Abstract
T helper 17 (Th17) cells produce IL-17 but can also make tumor necrosis factor, interleukin (IL)–6, IL-10, IL-21, and IL-22. These cytokines collectively contribute to the functional outcome of the Th response. IL-22 plays a critical role in some Th17-associated diseases, such as psoriasis, but its relationship to IL-17 remains controversial. Here, we used a systematic multiparametric analysis of Th-17-associated cytokines, which revealed the unexpected finding that the regulation pattern of IL-22 was most closely related to interferon-γ, the prototypical Th1 cytokine, and not to IL-17. To explain this observation, we systematically tested the role of Th1- and Th17-inducing cytokines. We could show that IL-12 and IL-23 induced high levels of IL-22 but no IL-17. Conversely, transforming growth factor-β inhibited IL-22 production but promoted IL-17. Thus, IL-17 and IL-22 are differentially regulated during cytokine-induced Th cell differentiation. This has important implications for the understanding and pharmacologic manipulation of Th17-associated pathologies.
Introduction
Naive T cells can develop into different T helper (Th) subsets with different cytokine profiles and distinct effector functions.1,2 The Th17 subset produces interleukin (IL)–17, which is particularly important in the activation of antimicrobial defense and autoimmunity.3 However, it became clear that Th17 cells can also produce other Th cytokines, such as tumor necrosis factor (TNF), IL-6, IL-10, IL-21, and IL-22.4,5 The interplay between IL-17 and coproduced cytokines could critically affect the global outcome of a Th17 response and modulate the balance between pathogenesis and protection.5
The relationship between IL-17 and IL-22 is of particular interest. IL-22 exerts similar functions to IL-17, both contributing to the control of extracellular bacterial infection by induction of a strong mucosal immunity.2 The IL-17 and IL-22 expression is often linked to proinflammatory processes, such as psoriasis and Crohn disease.2,6-8 These observations are consistent with a coregulation of IL-17 and IL-22. However, IL-22 can be produced by non-Th17 cell types9-11 independently of IL-17 production. Furthermore, IL-22 also has specific functions, such as induction of tissue-repair and wound-healing responses protecting from liver disease12,13 or myocarditis,14 where IL-17 is not implicated. This evidence suggests a different regulation between IL-17 and IL-22 production. Thus, the relationship between IL-17 and IL-22 remains unclear.
Methods
Purification of naive CD4+ T lymphocytes from adult blood
Peripheral blood naive CD4+ T cells (CD4+ CD45RA+ CD25− CD45RO−) or effector memory CD4 T cells (CD4+CD45RA−CD27−) were isolated from peripheral blood mononuclear cell using immunomagnetic depletion (Miltenyi Biotec) and FACSAria sorting (BD Biosciences) as previously described.4 All cells were used with the approval of the Institutional Review Board of Institut Curie, and blood donors gave their informed consent for research use of buffy coats in accordance with the Declaration of Helsinki.
T helper cell differentiation assay
Naive CD4+ T cells were cultured in Yssel medium (gift of Hans Yssel, Inserm) containing fetal calf serum as previously described.4 Stimulation was performed with cytokines (R&D Systems) and Dynabeads CD3/CD28 (Invitrogen) for 5 days. After 24 hours of restimulation, supernatants were harvested for cytokine detection and cells were collected for transcriptional analysis.
Analysis of cytokine production
IL-17 (R&D Systems), IL-21 (eBioscience), and IL-22 (PeproTech) were measured by enzyme-linked immunosorbent assay (ELISA), IL-4, IL-5, IL-6, IL-10, IL-13, interferon-γ (IFN-γ), and TNF by cytometric bead assay Flex Sets (BD Biosciences). The cells were stained with the red LIVE/DEAD (Invitrogen) to distinguish the living from the dead cells. Cytokine-producing cells were analyzed by intracellular staining after addition of brefeldin (10 μg/mL) during the last 3 hours of restimulation. Cells were fixed and permeabilized using the Staining Buffer Set (eBioscience), stained with anti–IL-22 Alexa 647 (eBioscience), anti–IFN-γ V-450 (BD Biosciences PharMingen), anti–IL-17 Alexa-488 (eBioscience), washed, and then analyzed by flow cytometry (Cyan; Dako North America).
Real-time quantitative RT-PCR
Total RNA was extracted by RNeasy Microkit (QIAGEN) and processed as previously described.4 The following probes were used: IL17A (Hs00174383_m1), IL22 (Hs00220924_m1), RORC (Hs01076112_m1), RORA (Hs00536545_m1), AHR (Hs00169233_m1), TBx21 (Hs00203436_m1), and IL26 (Hs00218189_m1). For each sample, mRNA abundance was normalized to the amounts of the ribosomal protein L34 (Hs00241560_m1).
Statistical analysis
Data used to perform the clustering were corrected for the donor effect by applying a linear model. To summarize the information, the replicates were aggregated within each condition to their barycentric value for each cytokine. Hierarchical clustering analysis was performed using a distance based on Pearson correlation and Ward criteria as an agglomerative method. For pairwise comparisons, we used a nonparametric 2-tailed Wilcoxon test. P values less than .05 were considered statistically significant. We used the Pearson correlation coefficient to assess the significance of correlation between IL-17A, IL-22, TNF, IFN-γ protein, or IL17 and IL22 mRNA with RORC, RORA, AHR, and TBx21.
Results and discussion
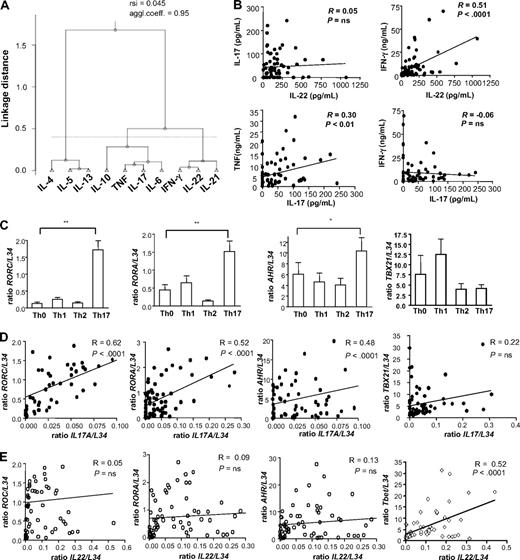
To assess the relationship between Th17-derived cytokines, we generated a variety of Th subsets, including Th0, Th1, and Th2 as controls, together with optimal and suboptimal Th17 subsets (supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article), as previously described.4 We analyzed the production of 10 Th-derived cytokines, and we studied the relation between them by clustering analysis. This strategy enables an unbiased insight into the similarities between Th cytokines, according to their pattern of regulation in different experimental conditions.15 The segregation of IFN-γ, IL-4, and IL-17 in 3 different clusters confirmed the distinct phenotype of Th1, Th2, and Th17 cells, respectively. IL-5 and IL-13 were most closely related to IL-4, consistent with a similar pattern of regulation of these Th2 cytokines. The cytokine showing the strongest association with IL-17 production was TNF (Figure 1A). Unexpectedly, we found that IL-22, which is often described as a Th17 cytokine,16 correlated most closely to IFN-γ, the prototypical Th1 cytokine (Figure 1A). These results were confirmed by pairwise correlation of cytokine levels: IL-22 and IL-17 production did not significantly correlate; IL-22 correlated with IFN-γ; and IL-17 positively correlated with TNF but not with IFN-γ (Figure 1B). Interestingly, the adjacent positioning of the human IL22 and IFNγ genes on chromosome 12q1417 also supports a similar regulation of IL-22 and IFN-γ production. In addition, we could also analyze the expression of IL26, which was recently associated with human Th17 lineage.18,19 We confirmed that IL26 is mainly expressed under Th17 conditions (supplemental Figure 1A), and we observed that it significantly correlated with IL17, but not with IL22 expression (supplemental Figure 1B).
IL-22 regulation is most closely related to IFN-γ rather than IL-17. Naive T cells were cultured under Th0 (no polarizing cytokines), Th1 (1 ng/mL IL-12), Th2 (25 ng/mL IL-4), Th17 (10 ng/mL IL-1β, 20 ng/mL IL-6, 10 ng/mL TNF, 1 ng/mL TGF-β, 100 ng/mL IL-23), suboptimal Th17 (absence of individual Th17-promoting cytokines) in the presence of anti-CD3 + anti-CD28. Protein and transcript analyses were performed after 24 hours of restimulation with anti-CD3 + anti-CD28. Supplemental Table 1 contains more details. (A) Clustering analysis of Th cytokines produced in all experimental conditions using a Pearson correlation–based distance. Cytokines were separated in clusters by comparing their linkage distance. The agglomerative coefficient reflects the structure of the data (values close to 1 indicate well-separated clusters), and resampling similarity index (rsi) evaluates the robustness of the clustering. (B) Graphs of amounts of IL-17 or IL-22 protein were correlated to IFN-γ or TNF levels, using Pearson correlation. R indicates correlation coefficient. (C) RT-PCR for expression of RORC, RORA, AHR, TBx21 in Th0, Th1, Th2, and Th17 conditions. Threshold cycle values were normalized to mRNA of ribosomal protein L34 gene. Data were normalized to the maximal value obtained for each donor. Data are mean ± SEM of 9 donors. (D-E) Graphs of IL17A and IL22 transcript levels, obtained from 9 independent experiments with cells cultured as previously described, were correlated to RORC, RORA, AHR, and TBx21 transcript levels, using Pearson correlation. R indicates correlation coefficient.
IL-22 regulation is most closely related to IFN-γ rather than IL-17. Naive T cells were cultured under Th0 (no polarizing cytokines), Th1 (1 ng/mL IL-12), Th2 (25 ng/mL IL-4), Th17 (10 ng/mL IL-1β, 20 ng/mL IL-6, 10 ng/mL TNF, 1 ng/mL TGF-β, 100 ng/mL IL-23), suboptimal Th17 (absence of individual Th17-promoting cytokines) in the presence of anti-CD3 + anti-CD28. Protein and transcript analyses were performed after 24 hours of restimulation with anti-CD3 + anti-CD28. Supplemental Table 1 contains more details. (A) Clustering analysis of Th cytokines produced in all experimental conditions using a Pearson correlation–based distance. Cytokines were separated in clusters by comparing their linkage distance. The agglomerative coefficient reflects the structure of the data (values close to 1 indicate well-separated clusters), and resampling similarity index (rsi) evaluates the robustness of the clustering. (B) Graphs of amounts of IL-17 or IL-22 protein were correlated to IFN-γ or TNF levels, using Pearson correlation. R indicates correlation coefficient. (C) RT-PCR for expression of RORC, RORA, AHR, TBx21 in Th0, Th1, Th2, and Th17 conditions. Threshold cycle values were normalized to mRNA of ribosomal protein L34 gene. Data were normalized to the maximal value obtained for each donor. Data are mean ± SEM of 9 donors. (D-E) Graphs of IL17A and IL22 transcript levels, obtained from 9 independent experiments with cells cultured as previously described, were correlated to RORC, RORA, AHR, and TBx21 transcript levels, using Pearson correlation. R indicates correlation coefficient.
To investigate whether the IL-22–producing cells preferentially coproduce IFN-γ rather than IL-17, we analyzed the IL-17 and IFN-γ production by IL-22+ cells using in vitro differentiated Th cells and ex vivo purified effector memory CD4 T cells. Interestingly, the majority of IL-22+ cells did not produce either IL-17 or IFN-γ under Th0 and Th17 conditions (86.8% ± 4.1% and 83.1% ± 5.0%, respectively; supplemental Figure 2A-C). In similar conditions, the IL-22+ cells mainly coproduced IFN-γ rather than IL-17 (33.47% ± 21.18% compared with 1.6% ± 1.1%; supplemental Figure 2A,C). This further supports a coregulation of IL-22/IFN-γ in cytokine-induced Th cells. In contrast, IL-22–producing cells among circulating effector memory T cells similarly produced IFN-γ and IL-17, although the majority were IL-22 single producers (supplemental Figure 2B-C).
Next, we assessed the relationship between IL-17 and IL-22 at the level of transcription factors. We confirmed in our human system that RORC, RORA, and AHR20,21 were specifically induced in Th17 conditions (Figure 1C). To address their relationship with IL-17 and IL-22 production, we analyzed their expression in several experimental conditions, which were inducing variable levels of IL-17 and IL-22 (supplemental Table 1). We confirmed a strong correlation between IL17A and RORC expression,4,19 and we showed, for the first time, that human IL17A correlated also with RORA and AHR expression (Figure 1D). We observed a similar correlation between these transcription factors and IL-17F (supplemental Figure 3). In contrast, IL-22 did not correlate with any of the Th17-related transcription factors that we have analyzed (Figure 1E). In the mouse, IL-22 production is dependent on AHR,21 suggesting a different transcriptional regulation of human and mouse IL-22 production. In our system, IL-22 and not IL-17 expression significantly correlated with TBx21, the major Th1 transcription factor, indicating a stronger relationship between IL-22 and IFN-γ compared with IL-17 (Figure 1E). Overall statistical and computational approaches revealed that IL-17 and IL-22 production was differentially regulated during cytokine-induced Th cell differentiation. This prompted us to investigate the underlying factors implicated in the differential regulation of IL-17 and IL-22.
We and others recently showed that human IL-17 induction requires IL-1β, IL-6, IL-23, and transforming growth factor-β (TGF-β).4,22 The previous clustering analysis (Figure 1A) showed that IL-22 behaved more similarly to Th1 than to Th17 cytokines, suggesting that Th1- or Th17-inducing cytokines may explain the differential regulation of IL-17 and IL-22. To address this hypothesis, we systematically tested the role of single cytokines promoting Th1 and/or Th17 differentiation in the induction of IL-22 and IL-17 by naive CD4 T cells. We found that individual inflammatory cytokines (IL-1β, IL-6, TNF) and TGF-β did not induce IL-22 (Figure 2A) or IL-17 production (Figure 2B). On the contrary, naive T cells cultured in the presence of IL-12 (Th1 condition) or IL-23 produced high levels of IL-22 but not IL-17 (Figure 2B). This indicated that IL-23 and IL-12 are 2 underlying factors explaining the differential regulation of IL-17 and IL-22 revealed by statistical methods. These findings are consistent with previous results reporting a role for IL-23 in dermal inflammation and acanthosis in a psoriasis mouse model.8 The importance of IL-12 in inducing IL-22 is supported by a previous study showing a reduced IL-22 secretion by T cells purified from IL-12- and IL-12 receptor-mutated patients.23 This also suggests that IL-22/IFN-γ coproduction could play an important role in Th1-associated diseases. In summary, different environmental conditions can mediate the production of IL-17 or IL-22, potentially explaining immune responses not characterized by the coexpression of IL-17 and IL-22.12-14
IL-12, IL-23, and TGF-β differentially regulate IL-17 and IL-22 production. (A-D) ELISA assay for IL-22 and IL-17 production by naive T cells differentiated for 5 days in the presence of anti-CD3 + anti-CD28 and different cytokines; IL-17 and IL-22 production in supernatant was measured after 24 hours of restimulation with anti-CD3 + anti-CD28. Data are mean ± SEM of 4 donors. ***P < .001, **P < .01, and *P < .05 (Wilcoxon test). (E-H) ELISA for production of IL-22 and IL-17 by naive T cells differentiated with IL-23 or IL-12 or proinflammatory cytokines + IL-23 and different concentrations of TGF-β. Cytokines were measured after 24 hours of restimulation with anti-CD3 + anti-CD28. Cytokine amounts detected were normalized to the maximum value obtained for that cytokine across the whole set of condition, for each donor. Data are mean ± SEM of 3 donors.
IL-12, IL-23, and TGF-β differentially regulate IL-17 and IL-22 production. (A-D) ELISA assay for IL-22 and IL-17 production by naive T cells differentiated for 5 days in the presence of anti-CD3 + anti-CD28 and different cytokines; IL-17 and IL-22 production in supernatant was measured after 24 hours of restimulation with anti-CD3 + anti-CD28. Data are mean ± SEM of 4 donors. ***P < .001, **P < .01, and *P < .05 (Wilcoxon test). (E-H) ELISA for production of IL-22 and IL-17 by naive T cells differentiated with IL-23 or IL-12 or proinflammatory cytokines + IL-23 and different concentrations of TGF-β. Cytokines were measured after 24 hours of restimulation with anti-CD3 + anti-CD28. Cytokine amounts detected were normalized to the maximum value obtained for that cytokine across the whole set of condition, for each donor. Data are mean ± SEM of 3 donors.
Notably, the IL-23–mediated IL-22 production was higher than the expression found in the optimal Th17 condition (Figure 2A). This suggested that some of the other Th17-inducing components could have an inhibitory role in IL-22 production. To test this hypothesis, we measured the IL-22 production in cells stimulated with IL-23 combined to inflammatory cytokines or TGF-β. The combination of inflammatory cytokines induced low levels of IL-22 and did not affect the IL-23–mediated IL-22 production. In contrast, IL-22 was significantly inhibited by TGF-β (Figure 2C). Inflammatory cytokines and not TGF-β could induce low levels of IL-17 in IL-23–stimulated cells (Figure 2D). IL-23– and IL-12–mediated IL-22 production was inhibited by TGF-β in a dose-dependent manner (Figure 2E-F). A comparable inhibition of IL-22 by TGF-β was also observed under Th17-promoting conditions (Figure 2G). In contrast, IL-17 production in the same culture conditions was promoted by TGF-β (Figure 2H), confirming the differential IL-17/IL-22 regulation. Notably, TGF-β did not affect IL-17 and IL-22 production by already differentiated Th17 cells (supplemental Figure 4), indicating that TGF-β acts during the early events of Th differentiation. The ability of TGF-β to inhibit IL-22 production (Figure 2G) as well as to induce apoptosis of Th1 cells24 may collectively contribute to enhancing Th17 differentiation.
In conclusion, we demonstrated that the production of human IL-17 and IL-22 is differentially regulated during cytokine-induced Th cell differentiation. Although our study does not exclude that IL-17 and IL-22 may be coregulated in other systems,16 it provides evidence that IL-22 is not a Th17-specific cytokine, and may be more broadly implicated in Th1- and IL-23–driven responses.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Zofia Maciorowsky, Annick Viguier, Coralie Guerin, and Adamo Diamantini for the cytofluorimetric sorting, and Claire Hivroz and Luca Battistini for helpful suggestions and critical reading of the manuscript.
This work was supported by a Marie Curie Excellence Grant (no. 014162). M.T. is supported by a fellowship from the Fondation pour la Recherche Médicale.
Authorship
Contribution: E.V. designed and performed the experiments and contributed to writing the paper; M.T. and M.-A.M.-P. performed some experiments; N.S. and P.H. did computational and statistical analysis; E.B. supervised the computational and statistical analysis; and V.S. supervised the study and contributed to experimental design and the writing of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Vassili Soumelis, Institut Curie, 26 rue d'Ulm, 75005 Paris, France; e-mail: vassili.soumelis@curie.net.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal