., J.P., R.B., and J.F. provided reagents.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael Bulger, Department of Pediatrics, Center for Pediatric Biomedical Research, University of Rochester Medical Center, Box 703, 601 Elmwood Ave, Rochester, NY 14642; e-mail: Michael_bulger@urmc.rochester.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal