Abstract
C-C chemokine receptor type 7 (CCR7) is a chemokine receptor playing a pivotal role in the induction of human natural killer (NK)–cell migration to lymph nodes. We show that “licensed” peripheral blood killer immunoglobulin-like receptor–positive (KIR+) NK-cell populations, as well as KIR+ NK-cell clones, de novo express CCR7 upon coculture with mature dendritic cells (mDCs) or Epstein-Barr virus (EBV)–transformed lymphoblastoid cell lines. As a consequence, they become capable of migrating in response to the CCR7-specific chemokines C-C chemokine ligand (CCL)–19 and/or CCL21. The acquisition of CCR7 by NK cells requires direct cell-to-cell contact, is detectable within a few minutes, and is due to receptor uptake from CCR7+ cells. This mechanism is tightly regulated by KIR-mediated recognition of human leukocyte antigen (HLA) class I as well as by adhesion molecules including leukocyte function-associated antigen 1 (LFA-1) and CD2. Analysis of NK-cell clones revealed that alloreactive (KIR-ligand mismatched) but not autologous NK cells acquire CCR7. These data have important implications in haploidentical hematopoietic stem cell transplantation (HSCT), in which alloreactive NK cells may acquire the ability to migrate to secondary lymphoid compartments (SLCs), where they can kill recipient antigen-presenting cells (APCs) and T cells thus preventing graft-versus-host (and host-versus-graft) reactions.
Introduction
The function of natural killer (NK) cells is regulated by a finely tuned balance of signals delivered by inhibitory and activating receptors.1,2 The inhibitory receptors specific for major histocompatibility complex (MHC) class I molecules can dampen NK-cell effector function upon appropriate ligation. These include killer immunoglobulin (Ig)–like receptors (KIRs) in humans, Ly49 in mouse, and CD94/NKG2A heterodimers in both species.3-8 In humans, KIRs can harbor 2 (KIR2D) or 3 (KIR3D) extracellular C2-type Ig-like domains. KIRs recognize groups of MHC class I molecules that are determined by amino acid position in the C-terminal portion of the MHC class I α1 helix.9 On the other hand, CD94/NKG2A recognizes the nonclassical MHC class I molecule human leukocyte antigen E (HLA-E) in humans10 and Qa1 in mice.11
The interactions between MHC class I molecules and their inhibitory receptors are thought to play a major role in the mechanisms of self-tolerance during NK-cell effector phases. Indeed, missing or low self-MHC expression results in NK-cell activation and induction of target cell lysis.
Early stages of tissue damage, secondary to invasion of pathogen, including viruses, are associated with the recruitment and local activation of both dendritic cells (DCs) and NK cells.12,13 The majority of peripheral blood NK cells are characterized by the CD56dull CD16+ KIR and/or NKG2A+ surface phenotype and express constitutively chemokine receptors such as CXCR1, CX3CR1, and ChemR23. On the other hand, the minor peripheral blood CD56bright CD16− KIR− NKG2A+ NK-cell subset expresses C-C chemokine receptor type 7 (CCR7).14-17 Based on their chemokine receptor profile it is conceivable that CD56dull CD16+ cells are recruited mainly into inflamed peripheral tissues, whereas CD56bright CD16− cells are attracted to secondary lymphoid compartments (SLCs) such as lymph nodes, in response to C-C chemokine ligand (CCL)–19 and CCL21.18 Accordingly, in inflamed peripheral tissues, NK cells are represented mostly by CD56dull CD16+ cells and express CXCR1 and ChemR23,17 whereas within normal noninflamed lymph nodes, NK cells are homogeneously characterized by the CD56bright CD16dull/− KIR− NKG2A+ surface phenotype.14,15,19,20 These NK cells are localized in the paracortical T-cell areas and are high interferon-gamma producers but display low cytolytic activity. Although these different pathways of NK-cell recruitment reflect the constitutive expression of different sets of chemokine receptors, another mode of NK-cell migration to lymph nodes has recently been described for the CD56dull CD16+ NK-cell subset. When exposed to exogenous interleukin-18 (IL-18) these cells de novo express surface CCR7 and respond to CCL19 and CCL21.21 On the basis of these observations it has been proposed that CD56dull CD16+ NK cells, recruited into inflamed tissues (where proinflammatory cytokines/chemokines favor their interaction with DCs),12,22-24 may exert an instructive activity on naive T-cell priming. This may occur by different mechanisms: (1) NK cells induce DC polarization resulting in the production of high levels of IL-12, which, in turn, would promote type 1 T-cell responses. This mechanism does not require that tissue-activated NK cells migrate to lymph nodes.25,26 (2) NK cells exposed to IL-18 (released by pathogen-activated APCs) migrate to lymph nodes, enter into the paracortex area where they can interact with both DCs and T cells and thus regulate T-cell responses.21,27,28
After recruitment and activation within inflamed peripheral tissues, NK cells acquire the ability to kill DCs (“editing process”) and to exert cytolytic activity against transformed or infected target cells. Therefore, they represent an early barrier against pathogen invasion. Notably, the editing process is selectively mediated by NK cells expressing the KIR− NKG2A+ phenotype, which kill DCs expressing low levels of HLA-E. On the other hand, mature DCs (mDCs) become resistant to lysis as a result of the up-regulation of HLA-E surface expression.29 In addition, KIR− NKG2A+ NK cells, upon interaction with DCs, release cytokines (tumor necrosis factor-alpha and granulocyte/macrophage colony stimulating factor) that facilitate DC maturation.16 On the contrary, KIR+ NK cells are unable to kill DCs and thus do not participate in the DC editing process. This reflects the interaction of the inhibitory KIR with classical HLA-class I molecules, expressed in sufficient amounts on both immature and mature DCs. On the other hand, in inflamed tissues, KIR+ NK cells can kill target cells expressing low levels of HLA class I as a result of viral infection or tumor transformation.
The definition of “alloreactive” applies to KIR+ NKG2A− NK cells that selectively express KIR that are not engaged by the HLA-class I alleles present on target cells.3,30-32 Alloreactive NK cells have been shown to play a crucial role in the successful therapy of high-risk acute myeloid leukemia or acute lymphoid leukemia in the haploidentical hematopoietic stem cell transplantation (HSCT) setting both in adults and in children.33-35
Patients who received a transplant from an NK alloreactive donor benefited from higher rates of engraftment and reduced incidence of graft-versus-host disease (GVHD). Both clinical and experimental data in a murine model suggested that alloreactive NK cells could play a crucial role in preventing donor T-cell priming and subsequent GVHD by killing recipient APCs within lymph nodes.33 However, because KIR+ NK cells normally do not express CCR7,16 it was unclear how donor NK cells could migrate to lymph nodes and kill recipient DCs.
In this study, we show that KIR+ CD56dull NK cells rapidly express surface CCR7 upon interaction with either mature monocyte-derived DCs (MoDCs) or Epstein-Barr virus (EBV)–transformed B cells, and acquire migratory properties toward lymph node–associated chemokines. This phenomenon that is due to uptake of CCR7 from target cells appears to be tightly regulated by KIR/HLA-class I interactions. Indeed, in the case of KIR/HLA-class I mismatches (as in haploidentical HSCT), CCR7 uptake is confined to alloreactive NK cells.
Methods
Cell preparations
Peripheral blood mononuclear cells were derived from healthy donors by Ficoll-Hypaque gradients (Sigma-Aldrich) and purified by negative selection, using NK cell isolation Kit II (Miltenyi Biotec GmbH). The purity of NK cells was more than 98% as assessed by flow cytometric analysis. Myeloid DCs were generated from monocytes purified by CD14MicroBeads human Isolation Kit (Miltenyi Biotec GmbH) derived from peripheral blood mononuclear cells of healthy donors.36 Monocytes were cultured in RPMI 1640 containing 10% fetal calf serum (FCS), in the presence of IL-4 and granulocyte/macrophage colony stimulating factor (Pepro Tech) at final concentrations of 20 ng/mL and 50 ng/mL, respectively. After 6 days of culture, cells were characterized by the CD14− CD1a+ CD83− phenotype corresponding to immature DCs (iDCs).29 To generate CD83+ CD86+ mDCs, iDCs were stimulated with lipopolysaccharide (LPS; Sigma-Aldrich) at a final concentration of 1 μg/mL.
To obtain polyclonally or clonally activated NK cells, freshly isolated NK cells were cultured on irradiated feeder cells in the presence of 100 U/mL recombinant human IL-2 (Proleukin; Chiron) and 1.5 ng/mL phytohemagglutinin (GIBCO Ltd).36
The medium used throughout the experiments was RPMI 1640 medium supplemented with 2 mM l-glutamine, 1% penicillin-streptomycin-neomycin mixture, and 10% heat-inactivated FCS.
Approval was obtained from the University of Genoa Review Board. Informed consent was provided according to the Declaration of Helsinki.
Freshly purified NK cells or polyclonal or clonally IL-2–activated NK cells were stimulated with different cell lines and with immature or mature MoDCs or with different cytokines such as recombinant human IL-18 at 0.1 μg/mL (Biosource International Inc) in the presence or in the absence of anti–HLA I monoclonal antibodies (mAbs). These experiments were also performed either in the presence or in the absence of the antagonistic anti–IL-18 mAb (0.5 μg/mL; MBL).
NK cells were plated at 106 cells/mL in round-bottom 96-well tissue culture plates (Corning Costar); in coculture experiments, NK cells were mixed with targets at an effector-to-target cell (E/T) ratio of 1:1, spun down for 3 minutes at 159g, and incubated for different time points at 37°C in 5% CO2. Next, NK cells were directly assessed for surface phenotype, cytolytic activity, and cytokine production.36
Migration assay
Chemotaxis of NK cells was measured by migration through a polycarbonate filter with 3.0-μm pore size in 24-well Transwell chambers (Corning Costar) with the use of RPMI/10% FCS as assay medium. Assay medium (600 μL) containing 10 or 250 ng/mL CCL19 or CCL21 (Pepro Tech) was added to the lower chamber. Assay medium alone (600 μL) was used as control for spontaneous migration. NK cells (2 × 105) stimulated or not were added to the upper chamber in a total volume of 100 μL. After 3 hours of incubation at 37°C, cells that had migrated to the bottom chamber were counted by flow cytometry, acquiring events for a fixed time period of 60 seconds, and counted by optical microscopy. The number of spontaneously migrated cells was subtracted from the total number of migrated cells. Values are given as the chemotactic index compared with migration of unstimulated NK cells.28
Reverse-transcription PCR analysis
Total RNA was extracted from 221 lymphoblastoid cell line, NK-cell populations, and NK-cell populations sorted after coculture with 221 cell line using RNAeasy Micro Kit (QIAGEN). Oligo (dT)–primed cDNA was prepared by standard technique using Transcriptor (Roche). Polymerase chain reaction (PCR) amplifications were performed with primers described in supplemental data (available on the Blood website; see the Supplemental Materials link at the top of the online article). Amplifications were performed for 30 cycles (30 seconds at 95°C, 30 seconds at 58°C, 1 minute at 68°C) using Platinum TAQ (Invitrogen). PCR products were run on a 0.8% agarose gel and visualized by ethidium bromide staining.
Confocal microscopy
NK-cell populations were mixed with HEK293 cells stably transfected with CCR7/green fluorescent protein (GFP) for 30 minutes at 37°C. After this period, cells were loaded on poly-lysine–coated slides (Menzel-Gläser), fixed with 2% paraformaldehyde in phosphate-buffered saline, and stained with ECM17 mAb (anti-CD11a, IgM) followed by AlexaFluor-546 goat anti–mouse IgM antibody (Molecular Probes). Samples were analyzed using a Leica TCS SL confocal system (Leica Microsystems) with a 63× objective. Samples were excited at 488 and 546 nm. Images were analyzed with the standard Leica confocal software.
Results
De novo expression of CCR7 in CD56dull CD16+ NK cells upon interaction with lymphoblastoid B-cell lines
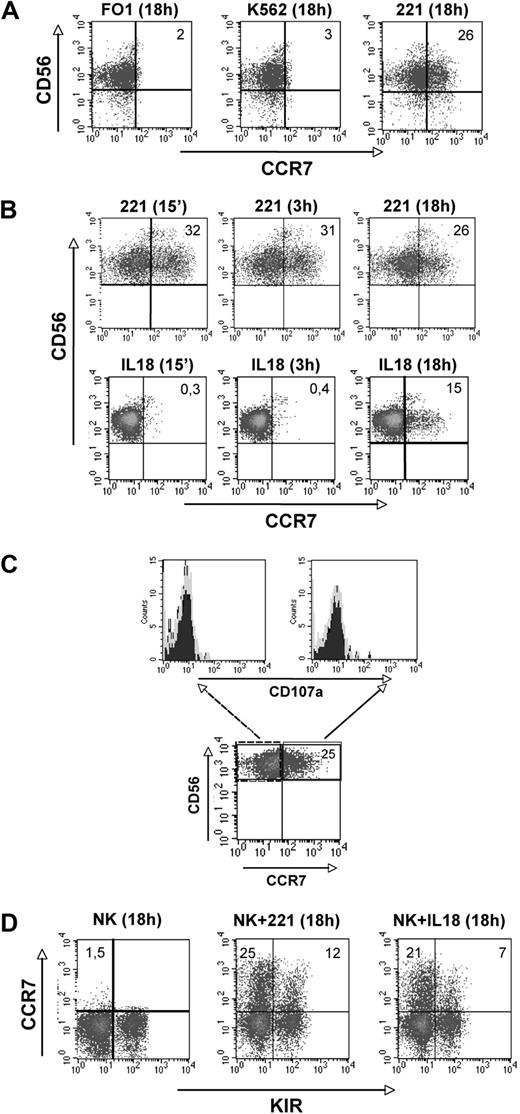
In these experiments we evaluated whether human NK-cell interaction with either tumors or virally infected cells would modify the pattern of expression of CCR7, a chemokine receptor crucial for conferring the ability to migrate to lymph nodes. Purified peripheral blood NK cells were cocultured with different HLA-class I–negative target cells including K562, FO1, and LCL 721.221 (referred to as 221) cell lines that are all susceptible to NK-mediated cytolysis. At different time intervals, NK cells were analyzed for the expression of informative surface markers including CCR7. As shown in Figure 1A, a fraction of NK cells expressed CCR7 upon interaction with 221, but not with the other target cells analyzed. In 12 different experiments, using different donors, the range of CCR7+ CD56dull NK cells after coculture was 12% to 49% (mean, 34.2%). Notably, this expression was confined to the CD56dull CD16+ subset. The levels of CCR7 expressed in this NK-cell subset were higher than those expressed by CD56bright CD16− NK cells (either constitutively or after coculture with 221 cells). The expression of CCR7 after NK-cell exposure to 221 occurred rapidly after as few as 15 minutes. For comparison, the same NK cells were analyzed for the expression of CCR7 induced upon culture in the presence of IL-18.21 In this case, CCR7 expression was detectable after approximately 18 hours of incubation (Figure 1B). Remarkably, the 221-induced CCR7 expression preceded the NK-cell degranulation induced upon interaction with 221 cells. Indeed surface CD107a was not detected in NK cells already expressing CCR7+ soon after exposure to 221 cells (Figure 1C). Analysis of the KIR surface phenotype revealed that the de novo expression of CCR7 in NK cells cultured with the 221 cell line was detectable in both KIR+ and KIR− CD56dull NK cells (Figure 1D). Similar results were obtained upon NK-cell exposure to IL-18, whereas no CCR7 expression was detectable when the same NK cells were cultured alone.
Acquisition of CCR7 surface expression by CD56dull NK cells. (A-D) Freshly isolated NK cells were purified and cultured for 18 hours either in medium alone or in the presence of the indicated HLA-class I–negative cell lines and then analyzed by 2-color immunofluorescence for the expression of C-C chemokine receptor type 7 (CCR7) in combination with CD56 (A). (B) Freshly isolated natural killer (NK) cells were cultured with the 221 cell line or in the presence of exogenous IL-18 for different time points and analyzed for CCR7 expression in combination with CD56. In both panels A and B, the values reported in the top right corners indicate the percentage of CD56+ CCR7+ NK cells. (C) Polyclonal IL-2–activated NK cells were cultured for 1 hour in the presence of the HLA-class I–negative 221 cell line and then analyzed by 2-color immunofluorescence for the expression of CCR7 in combination with CD56. Both CD56+ CCR7− and CD56+ CCR7+ NK cells were also analyzed for CD107a expression. Data are representative of 3 independent experiments performed using different donors. (D) CD56dull NK cells that had acquired the CCR7+ phenotype after 18 hours of coculture with the 221 cell line or exogenous IL-18 were analyzed for KIR expression using a mixture of anti-KIR mAbs. The values reported in the top left and in the top right corners indicate the percentage of CD56+ KIR− and of CD56+ KIR+ NK cells, respectively. These experiments are representative of 12 independent experiments performed using different donors.
Acquisition of CCR7 surface expression by CD56dull NK cells. (A-D) Freshly isolated NK cells were purified and cultured for 18 hours either in medium alone or in the presence of the indicated HLA-class I–negative cell lines and then analyzed by 2-color immunofluorescence for the expression of C-C chemokine receptor type 7 (CCR7) in combination with CD56 (A). (B) Freshly isolated natural killer (NK) cells were cultured with the 221 cell line or in the presence of exogenous IL-18 for different time points and analyzed for CCR7 expression in combination with CD56. In both panels A and B, the values reported in the top right corners indicate the percentage of CD56+ CCR7+ NK cells. (C) Polyclonal IL-2–activated NK cells were cultured for 1 hour in the presence of the HLA-class I–negative 221 cell line and then analyzed by 2-color immunofluorescence for the expression of CCR7 in combination with CD56. Both CD56+ CCR7− and CD56+ CCR7+ NK cells were also analyzed for CD107a expression. Data are representative of 3 independent experiments performed using different donors. (D) CD56dull NK cells that had acquired the CCR7+ phenotype after 18 hours of coculture with the 221 cell line or exogenous IL-18 were analyzed for KIR expression using a mixture of anti-KIR mAbs. The values reported in the top left and in the top right corners indicate the percentage of CD56+ KIR− and of CD56+ KIR+ NK cells, respectively. These experiments are representative of 12 independent experiments performed using different donors.
The acquisition of CCR7 upon interaction with lymphoblastoid B cells is regulated by HLA-class I/inhibitory NK receptor interaction
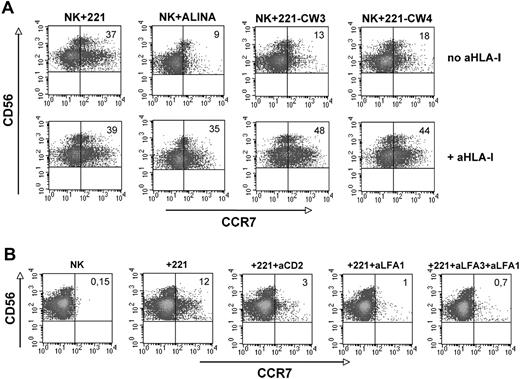
To assess whether the induction of CCR7 expression could be regulated by the interaction of inhibitory NK receptors with HLA-class I molecules NK cells were cocultured with another B-EBV-cell line (Alina) that, different from 221 cells, is characterized by the expression of surface HLA-class I molecules. In other experiments NK cells were cocultured with 221 cell transfectants expressing either HLA-CW3 or HLA-CW4.37 As shown in Figure 2A, all these EBV cell lines displayed a sharply reduced ability to induce CCR7 expression on NK cells compared with untransfected 221 cells. However, CCR7 expression on NK cells could be greatly enhanced when anti–HLA-class I mAbs were added to these cocultures. For example, only 9% of NK cells cultured with Alina cells expressed CCR7, whereas in the presence of anti–HLA-class I mAb CCR7+ NK cells were 35%. Similarly, CCR7 was acquired by only a small fraction of NK cells cocultured with 221 cells transfected with HLA-C, whereas its expression was highly increased when the same cocultures were performed in the presence of anti–HLA-class I mAb. Altogether these data indicate that the interaction between HLA-class I molecules expressed on B-EBV cell lines and HLA-specific inhibitory receptors on NK cells prevents the induction of CCR7 expression on the CD56dull CD16+ subset.
Role of inhibitory NK receptor/HLA-class I interactions in the induction of CCR7 expression. (A-B) Freshly isolated NK cells were purified, cultured for 1 hour with the indicated cell lines either in the presence or in the absence of anti–HLA-class I monoclonal antibodies (mAbs), and analyzed by 2-color immunofluorescence for the expression of CCR7 in combination with CD56 (A). Fresh NK cells were also cocultured for 1 hour with the HLA I–negative 221 cell line in the presence or in the absence of antibodies specific for the indicated surface molecules and then analyzed for CCR7 expression (B). In both panels A and B, the values reported in the top right corners indicate the percentage of CD56+ CCR7+ NK cells. The experiment shown is representative of 8 independent experiments performed using different donors.
Role of inhibitory NK receptor/HLA-class I interactions in the induction of CCR7 expression. (A-B) Freshly isolated NK cells were purified, cultured for 1 hour with the indicated cell lines either in the presence or in the absence of anti–HLA-class I monoclonal antibodies (mAbs), and analyzed by 2-color immunofluorescence for the expression of CCR7 in combination with CD56 (A). Fresh NK cells were also cocultured for 1 hour with the HLA I–negative 221 cell line in the presence or in the absence of antibodies specific for the indicated surface molecules and then analyzed for CCR7 expression (B). In both panels A and B, the values reported in the top right corners indicate the percentage of CD56+ CCR7+ NK cells. The experiment shown is representative of 8 independent experiments performed using different donors.
Next we investigated the receptor/ligand interaction(s) that is responsible for the induction of CCR7 expression in NK cells. In these experiments a panel of antibodies specific for different triggering receptors or coreceptors,38 or for adhesion molecules, was used to assess their ability to inhibit CCR7 expression in NK cells cocultured with 221 cells. As shown in Figure 2B, antibodies to CD2 (or leukocyte function-associated antigen 3 [LFA-3]) and LFA-1, alone or in combination, had a strong inhibitory effect, although some degree of inhibition was detected with anti-NKp46 mAb (not shown). Antibodies specific for other activating receptors including NKp30, 2B4, NTBA (NK-, T-, and B-cell Antigen), NKG2D, and DNAM-1 (DNAX accessory molecule 1) did not inhibit CCR7 expression (not shown).
Induction of CCR7 on NK cells cocultured with dendritic cells
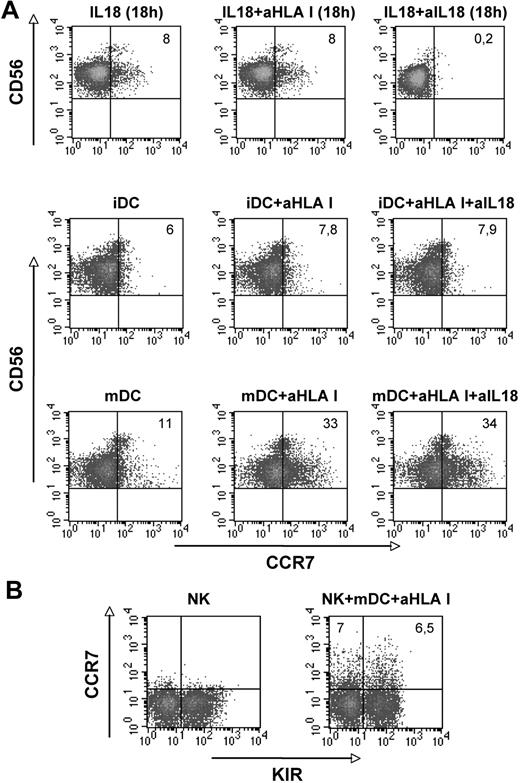
To evaluate whether NK cells would express CCR7 upon exposure to DCs, that is, a physiologic cellular interaction regulating both innate and adaptive immune responses, purified peripheral blood NK cells were cocultured with either immature or mature (LPS-induced) MoDCs.22-24 These experiments were performed both in the absence and in the presence of anti–HLA-class I mAb. As shown in Figure 3A, only few CD56dull NK cells expressed CCR7 when cocultured with immature DCs (iDCs) and no increased expression was detected in the presence of anti–HLA-class I mAb. In addition, coculture of NK cells with mDCs did not result in substantial CCR7 expression. However, when anti–HLA-class I mAb was added to the cultures, strong increase of CCR7+ NK cells was detected. The range of CCR7+ CD56dull NK cells after interaction with mDCs in the presence of anti–HLA-class I mAbs was 12% to 37% in 10 different donors (mean, 27.3). Thus, NK cells appear to express CCR7 upon interaction with mDCs (and not iDCs). Moreover, also in this case, HLA-specific inhibitory NK receptors appear to play a crucial regulatory role. Because DCs can release IL-18,39 to rule out that expression of CCR7 was induced by mDC-derived IL-18, control cultures were established in the presence of a neutralizing anti–IL-18 mAb (together with anti–HLA-class I mAb). As shown in Figure 3A, no inhibitory effect occurred. On the other hand, anti–IL-18 mAb inhibited the CCR7 expression induced by exogenous IL-18. Thus the expression of CCR7 induced by DCs is not due to IL-18 released by DCs themselves. This conclusion is further supported by the finding that analysis of the time course of CCR7 induction revealed a rapid expression (similar to that observed in cocultures with EBV cell lines), not compatible with the time interval required for IL-18–dependent expression. Indeed, CCR7+ NK cells were detected after only 1 hour of coculture with mDCs (Figure 3A). In addition, in this case, the cells that acquired CCR7 belonged to both KIR+ and KIR− CD56dull NK cells (Figure 3B).
Analysis of the CCR7 surface expression on CD56dull NK cells upon interaction with either immature or mature DCs. (A-B) Freshly isolated NK cells were purified and cocultured for 1 hour with immature DCs (iDCs) or mature dendritic cells (mDCs) in the absence or in the presence of anti–HLA I mAbs or in the presence of anti–HLA I mAbs plus neutralizing anti–IL-18 mAb (A middle and bottom lines) and assessed for CCR7 expression. Control cultures were performed with exogenous IL-18 for 18 hours to verify the efficiency of the neutralizing anti–IL-18 mAb (A top line). The values reported in the top right corners indicate the percentage of CD56+ CCR7+ NK cells. (B) NK cells that had acquired the CCR7+ phenotype after 1-hour coculture with mDCs, in the presence of anti–HLA-class I mAb, were analyzed for KIR expression using a mixture of anti-KIR mAbs. The values reported in the top left and in the top right corners indicate the percentage of CD56+ KIR− and of CD56+ KIR+ NK cells, respectively. Data are representative of 10 independent experiments performed using different donors.
Analysis of the CCR7 surface expression on CD56dull NK cells upon interaction with either immature or mature DCs. (A-B) Freshly isolated NK cells were purified and cocultured for 1 hour with immature DCs (iDCs) or mature dendritic cells (mDCs) in the absence or in the presence of anti–HLA I mAbs or in the presence of anti–HLA I mAbs plus neutralizing anti–IL-18 mAb (A middle and bottom lines) and assessed for CCR7 expression. Control cultures were performed with exogenous IL-18 for 18 hours to verify the efficiency of the neutralizing anti–IL-18 mAb (A top line). The values reported in the top right corners indicate the percentage of CD56+ CCR7+ NK cells. (B) NK cells that had acquired the CCR7+ phenotype after 1-hour coculture with mDCs, in the presence of anti–HLA-class I mAb, were analyzed for KIR expression using a mixture of anti-KIR mAbs. The values reported in the top left and in the top right corners indicate the percentage of CD56+ KIR− and of CD56+ KIR+ NK cells, respectively. Data are representative of 10 independent experiments performed using different donors.
The de novo expression of CCR7 confers to NK cells the ability to migrate in response to SLC chemokines
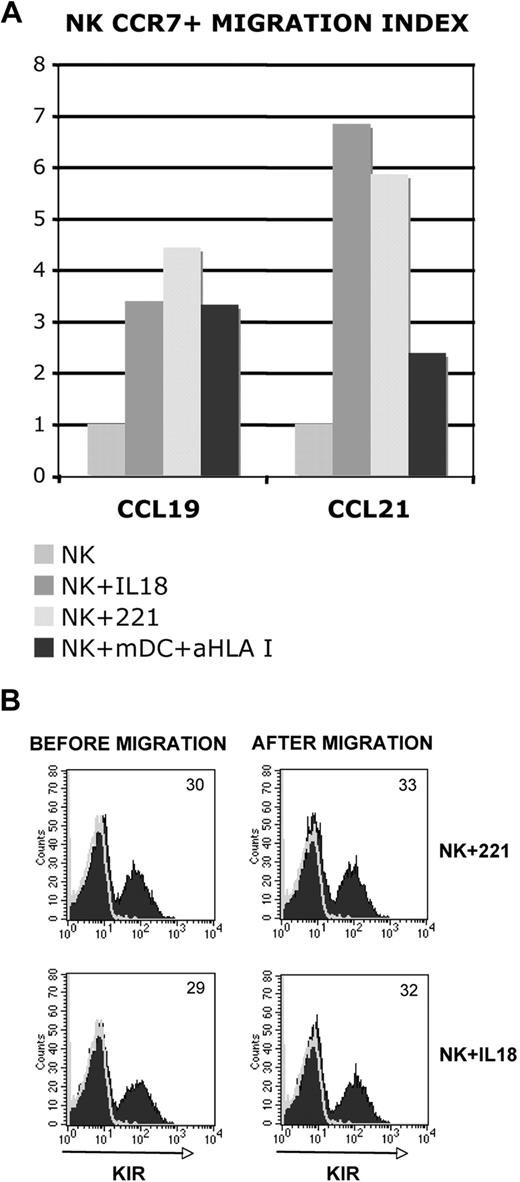
The expression of CCR7 was previously shown to correlate with the ability of NK cells to migrate in response to CCL19 and/or CCL21, that is, typical chemokines released within SLCs.15 We thus analyzed whether also NK cells that had acquired CCR7 after coculture with either 221 cells or mDCs would display this functional property. NK cells in which CCR7 was induced by IL-18 were used as positive control.21 As shown in Figure 4A, both CCR7+ NK-cell populations migrated in a similar fashion in response to CCL19 or CCL21. Because both KIR+ and KIR− NK cells de novo expressed CCR7, we further analyzed the KIR phenotype of NK cells before and after migration. As shown in Figure 4B, approximately the same proportion of KIR+ cells could be detected in NK cells before or after migration. Similar results were obtained with NK cells exposed to mDCs (not shown). Thus, the de novo expression of CCR7 by NK cells is accompanied by the acquisition of migratory properties in response to SLC chemokines.
Correlation between CCR7 expression and ability of NK cells to migrate in response to CCL19 and/or CCL21. (A-B) NK cells that had been cultured alone or cocultured with either 221 cells or mDCs were analyzed for their migratory properties in response to either CCL19 or CCL21 (A). NK cells cultured in the presence of IL-18 were also analyzed as positive control. (B) The KIR phenotype of NK cells that had been exposed to either the 221 cell line or IL-18 was evaluated before and after migration. In panel B, the values reported in the top right corner indicate the percentage of KIR+ NK cells. Data are representative of 8 independent experiments performed using different donors.
Correlation between CCR7 expression and ability of NK cells to migrate in response to CCL19 and/or CCL21. (A-B) NK cells that had been cultured alone or cocultured with either 221 cells or mDCs were analyzed for their migratory properties in response to either CCL19 or CCL21 (A). NK cells cultured in the presence of IL-18 were also analyzed as positive control. (B) The KIR phenotype of NK cells that had been exposed to either the 221 cell line or IL-18 was evaluated before and after migration. In panel B, the values reported in the top right corner indicate the percentage of KIR+ NK cells. Data are representative of 8 independent experiments performed using different donors.
Alloreactive NK cells express CCR7 after exposure to allogeneic DCs
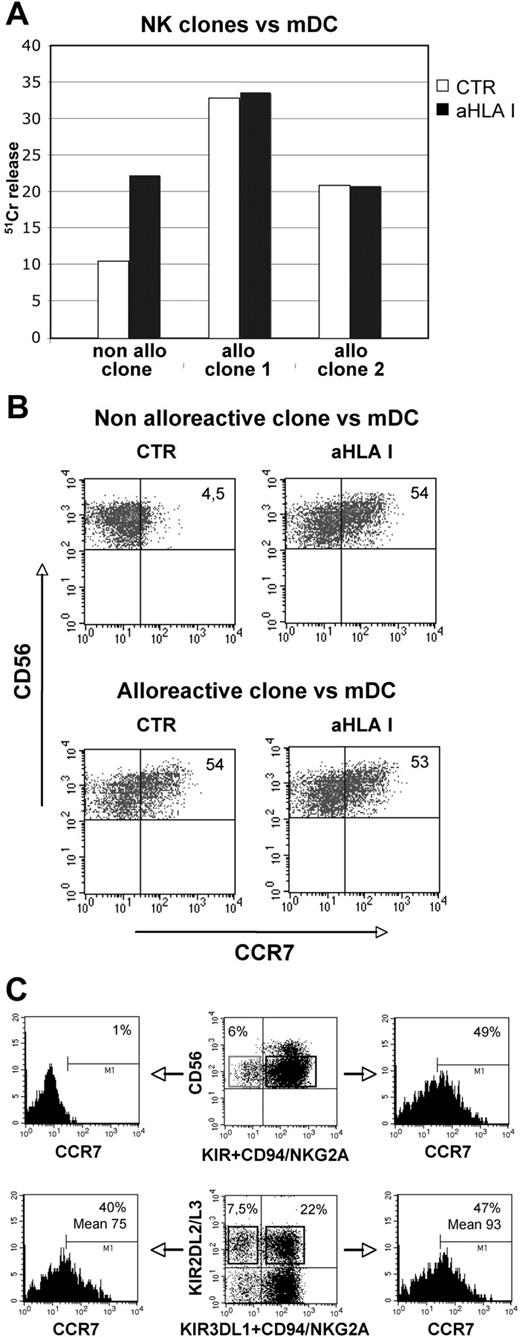
As shown in Figures 2A and 3A, the acquisition of CCR7 by NK cells interacting with either B-EBV cells or mDCs is regulated by the interactions between inhibitory receptors and HLA-class I molecules (Figures 2–3). A previous study29 indicated that in an autologous setting only a small percentage of NK cells characterized by the KIR− NKG2A+ phenotype was capable of killing mDCs. However, in the presence of anti–HLA-class I antibodies, most NK cells including the KIR+ ones could kill autologous mDCs. On the other hand, when alloreactive NK cells were used as effectors, killing of mDCs occurred also in the absence of inhibitory mAb. To better understand the correlation between CCR7 expression and inhibitory receptor-mediated signaling, we selected NK-cell clones expressing the appropriate KIR phenotype and analyzed whether they displayed alloreactivity against mDCs from allogeneic donors and whether this function correlated with the acquisition of CCR7. To this end, we used mDCs from donors expressing HLA-C alleles belonging to the C1 specificity and NK-cell clones expressing either C1-specific or C2-specific KIRs.34 As shown in Figure 5A, 2 different clones expressing KIR2DL1 (C2 specific) displayed alloreactivity against (ie, lysed) the allogeneic C1+/C1+ mDCs used as target cells. On the other hand, no DC killing occurred with autologous NK clones expressing KIR2DL2 or KIR2DL3 (C1 specific). Killing, however, was restored in the presence of anti–HLA-class I mAb.34 The same NK clones were analyzed for CCR7 expression after coculture with the same C1+/C1+ mDCs. As shown in Figure 5B and Table 1, both KIR2DL1+ and KIR2DL2+ clones acquired CCR7 in the presence of anti–HLA-class I mAb. However, only KIR2DL1+ clones expressed CCR7 in the absence of anti–HLA-class I mAb. These experiments clearly indicate that, upon interaction with allogeneic mDCs, the expression of CCR7 (and the ability to mediate cytotoxicity) is a peculiarity of alloreactive NK cells, although it does not occur with autologous KIR+ NK cells.
CCR7 expression on alloreactive and “licensed” NK cells after exposure to allogeneic DCs or HLA-class I–negative Epstein-Barr virus cell line. (A-C) Alloreactive and nonalloreactive NK-cell clones were assessed for their ability to kill mDCs in a 51Cr-release assay at an E/T ratio of 5:1. The cytolytic activity of NK-cell clones against mDCs was assessed either in the absence or in the presence of anti–HLA-class I mAb (A). The SD did not exceed 4% in the 51Cr-release assays. The same NK clones were also analyzed for CCR7 acquisition after 1-hour coculture with mDCs in the absence or in the presence of anti–HLA-class I mAb (B). In panel B, the values reported in the top right corners indicate the percentage of CD56+, CCR7+ NK cells. Data are representative of 7 independent experiments performed using different NK clones. (C) Peripheral blood NK cells from a C1+/C1+ donor were cultured with 221 cells and subsequently stained with anti-CCR7, anti-KIR, and anti-NKG2A mAb. (Top panels) unlicensed NK cells (top left quadrant) lacking KIR2DL2/3, KIR3DL1, and NKG2A were compared for CCR7 acquisition with licensed NK cells (top right quadrant). (Bottom panels) Cells expressing KIR2DL2/3 alone (top left quadrant) or in combination with either KIR3DL1 or NKG2A (top right quadrant) were evaluated for CCR7 acquisition. Similar results were obtained in 2 additional experiments.
CCR7 expression on alloreactive and “licensed” NK cells after exposure to allogeneic DCs or HLA-class I–negative Epstein-Barr virus cell line. (A-C) Alloreactive and nonalloreactive NK-cell clones were assessed for their ability to kill mDCs in a 51Cr-release assay at an E/T ratio of 5:1. The cytolytic activity of NK-cell clones against mDCs was assessed either in the absence or in the presence of anti–HLA-class I mAb (A). The SD did not exceed 4% in the 51Cr-release assays. The same NK clones were also analyzed for CCR7 acquisition after 1-hour coculture with mDCs in the absence or in the presence of anti–HLA-class I mAb (B). In panel B, the values reported in the top right corners indicate the percentage of CD56+, CCR7+ NK cells. Data are representative of 7 independent experiments performed using different NK clones. (C) Peripheral blood NK cells from a C1+/C1+ donor were cultured with 221 cells and subsequently stained with anti-CCR7, anti-KIR, and anti-NKG2A mAb. (Top panels) unlicensed NK cells (top left quadrant) lacking KIR2DL2/3, KIR3DL1, and NKG2A were compared for CCR7 acquisition with licensed NK cells (top right quadrant). (Bottom panels) Cells expressing KIR2DL2/3 alone (top left quadrant) or in combination with either KIR3DL1 or NKG2A (top right quadrant) were evaluated for CCR7 acquisition. Similar results were obtained in 2 additional experiments.
Surface expression of CCR7 on alloreactive and nonalloreactive NK-cell clones after coculture with mDCs
| Clone . | CCR7+,% . | % CCR7+ in the presence of anti–HLA I mAb, % . |
|---|---|---|
| Clone 3, alloreactive | 40 | 45 |
| Clone 4, alloreactive | 60 | 61 |
| Clone 5, alloreactive | 55 | 54 |
| Clone 6, alloreactive | 80 | 82 |
| Clone 7, nonalloreactive | 8 | 51 |
| Clone 8, nonalloreactive | 3.6 | 60 |
| Clone 9, nonalloreactive | 2 | 55 |
| Clone 10, nonalloreactive | 4 | 49 |
| Clone . | CCR7+,% . | % CCR7+ in the presence of anti–HLA I mAb, % . |
|---|---|---|
| Clone 3, alloreactive | 40 | 45 |
| Clone 4, alloreactive | 60 | 61 |
| Clone 5, alloreactive | 55 | 54 |
| Clone 6, alloreactive | 80 | 82 |
| Clone 7, nonalloreactive | 8 | 51 |
| Clone 8, nonalloreactive | 3.6 | 60 |
| Clone 9, nonalloreactive | 2 | 55 |
| Clone 10, nonalloreactive | 4 | 49 |
Natural killer (NK)–cell clones expressing either C1-specific or C2-specific killer immunoglobulin-like receptors were analyzed by immunofluorescence and fluorescence-activated cell-sorting analysis for the expression of C-C chemokine receptor type 7 (CCR7) molecule after 1 hour of coculture with mature dendritic cells (mDCs) from donors expressing human leukocyte antigen (HLA)–C alleles belonging to the C1 specificity, in the absence or presence of anti–HLA class I monoclonal antibody.
Next, to understand whether NK cells acquiring CCR7 were those undergoing the licensing/education process,40-42 we analyzed peripheral blood NK cells from C1+/C1+ donors, lacking most activating KIRs (KIR haplotype A),7 that had been cocultured with 221 cells. As shown in Figure 5C, uptake of CCR7 was virtually absent in unlicensed cells, lacking KIR2DL2/3, KIR3DL1, and NKG2A (ie, the inhibitory receptors for self-HLA-class I molecules). Unlicensed cells represented 6% of NK cells and included both KIR2DL1 single-positive (42%) and KIR/NKG2A double-negative (58%) cells. Comparison between NK cells expressing a single self-reacting KIR (KIR2DL2/3) and cells expressing more than 1 inhibitory receptor did not reveal significant differences in CCR7 uptake (Figure 5C).
NK cells acquire CCR7 by a mechanism of trogocytosis
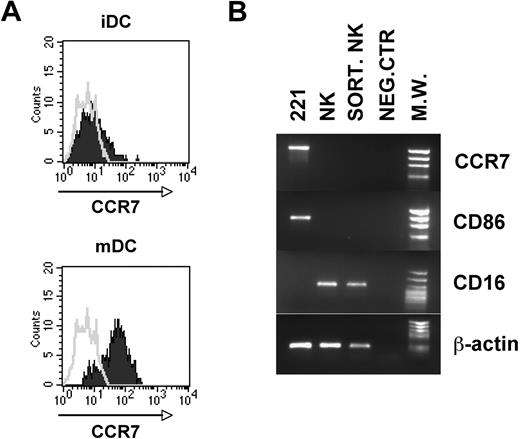
The finding that NK cells acquire CCR7 upon interaction with certain cell types prompted us to analyze whether this event could be related to peculiar properties of the cells interacting with NK cells. The EBV cell lines capable of inducing CCR7 expression on NK cells were characterized by high levels of surface CCR7 (not shown). On the contrary, cell lines such as K562 and FO1, unable to induce CCR7 expression, were CCR7 negative. In addition, comparison of the CCR7 expression by iDCs or mDCs revealed that only mDCs express high levels of CCR7 (Figure 6A). These data suggested that the acquisition of CCR7 by NK cells might reflect a mechanism of uptake from CCR7+ cells. In line with this possibility, the de novo expression of CCR7 by NK cells was detectable after only few minutes of coculture with EBV cell lines (Figure 1B) or mDCs (Figure 3A). Direct evidence that CCR7 surface expression is due to uptake from other cells and not to de novo transcription/synthesis was obtained in experiments in which CCR7-negative NK cells cocultured with 221 cells were isolated by cell sorting and analyzed by reverse-transcription (RT)–PCR for CCR7 mRNA. As shown in Figure 6B, these NK cells acquired CCR7 surface molecules but did not express CCR7 mRNA.
Analysis of surface CCR7 expression on DCs and of CCR7 mRNA on NK cells after coculture with 221 cell line. (A-B) Both immature and mature DCs were analyzed by immunofluorescence for the expression of CCR7 (A). (B) NK cells that had been cocultured with the 221 cell line were isolated by cell sorting and analyzed by RT-PCR for CCR7 expression compared with the 221 cell line and polyclonal NK cells. NK cells used for cocultures in these experiments were represented by polyclonal IL-2–activated populations that were homogeneously characterized by a CCR7-negative surface phenotype. PCR products were run on a 0.8% agarose gel and visualized by ethidium bromide staining. RT-PCR was also performed with primers specific for CD86, CD16, and β-actin as positive control.
Analysis of surface CCR7 expression on DCs and of CCR7 mRNA on NK cells after coculture with 221 cell line. (A-B) Both immature and mature DCs were analyzed by immunofluorescence for the expression of CCR7 (A). (B) NK cells that had been cocultured with the 221 cell line were isolated by cell sorting and analyzed by RT-PCR for CCR7 expression compared with the 221 cell line and polyclonal NK cells. NK cells used for cocultures in these experiments were represented by polyclonal IL-2–activated populations that were homogeneously characterized by a CCR7-negative surface phenotype. PCR products were run on a 0.8% agarose gel and visualized by ethidium bromide staining. RT-PCR was also performed with primers specific for CD86, CD16, and β-actin as positive control.
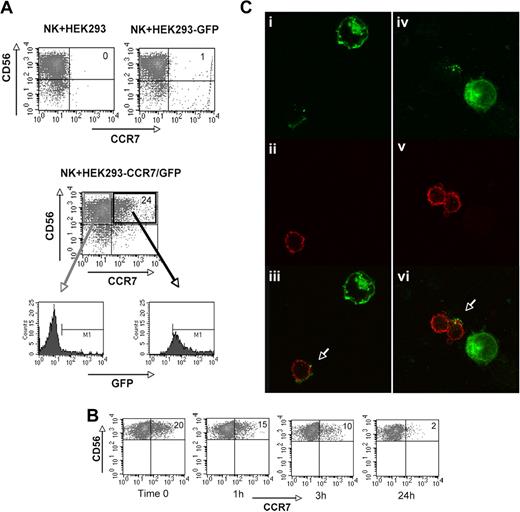
To obtain further evidence that CCR7 was indeed passively acquired from other cells by a phenomenon termed “trogocytosis,”43-47 we analyzed cell transfectants generated using the CCR7-negative human HEK293 cell line. These cell transfectants, expressing either GFP or CCR7/GFP, were cocultured with IL-2–activated NK cells. As shown in Figure 7A, when cocultured with untransfected or GFP-transfected HEK293 cells, NK cells did not express CCR7 (top panels). In contrast, they expressed CCR7 when cultured with CCR7/GFP+ HEK293 cells (Figure 7A middle panel). The finding that CCR7+ NK cells were also GFP+ (Figure 7A bottom panels) indicates that CCR7 was transferred to NK cells from CCR7/GFP+ HEK293 cells. Similar results were obtained also in cocultures of NK cells and CCR7/GFP+ Chinese hamster ovary (CHO) cell transfectants. Because CHO cells grow as adherent cells, NK cells could be easily isolated after coculture and analyzed for the persistence of CCR7 at different time intervals. As shown in Figure 7B, the surface expression of CCR7 was decreased after only 1 hour and was undetectable after 24 hours. Further support for the concept that CCR7 was taken up by NK cells was provided by experiments of confocal microscopy. As shown in Figure 7C, polyclonal IL-2–activated NK cells that had been cocultured with HEK293-CCR7/GFP+ cells for 15 to 30 minutes became GFP+, thus indicating that CCR7/GFP was transferred from HEK293 cell transfectants to NK cells by a mechanism of uptake.
Acquisition and loss of CCR7 by NK cells cocultured with CCR7/GFP cell transfectants. (A-C) In these experiments, polyclonal IL-2–activated NK cells that had been cultured for 1 hour either in the presence of HEK293 or HEK293-GFP (top panel) or in the presence of HEK293-CCR7/GFP (middle panel) were assessed for CCR7 expression by cytofluorimetric analysis. NK cells that had acquired the CCR7+ phenotype upon interaction with HEK293-CCR7/GFP transfectants were analyzed by immunofluorescence for the expression of GFP (bottom panels). Data are representative of 5 independent experiments performed using different donors. (B) Adherent CCR7/GFP+ CHO cell transfectants were used for induction of CCR7 on polyclonal IL-2–activated NK cells in a 1-hour coculture. Then NK cells were isolated, cultured in the absence of CCR7+ cells, and analyzed at different time intervals for the expression of surface CCR7. Data are representative of 3 independent experiments performed using different donors. (C) Confocal images of CCR7/GFP transfer from CCR7/GFP+ HEK293 cells to NK cells. Polyclonal IL-2–activated NK cells were mixed with CCR7/GFP+ HEK293 cells; after 15- to 30-minute incubation at 37°C, cells were collected, laid onto poly-lysine slides, fixed, and stained with anti-CD11a mAb. Samples were analyzed by dual-image confocal microscopy. (Ci-iv) Green fluorescence (GFP); (Cii,v) red fluorescence (CD11a); (Ciii,Cvi) merge.
Acquisition and loss of CCR7 by NK cells cocultured with CCR7/GFP cell transfectants. (A-C) In these experiments, polyclonal IL-2–activated NK cells that had been cultured for 1 hour either in the presence of HEK293 or HEK293-GFP (top panel) or in the presence of HEK293-CCR7/GFP (middle panel) were assessed for CCR7 expression by cytofluorimetric analysis. NK cells that had acquired the CCR7+ phenotype upon interaction with HEK293-CCR7/GFP transfectants were analyzed by immunofluorescence for the expression of GFP (bottom panels). Data are representative of 5 independent experiments performed using different donors. (B) Adherent CCR7/GFP+ CHO cell transfectants were used for induction of CCR7 on polyclonal IL-2–activated NK cells in a 1-hour coculture. Then NK cells were isolated, cultured in the absence of CCR7+ cells, and analyzed at different time intervals for the expression of surface CCR7. Data are representative of 3 independent experiments performed using different donors. (C) Confocal images of CCR7/GFP transfer from CCR7/GFP+ HEK293 cells to NK cells. Polyclonal IL-2–activated NK cells were mixed with CCR7/GFP+ HEK293 cells; after 15- to 30-minute incubation at 37°C, cells were collected, laid onto poly-lysine slides, fixed, and stained with anti-CD11a mAb. Samples were analyzed by dual-image confocal microscopy. (Ci-iv) Green fluorescence (GFP); (Cii,v) red fluorescence (CD11a); (Ciii,Cvi) merge.
Discussion
This study demonstrates that NK cells may take up CCR7 molecules upon interaction with CCR7+ cells and acquire the ability to migrate to secondary lymphoid organs. Because both mDCs and certain tumor- or virus-infected cells express CCR7, the encounter of NK lymphocytes with these cells in inflamed peripheral tissues may redirect NK cells to lymph nodes. The finding that the acquisition of CCR7 is regulated by the interaction between MHC class I and inhibitory NK receptors confers to these data important implications both in antitumor responses and in allogeneic HSCT.
Upon coculture with EBV-transformed cells or LPS-induced mDCs, both fresh and in vitro CD56dull expanded NK cells de novo expressed surface CCR7 receptors and acquired migratory properties in response to SLC chemokines. This phenomenon was induced upon NK-cell activation by several surface NK receptors displaying adhesion/activating properties including LFA-1, CD2, and in part NKp46. On the other hand, the interaction between KIR and HLA-class I molecules exerted a marked inhibitory effect. Accordingly, NK-cell interaction with CCR7+ target cells lacking surface HLA class I (such as the 221 cell line) resulted in high levels of CCR7 expression. Moreover, when KIR+ NK cells interacted with mDCs, CCR7 was acquired only in the presence of KIR/KIR-ligand mismatches. This condition occurs in haploidentical HSCT to cure high-risk leukemias.33 Although recipient DCs express high levels of HLA-class I molecules, they express HLA-class I alleles that are not recognized by KIRs expressed by a subset of donor NK cells. These KIR+ NK cells referred to as alloreactive do not express the CD94/NKG2A receptor, specific for HLA-E (present on all HLA-class I+ cells), and/or other KIR recognizing HLA-class I alleles expressed by the recipient.34 Alloreactive NK cells kill KIR-ligand–mismatched leukemic blasts and play a crucial role in eradicating high-risk acute myeloid as well as lymphoid leukemias in the T cell–depleted haploidentical HSCT. It is of note that in this transplantation setting, all cases are at high risk of T cell–mediated alloreactivity both in the host-versus-graft and in the graft-versus-host direction. Remarkably, however, patients who received a transplant from an NK-alloreactive donor benefit from higher rates of engraftment and reduced incidence of GvHD.34,35 It has been proposed that the low rate of GVHD is consequent to the inefficient priming of (the few) alloreactive donor T cells due to the NK cell–mediated killing of recipient APCs. In agreement with this concept, in vitro studies showed that alloreactive NK cell clones kill allogeneic mature MoDCs.29 Importantly, alloreactive NK cells predominantly attack the hematopoietic cells of the host (including DCs, T cells, and leukemic cells) while sparing other tissues that are common targets for T cell–mediated GVHD. Some of these target cells might be killed within peripheral tissues but conceivably the elimination of DCs should occur primarily within SLCs and should play a major role in preventing graft-versus-host reactions. Indeed, it is mainly at these sites that patient DCs would prime donor allospecific naive T cells. Thus, as suggested by our present study, the newly acquired migratory capability of alloreactive NK cells to lymph nodes may greatly enhance the likelihood of their interaction with DCs and T cells.
We show that the acquisition of CCR7 by NK cells from CCR7+ targets occurs rapidly and precedes cytolysis. This might be important to allow the uptake of CCR7 by bystander NK cells before elimination of target cells. The encounter and the close contact between KIR+ NK cells and DCs would take place within inflamed peripheral tissues.26 Notably in an autologous setting, under physiologic conditions, KIR+ NK cells are unable to kill HLA-class I+ mDCs.29 Along this line, our experiments on NK cell clones show that these NK cells do not kill mDCs and do not acquire CCR7 from them. Because the mechanisms of KIR/HLA recognition regulating the NK cell–mediated cytolysis and the uptake of CCR7 are largely overlapping, it is likely that, in the absence of alloreactivity, only CCR7+ target cells that have undergone tumor transformation or viral infection would deliver CCR7 to NK cells. Indeed, we show that KIR+ NK cells can efficiently acquire CCR7 from either HLA-negative or KIR-ligand–mismatched EBV-transformed cells. In the first case, most licensed NK cells (including both KIR+ and NKG2A+ ones) acquired migratory properties because none of the inhibitory receptors was engaged (Figure 5C). In the second case (221 cell line transfected with different HLA-C alleles) only NK cells lacking the relevant KIR and/or NKG2A acquired CCR7 (Figure 5B and Table 1).
The analysis of NK-cell clones revealed that, upon NK/DC interaction, the acquisition of CCR7 was confined to classical alloreactive NK cells. Thus, it is conceivable that, in vivo, alloreactive NK cells, upon migration into SLCs, kill recipient mDCs present in these compartments. Indeed, lymph node–resident NK cells (CD56bright, KIR−, NKG2A+) different from alloreactive NK cells (CD56dull, KIR+, NKG2A−) have a reduced perforin content and are poorly cytolytic.14 Notably however, an increased number of KIR+ NK cells have been described within lymph nodes undergoing paracortical/follicular hyperplasia. This finding was interpreted as a de novo expression of KIRs by lymph node–resident NK cells after inflammation.48 On the other hand, based on our present data as well as on the report by Mailliard et al,21 it is also conceivable that at least some KIR+ NK cells detected in lymph nodes may not have acquired KIRs at these sites.
Previous studies showed that the KIR− NKG2A+ CD56dull NK-cell subset is responsible for the process of DC editing based predominantly on killing of iDCs.29 However, this NK-cell subset in most instances fails to kill mDCs. In agreement with these data, we show that KIR− NKG2A+ cells cocultured with either autologous or allogeneic mDCs could acquire CCR7 only in the presence of anti-NKG2A mAbs (ie, upon disruption of the inhibitory interaction between NKG2A and HLA-E).
Regarding the mechanism of CCR7 transfer from CCR7+ cells to NK cells, the capture of cell membrane molecules from bystander cells was previously reported to occur both in vitro43-47 and in vivo.49 This intercellular transfer involved several cell surface molecules in different cell types including T and B lymphocytes and NK cells as well as APCs and tumor cells. This mechanism, also termed trogocytosis (from the ancient Greek trogo, meaning gnaw), may represent a vector for rapid intercellular communication.45 Importantly, surface molecules acquired from other cell types could modify not only phenotypic but also functional characteristics of the recipient cells. It has also been proposed that the capture of target cell membrane fragments by NK cells may reflect recognition of specific ligands by NK receptors and the formation of an immunologic NK synapse.44,46 In line with this concept, we show that both activating and inhibitory NK receptors seem to control this process. On the other hand, although the NK receptors that control the CCR7 uptake or the cytolytic activity may be largely overlapping, the transfer of target cell molecules to NK cells does not appear to require killing of target cells as suggested by studies in perforin-deficient mice50 and by the time required for acquisition of CCR7 and for expression of CD107a.
In conclusion, our findings suggest a novel mode by which alloreactive KIR+ NK cells can migrate to lymph nodes, kill recipient DCs, and thus prevent priming of alloreactive donor T cells and induction of GVHD.
Finally, it is also possible that alloreactive NK cells may play a relevant role in preventing host-versus-graft reactions by killing recipient T cells both in lymph nodes and in peripheral tissues.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants awarded by Associazione Italiana per la Ricerca sul Cancro (AIRC); Istituto Superiore di Sanità (ISS), agreement no. 40G.41; Fondazione CARIPLO, proceeding no. 20060034085; Ministero della Sanità, Ministero dell'Istruzione, dell'Università e della Ricerca Scientifica e Tecnologica (MIUR-PRIN 2006/project 2006061378_003; MIUR-FIRB 2003 project RBLA039LSF_001); Ministero della Salute: Ricerca Finalizzata 2005/agreement no. 57; and Ricerca Oncologica-Project of Integrated Program 2006-08, agreement no. RO strategici 3/07 and Progetto di Ricerca di Ateneo 2008.
Authorship
Contribution: E.M. participated in designing and performing the research and in writing the article; C.C. and C.P. did RT-PCR analysis, produced transient and stable cell transfectants, and analyzed confocal microscopy data; S.P. participated in performing the research; D.P. and S.A. participated in generating KIR+ NK-cell clones and dendritic cells, respectively; L.M. analyzed data and participated in writing the article; and A.M. participated in searching for funds, designing the research, and writing the article.
Conflict-of-interest disclosure: A.M. is founder and shareholder of Innate-Pharma. The remaining authors declare no competing financial interests.
Correspondence: Alessandro Moretta, Dipartimento di Medicina Sperimentale, Sezione di Istologia, Via GB Marsano 10, 16132 Genova, Italy; e-mail: alemoret@unige.it.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal