Abstract
MLL-AF4 acute lymphocytic leukemia (ALL) has a poor prognosis. MicroRNAs (miRNA) are small noncoding RNAs that posttranscriptionally regulate expression of target mRNAs. Our analysis of previously published data showed that expression of miR-128b and miR-221 is down-regulated in MLL-rearranged ALL relative to other types of ALL. Reexpression of these miRNAs cooperatively sensitizes 2 cultured lines of MLL-AF4 ALL cells to glucocorticoids. Target genes down-regulated by miR-128b include MLL, AF4, and both MLL-AF4 and AF4-MLL fusion genes; miR-221 down-regulates CDKN1B. These results demonstrate that down-regulation of miR-128b and miR-221 is implicated in glucocorticoid resistance and that restoration of their levels is a potentially promising therapeutic in MLL-AF4 ALL.
Introduction
MicroRNAs (miRNAs) are a recently discovered class of small noncoding RNAs that are 18 to 24 nucleotides long and that down-regulate target genes at a posttranscriptional level. The majority of miRNA genes are transcribed by RNA polymerase II into long primary (pri) miRNA transcripts, processed by the nuclear nuclease Drosha into approximately 60-bp hairpins termed precursor (pre) miRNAs, and further cleaved in the cytosol by the Dicer nuclease into mature miRNAs. Mature miRNAs are then incorporated into the multiprotein RNA-induced silencing complex, exerting posttranscriptional repression of target mRNAs,1 either by inducing mRNA degradation or blocking mRNA translation.2-4
Each miRNA is thought to have several target mRNAs, and computational predictions suggest that more than a third of all human genes are targets of miRNAs.5,6 In animals, miRNAs control many developmental and physiologic processes. For example, abnormal expression of certain miRNAs leads to a developmental arrest in Caenorhabditis elegans.7 In the hematopoietic system, miR-181a down-regulates several phosphatases that regulate the sensitivity of T cells to antigens, and overexpression of miR-181 in hematopoietic stem/progenitor cells significantly increases B-cell production. Overexpression of miR-150 leads to a block in B-cell formation at the proB to preB cell transition by down-regulating c-myb, among other targets.8-11
Many studies show specific alterations in miRNA expression profiles that correlate with particular human tumor phenotypes.12,13 Down-regulation of specific miRNAs in certain cancers implies that some miRNAs may act as tumor suppressors. For example, reduced expression of let-7 family members, which directly down-regulate the expression of Ras and other proto-oncogenes, occurs in lung cancer. miR-15 and -16, negative regulators of bcl-2, exhibit reduced expression in B-cell chronic lymphocytic leukemia.13-15 On the other hand, increased expression of miR-17-92 and miR-155 often occurs in B-cell lymphomas,16,17 implying that these miRNAs are oncogenes.17,18
The MLL gene is located at 11q23, a site frequently involved in chromosomal translocations in aggressive human lymphoid and myeloid leukemias. As a result of these chromosomal translocations, a portion of MLL becomes fused with one of more than 40 different partner proteins, yielding a diverse collection of chimeric fusion proteins. MLL rearrangements are a common genetic alteration found in human leukemia. Of these MLL-associated leukemias, MLL-AF4 acute lymphocytic leukemia (ALL), resulting from a balanced translocation between MLL and AF4, occurs in approximately 50% of ALL cases in infants, 2% in children, and 5% to 6% in adults.19 MLL-AF4 ALL is associated with steroid resistance and has a poor prognosis.19,20 Overexpression of MLL-AF4 reduces cell growth and induces resistance to etoposide-mediated cytotoxicity.21,22 The reciprocal fusion protein, AF4-MLL, may also have oncogenic activity and may be an important hit for leukemogenesis.23,24 The detailed roles of these fusion proteins in leukemogenesis are not well understood.
A recent survey showed that miRNA expression patterns differ among ALL subtypes.12 We began by reanalyzing the published12 raw data (available on the web at http://www.broad.mit.edu/mpr/publications/projects/microRNA/ALL.gct) and discovered that many miRNAs were down-regulated in the ALL samples with MLL rearrangements compared with ALL samples that do not harbor MLL rearrangements (supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Importantly, some miRNAs that have been reported to be tumor suppressors are down-regulated to a considerable degree, raising the question of whether these miRNAs are involved in the biology of MLL-rearranged ALL, especially in its poor prognosis.
Here we focus on miR-128b and miR-221, 2 miRNAs commonly down-regulated in MLL-rearranged ALL compared with other types of ALL (supplemental Table 1)12 because, as we detail in “Discussion,” we hypothesized that these down-regulate several proteins that are thought to contribute to leukemia development. We show that reexpression of miR-128b sensitizes 2 MLL-AF4 ALL cell lines to killing by both low and high concentrations of glucocorticoids, as well as by etoposide and serum depletion. Expression of miR-221 sensitizes cells to killing by high concentrations of glucocorticoids and cooperates with miR-128b in this regard. Further, we show that miR-128b down-regulates MLL, AF4, as well as both MLL-AF4 and AF4-MLL fusion genes, and that miR-221 down-regulates CDKN1B, a downstream transcriptional target of MLL and MLL-AF4.21,25
Methods
Cell culture and retroviral transfer
RS4;11 and SEM cell lines carrying the chromosomal translocation t(4;11)(q21;q23) were obtained from ATCC and DSMZ, respectively. The cells were maintained in RPMI 1640 containing 10% fetal calf serum. High-titer retroviral supernatant was created after the cotransfection of a miRNA expression vector and the pCLamph viral packaging construct into 293T cells using FuGENE 6 (Roche Diagnostic). RS4;11 cells were plated at 4 to 5 × 105/mL in a 24-well plate and spin-infected with the desired retroviral supernatant for 2 hours.26
Constructs
Genomic DNA from RS4;11 was extracted using DNeasy Tissue extraction kit (QIAGEN). The genomic sequences of the segments in pri-miR-128b, pri-miR-221, and pri-let7b were amplified by genomic polymerase chain reaction (PCR) using Pfx polymerase (Invitrogen). The oligonucleotide sequences used for the PCR are shown in supplemental Table 2. The reaction parameters were 35 cycles of 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 1 minute and 30 seconds. PCR products were purified using a PCR purification kit (QIAGEN) and sequenced. The purified genomic PCR product was cloned into the Xho1-EcoR1 site of a mammalian retroviral expression vector, pMDH.9 miR-221mut and miR-128bmut were generated using site-directed mutagenesis (QuikChange-XL, Stratagene) and confirmed by full-length DNA sequencing. The oligonucleotide sequences used for mutagenesis are shown in supplemental Table 2.
Segments in 3′untranslated region (UTR) of the human MLL, AF4, and CDKN1B were amplified by genomic PCR and cloned in between the Xho1-Not1 sites of psicheck-2 (Promega). MLL-UTR-128b contains 3 potential miR-128b binding sites, and AF4-UTR-128b and CDKN1B-UTR-128b contains 2 miR-128b binding sites. These sites are located at nucleotides 2324 to 2330, 2489 to 2495, and 2691 to 2697 of the 3′UTR of MLL mRNA (NM_005933), at nucleotides 174 to 180 and 545 to 551 of the 3′UTR of AF4 mRNA(NM_005935), and at nucleotides 201 to 208 and 274 to 281 of the 3′UTR of CDKN1B mRNA(NM_004064). The controls for MLL and AF4 contain a segment of 3′ UTR of MLL and AF4, which do not include the predicted miR-128b binding sites. The oligonucleotide sequences used for the PCR are shown in supplemental Table 2. The PCR condition was 35 cycles of 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 1 minute and 30 seconds.
Quantitative PCR
Total RNA was isolated with Trizol (Invitrogen) and used to synthesize cDNA with the superscript II cDNA synthesis kit (Invitrogen) and oligo dT primer (Invitrogen). The expression levels of pri-miR-128b, MLL,AF4, CDKN1B, and HOXA9 were measured by quantitative PCR using SYBR green master mix (ABI). The oligonucleotide sequences used for the PCR are shown in supplemental Table 2. The expressions of mature miR-128b and miR-221 were measured using a TaqMan MicroRNA Assay (ABI). The PCR condition was as follows: 40 cycles of 95°C for 15 seconds, and 60°C for 1 minute. All reactions were performed in triplicate.
Viability and cell-cycle assays
Fluorescence-activated cell sorter (FACS) analysis of apoptotic cells were performed as described.27 Cells were plated at 106/mL in a 24-well plate with or without dexamethasone (DEX; Sigma-Aldrich) for 40 hours. The cell population was then analyzed by FACS using allophycocyanin-conjugated annexin V staining (BD Biosciences) or propidium iodide. The percentage of viable cells was determined by dividing the number of annexin V− green fluorescent protein+ (GFP+) cells in the treated sample by the number of total cells at each time point or concentration (supplemental Figure 1). Error bars represent the mean plus or minus SD of the triplicates. Cell-cycle analyses were done as described by Xia et al.21
Dual luciferase assay
In each well of a 96-well plate, 293T cells were cotransfected with 20 ng psiCHECK-2s (Promega) and 100 ng miRNA expression vectors. After 48 hours of transfection, the relative amounts of Renilla and firefly luciferase were analyzed by a dual luciferase assay (Promega) using Safire2R (TECAN). The Renilla/firefly luciferase ratio was calculated and normalized against the control.
Western blot analysis
RS4;11 cells were lysed in sample loading buffer and directly separated through sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Immunoblotting was performed after transferring the sodium dodecyl sulfate–polyacrylamide gel electrophoresis to a nitrocellulose membrane. The following antibodies were used: anti-N-MLL, anti-C-MLL,28 anti-p27 (Santa Cruz Biotechnology), and peroxidase-conjugated antirabbit immunoglobulin (GE Healthcare).
Statistical analysis
We used the Wilcoxon rank-sum test and the modified t test to determine the significance of the differences in miRNA expression in MLL-rearranged relative to non–MLL-rearranged ALL, and used the paired Student t tests to analyze the effect of the miRNA overexpression by comparing the miRNA-induced change with that of the respective controls. The analysis of variance or Student t test was used in apoptosis assays, dual luciferase assays, and quantitative PCR, and values of P less than .05 were considered statistically significant. All analyses were performed with Microsoft Excel.
Results
Expression of miR-128b and miR-221 is reduced in primary MLL-rearranged cells relative to non–MLL-rearranged ALL cells
The resistance of MLL-AF4 ALL cells to glucocorticoid-induced apoptosis is associated with its notoriously poor prognosis.19,20 By analysis of the raw data published on the web,12 we found that, in MLL-rearranged ALL, many miRNAs are down-regulated compared with other types of ALL (supplemental Table 1; http://www.broad.mit.edu/mpr/publications/projects/microRNA/ALL.gct). Our initial attention focused on 2 down-regulated miRNAs, miR-128b and miR-221, because, as we detail below, these are hypothesized to down-regulate mRNAs encoding CDKN1B, MLL, AF4, and both MLL-AF4 and AF4-MLL fusion genes that are thought to contribute to leukemia development. The data for expression of these miRNAs in the 6 MLL-rearranged primary samples and the 67 non–MLL-rearranged ALL samples are depicted in supplemental Figure 2 and summarized in supplemental Table 1. As judged by the mean value, the levels of expression of miR-128b and miR-221 in MLL-rearranged relative to non–MLL-rearranged ALL (others) are 0.36 and 0.46, respectively; as judged by the median values, the expression levels of miR-128b and miR-221 in MLL-rearranged relative to non–MLL-rearranged ALL are 0.11 and 0.13. Using the modified t test, one appropriate statistical analysis tool, the P values for the significance of the differences in expression of miR-128b and miR-221 in MLL-rearranged relative to non–MLL-rearranged ALL are significant (P = .033 and P = .016, respectively). Using another appropriate statistical test, the nonparametric Wilcoxon rank-sum test, for miR-128b the P value is more than .023, which is clearly significant. For miR-221, the P value is more than .08, just above the usual cutoff for significance. Thus, there is no doubt that the reduced level of expression of miR-128b in primary samples of MLL-rearranged relative to non–MLL-rearranged ALL is indeed significant. The reduced level of expression of miR-221 in primary samples of MLL-rearranged relative to non–MLL-rearranged ALL is also significant as judged by one of the 2 appropriate statistical tests.
Expression of exogenous miR-128b and miR-221 restores glucocorticoid sensitivity to RS4;11 cells
In preliminary experiments, we examined whether the low levels of 6 of these down-regulated miRNAs (let7b, let7c, miR-221, miR-126, miR-195, and miR-128b) in MLL-AF4 ALL cells are responsible for the inability of glucocorticoids to inhibit cell division and induce apoptosis. Using retrovirus infection of the RS4;11 cell line, derived from an MLL-AF4 ALL patient, we overexpressed each of these miRNAs in turn; only ectopic expression of miR-221 and miR-128b sensitized cells to glucocorticoids (data not shown).
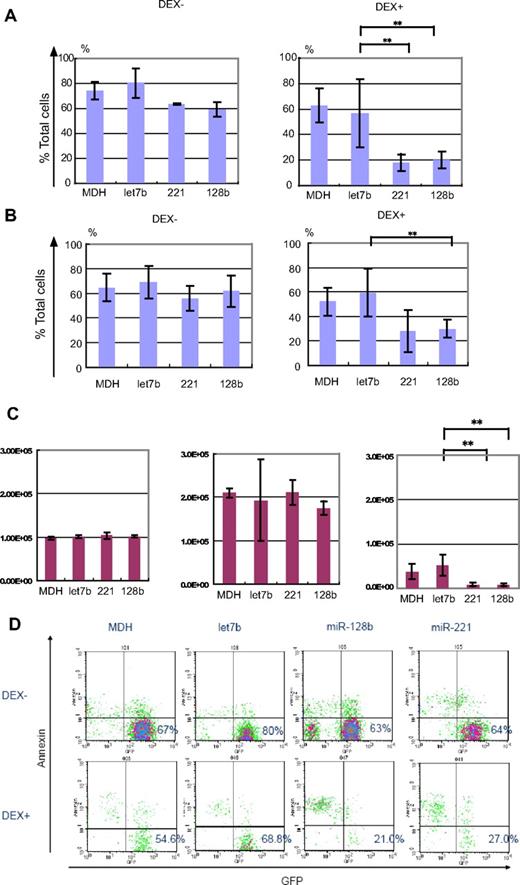
Next, RS4;11 cells were transduced with a retrovirus expression vector, MDH, which expresses let7b (a control miRNA), miR-221, miR-128b, or no miRNA (control). The transduced cells were sorted for those expressing GFP (a marker gene in the MDH vector) and treated with 10 μM DEX for 40 hours. Annexin V staining was used to measure apoptosis,27 and viable cells were defined as annexin V− (supplemental Figure 1). Figure 1A shows that approximately 70% of cells expressing let7b, miR-128b, or miR-221 were viable in the absence of DEX treatment, similar to sorted control MDH cells. After 40 hours of 10 μM DEX treatment, approximately 40% of control cells or cells expressing let7b were viable. A striking reduction of the percentage of viable cells to 20% was observed in RS4;11 cells expressing miR-128b or miR-221 (Figure 1A), indicating that enhanced apoptosis was induced by miR-221 or miR-128b expression. A FACS profile of one representative experiment is shown in Figure 1C, again demonstrating enhanced killing of cells expressing miR-128b or miR-221. A similar effect of miRNA overexpression was obtained by monitoring cell number. After 40 hours of culture without DEX treatment (Figure 1B left and middle panels), the numbers of viable cells had increased approximately 1.5-fold and to the same level regardless of whether the cells were expressing let7b, miR-128b, or miR-221. This establishes that neither miRNA significantly affects cell proliferation in the absence of DEX. In contrast, after 40 hours of 10 μM DEX treatment (Figure 1B right panel), the number of viable miR-128b– and miR-221–transduced RS4;11 cells was less than half of that in cells expressing let7b or control cells. These results demonstrate that neither let-7b, nor miR-128b, nor miR-221 overexpression affects cell growth in the absence of steroid treatment, but that miR-128b and miR-221 overexpression enhances DEX-induced apoptosis and cell death.
Ectopic expression of miR-128b and miR-221 restores glucocorticoid sensitivity to RS4;11 cells. (A) Sorted GFP+ control– (termed MDH), let7b–, miR-221–, or miR-128b–transduced cells are treated (DEX+) or not (DEX−) with 10 μM DEX for 40 hours, followed by FACS analysis. The percentage of viable cells is determined by dividing the number of annexin V− GFP+ cells by the number of total cells analyzed (n = 3) **P < .05. (B) The absolute cell number of annexin V− GFP+ cells before treatment (left panel), without (middle panel), or with 10 μM DEX treatment (right panel) for 40 hours (n = 3). **P < .05. (C) Representative FACS profiles of the sorted control–, let7b–, miR-128b–, or miR-221–transduced RS4;11 cells with (DEX+) or without (DEX−) treatment with 10 μM DEX. The GFP− cells in all panels represent dead cells in which GFP has been released; the majority are positive for annexin V staining (supplemental Figure 1). The percentage of viable cells is determined by dividing the number of annexin V− GFP+ cells by the number of total cells analyzed. (D) Sorted GFP+ miR-128b–, miR-221–, or control–transduced (MDH and let7b) cells are treated with different concentrations of DEX for 40 hours, followed by FACS analysis. GFP+ cells are assessed for viability by annexin V staining in the same manner as in panel A. The vertical axis shows the percentage of the number of GFP+ annexin V− cells after DEX treatment relative to the number of GFP+annexin V− cells without DEX treatment. The 50% inhibitory concentration values for DEX killing of cells expressing let7b, MDH, miR-221, and miR-128b are 367, 31.9, 0.6, and 0.00033 μM, respectively. *P < .05, let7b vs miR-128b. **P < .05, let7b vs miR-221 and miR-128b.
Ectopic expression of miR-128b and miR-221 restores glucocorticoid sensitivity to RS4;11 cells. (A) Sorted GFP+ control– (termed MDH), let7b–, miR-221–, or miR-128b–transduced cells are treated (DEX+) or not (DEX−) with 10 μM DEX for 40 hours, followed by FACS analysis. The percentage of viable cells is determined by dividing the number of annexin V− GFP+ cells by the number of total cells analyzed (n = 3) **P < .05. (B) The absolute cell number of annexin V− GFP+ cells before treatment (left panel), without (middle panel), or with 10 μM DEX treatment (right panel) for 40 hours (n = 3). **P < .05. (C) Representative FACS profiles of the sorted control–, let7b–, miR-128b–, or miR-221–transduced RS4;11 cells with (DEX+) or without (DEX−) treatment with 10 μM DEX. The GFP− cells in all panels represent dead cells in which GFP has been released; the majority are positive for annexin V staining (supplemental Figure 1). The percentage of viable cells is determined by dividing the number of annexin V− GFP+ cells by the number of total cells analyzed. (D) Sorted GFP+ miR-128b–, miR-221–, or control–transduced (MDH and let7b) cells are treated with different concentrations of DEX for 40 hours, followed by FACS analysis. GFP+ cells are assessed for viability by annexin V staining in the same manner as in panel A. The vertical axis shows the percentage of the number of GFP+ annexin V− cells after DEX treatment relative to the number of GFP+annexin V− cells without DEX treatment. The 50% inhibitory concentration values for DEX killing of cells expressing let7b, MDH, miR-221, and miR-128b are 367, 31.9, 0.6, and 0.00033 μM, respectively. *P < .05, let7b vs miR-128b. **P < .05, let7b vs miR-221 and miR-128b.
In a separate experiment, the effect of miR-128b and miR-221 expression on glucocorticoid resistance was analyzed at several DEX concentrations (Figure 1D). miR-128b–overexpressing cells exhibited enhanced killing at all DEX concentrations tested, as measured by the ratio of GFP+ annexin V− cells after DEX treatment to GFP+ annexin V− cells without DEX treatment (Figure 1D). In contrast, cells expressing miR-221 exhibited enhanced killing by 10 μM DEX but not at 0.001 or 0.1 μM (Figure 1D). Therefore, miR-128b dramatically restored sensitivity to DEX-induced apoptosis even at a low concentration of DEX, whereas miR-221 did so only at a high concentration of DEX.
Next, we analyzed the effect of miR-128b and miR-221 expression on DEX-induced apoptosis in SEM cells, another cell line derived from an MLL-AF4 ALL patient. This cell line exhibited greater resistance to DEX killing than did RS4;11. However, ectopic expression of either miR-221 or miR-128b caused increased apoptosis after treatment with 100 μM DEX, compared with control or let7b-transduced cells. Figure 2A shows that approximately 70% of cells expressing let7b, miR-128b, or miR-221 were viable in the absence of DEX treatment, similar to control MDH cells. After 40 hours of 100 μM DEX treatment, approximately 60% of control cells or cells expressing let7b were viable. A striking reduction of the percentage of viable cells to 20% was observed in SEM cells expressing miR-128b or miR-221 (Figure 2A), indicating that enhanced apoptosis was induced by miR-221 or miR-128b expression. A FACS profile of one representative experiment is shown in Figure 2D, again demonstrating enhanced killing of cells expressing miR-128b or miR-221. An independent means of detecting apoptotic cells used staining by propidium iodide. As Figure 2B shows, the fraction of viable cells was the same as those determined by annexin V staining (Figure 2B).
Ectopic expression of miR-128b and miR-221 restores glucocorticoid sensitivity to SEM cells. (A) Sorted GFP+ control– (termed MDH), let7b–, miR-221–, or miR-128b–transduced cells are treated (DEX+) or not (DEX−) with 100 μM DEX for 40 hours, followed by FACS analysis. As in Figure 1, the percentage of viable cells is determined by dividing the number of annexin V− GFP+ cells by the number of total cells analyzed (n = 4). **P < .05. (B) The cells were stained with propidium iodide (PI), and the percentage of viable cells is determined by dividing the number of PI− GFP+ cells by the number of total cells analyzed (n = 4). **P < .05. (C) The absolute cell number of annexin V− GFP+ cells before treatment (left panel), without (middle panel), or with 100 μM DEX treatment (right panel) for 40 hours. (n = 4). **P < .05. (D) Representative FACS profiles of the sorted control–, let7b–, miR-128b–, or miR-221–transduced RS4;11 cells with (DEX+) or without (DEX−) treatment with 100 μM DEX. The GFP− cells in all panels represent dead cells in which GFP has been released; the majority are positive for annexin V staining. The percentage of viable cells is determined by dividing the number of annexin V− GFP+ cells by the number of total cells analyzed.
Ectopic expression of miR-128b and miR-221 restores glucocorticoid sensitivity to SEM cells. (A) Sorted GFP+ control– (termed MDH), let7b–, miR-221–, or miR-128b–transduced cells are treated (DEX+) or not (DEX−) with 100 μM DEX for 40 hours, followed by FACS analysis. As in Figure 1, the percentage of viable cells is determined by dividing the number of annexin V− GFP+ cells by the number of total cells analyzed (n = 4). **P < .05. (B) The cells were stained with propidium iodide (PI), and the percentage of viable cells is determined by dividing the number of PI− GFP+ cells by the number of total cells analyzed (n = 4). **P < .05. (C) The absolute cell number of annexin V− GFP+ cells before treatment (left panel), without (middle panel), or with 100 μM DEX treatment (right panel) for 40 hours. (n = 4). **P < .05. (D) Representative FACS profiles of the sorted control–, let7b–, miR-128b–, or miR-221–transduced RS4;11 cells with (DEX+) or without (DEX−) treatment with 100 μM DEX. The GFP− cells in all panels represent dead cells in which GFP has been released; the majority are positive for annexin V staining. The percentage of viable cells is determined by dividing the number of annexin V− GFP+ cells by the number of total cells analyzed.
A similar effect of miR-221 or miR-128b overexpression was obtained by monitoring cell number. After 40 hours of culture without DEX treatment (Figure 2C left and middle panels), the numbers of viable cells had increased approximately 2.0-fold and to the same level regardless of whether the cells were expressing let7b, miR-128b, or miR-221. In contrast, after 40 hours of 100 μM DEX treatment (Figure 2C right panel), the number of viable miR-128b– and miR-221–transduced SEM cells was less than half of that of cells expressing let7b or control cells. These results demonstrate that neither let-7, nor miR-128b, nor miR-221 overexpression affects cell growth in the absence of steroid treatment, but that miR-128b and miR-221 overexpression enhances DEX-induced apoptosis and cell death.
Thus, our results using 2 different MLL-AF4 ALL cell lines (RS4;11 and SEM) established that reexpression of miR-221 and miR-128b restored steroid sensitivity and thus that the steroid resistance of these cells, and presumably that of MLL-AF4 ALL patients, might be the result of low levels of expression of these miRNAs.
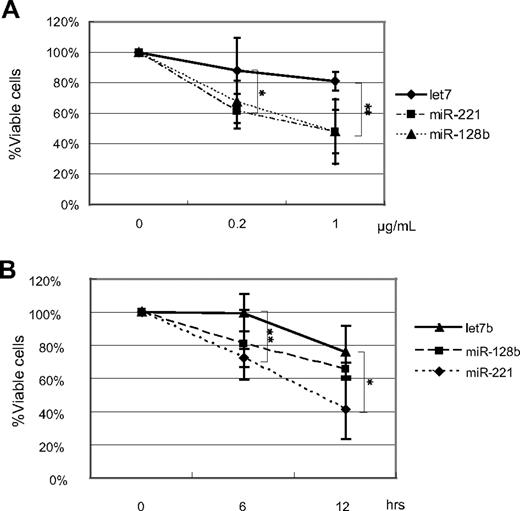
Furthermore, ectopic expression of miR-128b and miR-221 in RS4;11 enhanced sensitivity to killing by another anticancer drug, etoposide, as well as by serum deprivation (Figure 3); RS4;11 cells were reported to be resistant to killing by serum depletion.29 miR-221–overexpressing RS4;11 cells exhibited significantly enhanced killing at 0.2 μg etoposide/mL; miR-128b–overexpressing RS4;11 cells also exhibited significantly enhanced killing at 1 μg etoposide/mL. (Figure 3A)
The percentage of viable cells in miR-128b–, miR-221–, or control–transduced RS4;11 cells treated with different concentrations of etoposide and by serum depletion. (A) Sorted GFP+ miR-128b–, miR-221–, or let7b–transduced cells are treated with different concentrations of etoposide for 24 hours, followed by FACS analysis. GFP+ cells were assessed for viability by annexin V staining in the same manner as in Figure 1A. The vertical axis represents the percentage of the number of GFP+ annexin V− cells after etoposide treatment relative to the number of GFP+annexin V− cells without etoposide treatment. (B) Sorted GFP+ miR-128b–, miR-221–, or let7b–transduced cells are treated with serum depletion for 6 and 12 hours, followed by FACS analysis. GFP+ cells are assessed for viability by annexin V staining in the same manner as in Figure 1A. The vertical axis shows the percentage of the number of GFP+ annexin V− cells after serum depletion relative to the number of GFP+annexin V− cells at 0 hour. The absolute percentage of the viable cells in control-transduced cells is 60% to 70% after sorting. Three independent experiments were done. *P < .05, let7b vs miR-221. **P < .05, let7b vs miR-128b.
The percentage of viable cells in miR-128b–, miR-221–, or control–transduced RS4;11 cells treated with different concentrations of etoposide and by serum depletion. (A) Sorted GFP+ miR-128b–, miR-221–, or let7b–transduced cells are treated with different concentrations of etoposide for 24 hours, followed by FACS analysis. GFP+ cells were assessed for viability by annexin V staining in the same manner as in Figure 1A. The vertical axis represents the percentage of the number of GFP+ annexin V− cells after etoposide treatment relative to the number of GFP+annexin V− cells without etoposide treatment. (B) Sorted GFP+ miR-128b–, miR-221–, or let7b–transduced cells are treated with serum depletion for 6 and 12 hours, followed by FACS analysis. GFP+ cells are assessed for viability by annexin V staining in the same manner as in Figure 1A. The vertical axis shows the percentage of the number of GFP+ annexin V− cells after serum depletion relative to the number of GFP+annexin V− cells at 0 hour. The absolute percentage of the viable cells in control-transduced cells is 60% to 70% after sorting. Three independent experiments were done. *P < .05, let7b vs miR-221. **P < .05, let7b vs miR-128b.
Serum contains critical growth factors for many cells, and serum deprivation induces death of many types of cells. Let7b-transduced RS4;11 cells (ie, GFP+ cells) are relatively resistant to serum deprivation; at 12 hours in the absence of serum, the relative percentage of viable transduced cells (as measured by the ratio of the number of GFP+ annexin V− cells after DEX treatment relative to the number of GFP+ annexin V− cells without DEX treatment) remained between 80% and 100%. In contrast, cells expressing miR-128b displayed a significant reduction in viability at 6 hours in the absence of serum, and cells expressing miR-221 displayed a significant reduction in viability at 12 hours of serum starvation (Figure 3B).
Thus, our results showed that reexpression of miR-221 and miR-128b partly restored sensitivity to killing by etoposide and serum deprivation, although the enhancement of cell killing was less than that seen for killing by glucocorticoids.
miR-128b and miR-221 cooperatively restore glucocorticoid sensitivity
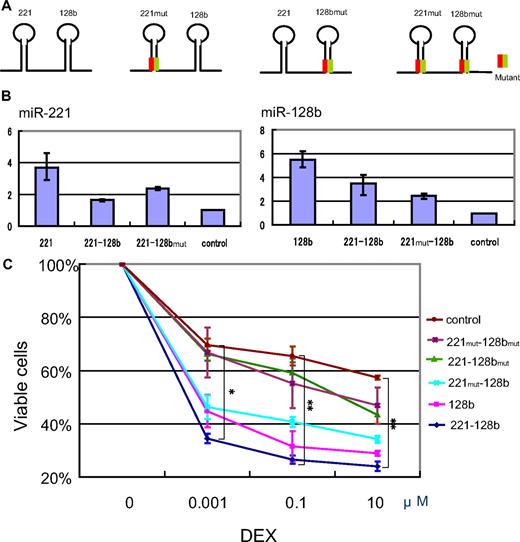
Figure 4 shows that the expression of miR-128b together with that of miR-221 results in an increase in steroid-induced apoptosis over that of either miRNA alone. To this end, we prepared 4 kinds of vectors coexpressing wild-type and mutant versions of miR-128b and miR-221. The mutant miR-128bmut was generated by altering the second to fourth nucleotides from the 5′end of mature miR-128b and miR-221mut by altering the third to fifth nucleotides from the 5′end of mature miR-221 (Figure 4A green). Compensatory mutations (orange) were introduced to the complementary loop strands to preserve the secondary structure of pre-miR-128b and pre-miR-221 and thus allow normal processing. The 4 vectors expressed miR-221-miR-128b, miR-221-miR-128bmut, miR-221mut -miR-128b, or miR-221mut–miR-128bmut (Figure 4A). Expression of mature miR-128b and miR-221 was analyzed by use of quantitative PCR to confirm that the processing efficiency of each miRNA was comparable among these constructs. Curiously, and for unknown reasons, the constructs containing double stem loops generated lower levels of mature miRNAs than those containing a single stem loop. Whereas the level of mature miR-128b generated by the single miR-128b construct was approximately 5 times that of control cells, the level of mature miR-128b generated by the miR-221–miR-128b and the miR-221mut–miR-128b constructs was only 3 times and twice that of the control, respectively. Similarly, the level of mature miR-221 generated by the single miR-221 construct was 4 times that of the control, whereas the level generated by the miR-221–miR-128b and miR-221–miR-128bmut constructs was 1.5 and twice as much as the control (Figure 4B).
Cooperative effect of miR-128b and miR-221 on restoration of glucocorticoid sensitivity. (A) Schematic of the constructs coexpressing wild-type and mutant versions of miR-128b and miR-221. Green and orange lines represent miR-221mut or miR-128mut and its complement, respectively. (B) Expression levels of mature miR-221 (left panel) and miR-128b (right panel) in sorted GFP+ cells that are overexpressing a construct expressing just miR-221 (221) or miR-128b (128b), or the double expression constructs miR-221–miR-128b (221-128b), miR-221–miR-128bmut (221-128bmut), or miR-221mut–miR-128b (221mut-128b). Control cells express the vacant vector. (C) Sorted GFP+ cells expressing just miR-128b or the double expression constructs miR-221–miR-128b, miR-221–miR-128bmut, miR-221mut–miR-128b, or miR-221mut–miR-128bmut, or control (vacant vector) were treated with different concentrations of DEX for 40 hours, followed by FACS analysis as in Figure 1. The vertical axis is plotted as in Figure 1D. Three independent experiments were done. **P < .05, miR-221-miR-128b versus all other constructs, and miR-221mut–miR-128b versus miR-221mut-miR–128bmut. *P < .05, miR-221–miR-128b versus all other constructs except for miR-128b, and miR-221mut–miR-128b versus miR-221mut–miR-128bmut.
Cooperative effect of miR-128b and miR-221 on restoration of glucocorticoid sensitivity. (A) Schematic of the constructs coexpressing wild-type and mutant versions of miR-128b and miR-221. Green and orange lines represent miR-221mut or miR-128mut and its complement, respectively. (B) Expression levels of mature miR-221 (left panel) and miR-128b (right panel) in sorted GFP+ cells that are overexpressing a construct expressing just miR-221 (221) or miR-128b (128b), or the double expression constructs miR-221–miR-128b (221-128b), miR-221–miR-128bmut (221-128bmut), or miR-221mut–miR-128b (221mut-128b). Control cells express the vacant vector. (C) Sorted GFP+ cells expressing just miR-128b or the double expression constructs miR-221–miR-128b, miR-221–miR-128bmut, miR-221mut–miR-128b, or miR-221mut–miR-128bmut, or control (vacant vector) were treated with different concentrations of DEX for 40 hours, followed by FACS analysis as in Figure 1. The vertical axis is plotted as in Figure 1D. Three independent experiments were done. **P < .05, miR-221-miR-128b versus all other constructs, and miR-221mut–miR-128b versus miR-221mut-miR–128bmut. *P < .05, miR-221–miR-128b versus all other constructs except for miR-128b, and miR-221mut–miR-128b versus miR-221mut–miR-128bmut.
As shown in Figure 4C, cells expressing miR-221–miR-128b exhibited a significant increase in sensitivity to apoptosis at all DEX concentrations tested, compared with miR-221mut–miR-128bmut–overexpressing cells or control cells (P < .05; Figure 4C). In contrast, cells expressing miR-221–miR-128bmut showed no increase in DEX sensitivity; we attribute this difference in glucocorticoid sensitivity with that seen in Figure 1 in cells expressing miR-221 alone, to the reduced level of miR-221 expression. Correspondingly, cells expressing miR-221mut–miR-128b, which contain twice as much as mature miR-128b as control cells, showed an increase in DEX-induced apoptosis, but the extent of DEX-induced apoptosis was significantly lower (P < .05; Figure 4C) than in cells expressing miR-128 alone or the double expression construct miR-221–miR-128b. Again, the degree of sensitivity of cells to DEX treatment correlated with the level of expression of mature miR-128b (P < .05 at 0.1 and 10 μM DEX; Figure 4C). A crucial comparison is between cells expressing miR-128b alone and cells expressing miR-221–miR-128b. The latter contain less mature miR-128b than the former yet show increased sensitivity to DEX at all 3 concentrations tested. This result, which is statistically significant and was repeated in 3 experiments, means that miR-221, which is also made in enhanced amounts in cells expressing miR-221–miR-128b, must synergize with miR-128b in inducing glucocorticoid sensitivity. We conclude that miR-128b is a potent sensitizer for steroid resistance in these MLL-AF4 cells; a small 2- to 3-fold increase in the level of mature miR-128b was sufficient to sensitize cells to glucocorticoids. Whereas a small increase of miR-221 expression alone had no effect on steroid sensitivity, miR-221 enhanced the ability of miR-128b to support steroid-induced apoptosis of MLL-AF4 ALL cells.
MLL and AF4 mRNAs possess several putative miR-128b binding sites that are responsible for down-regulation by miR-128b
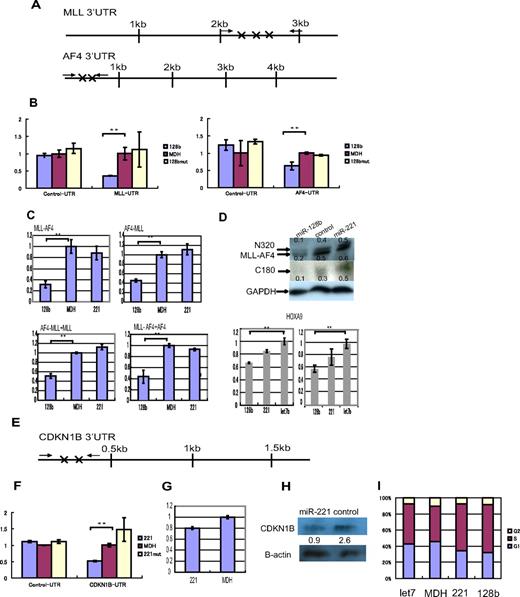
Three highly conserved 7-nucleotide sequences in the 3′UTR of human MLL mRNA and 2 nonconserved 7 nucleotide sequences in the AF4 3′UTR are complementary to the miR-128b “seed” region (Figure 5A). To determine whether these sites allow down-regulation by miR-128b, and thus whether miR-128 can down-regulate MLL and AF4, as well as the chimeric MLL-AF4 and AF4-MLL mRNAs, the 3′UTR regions containing these miR-128b binding sites were cloned downstream of the reporter Renilla luciferase open reading frame. Overexpression of miR-128b significantly reduced reporter luciferase expression of vectors containing the segments of the MLL-UTR and the AF4-UTR that contain the putative miR-128b binding sites, but not the expression of control vectors containing segments of the 3′UTRs of MLL and AF4 that are not predicted to interact with miR-128b (Figure 5B). Quantitative PCR showed that the expression of both endogenous MLL-AF4 and AF4-MLL mRNAs was markedly reduced (between 2- and 3-fold) in RS4;11 stably overexpressing miR-128b, compared with cells transduced with miR-221 or control cells (Figure 5C). Additional quantitative PCR, designed to detect both chimeric genes (AF4-MLL and MLL-AF4) and nonchimeric genes (MLL and AF4), showed that the expressions of these 4 genes were markedly reduced in RS4;11 cells overexpressing miR-128b as well (Figure 5C). MLL is often cleaved by Taspase at 2 conserved sites, generating MLL N-terminal (N320) and C-terminal (C180) fragments.28 Western blotting showed a clear reduction in the level of the endogenous N320 protein, as well as of MLL-AF4, and C180, in miR-128b–overexpressing cells (Figure 5D). Moreover, quantitative PCR showed that the expression of HOXA9 mRNAs, a transcriptional target of MLL-AF4, was reduced in both SEM and RS4;11 cells stably overexpressing miR-128b, compared with cells transduced with let7b (Figure 5D bottom panels).
The target genes of miR-128b and miR-221. (A) Positions of the predicted (target scan) miR-128b target sequences are marked (X) in the 3′ UTRs of human MLL and AF4 mRNAs. The solid arrows represent the position of primers used for cloning the 3′UTR segments that contains the predicted miR-128b. (B) Expression of the Lenilla luciferase reporter is significantly reduced when MLL-UTR-128b and AF4-UTR-128b reporter vectors, containing the miR-128b binding sites of the 3′-UTR of MLL and AF4, respectively, are cotransfected together with miR-128b. This reduction is not observed when control or miR-128bmut expression vectors are used. The Renilla/firefly luciferase ratio is calculated and normalized against the control (MDH) (n = 3). **P < .05. (C) Relative expression levels of MLL-AF4, AF4-MLL, AF4-MLL+MLL, and MLL-AF4+AF4 mRNAs are normalized to U1A expression levels using quantitative PCR in RS4;11 cells, which are overexpressing miR-128b, control vector, or miR-221 (n = 3). **P < .05. (D) Western blot analyses for N320 (MLL N-terminal protein), MLL-AF4, and C180 (MLL C-terminal protein) from total protein extracts of RS4;11 cells, which are overexpressing miR-128b, control vector, or miR-221. The relative intensities of each band, N320 (top in top panel), MLL-AF4 (bottom in top panel), or C180 (middle panel), were normalized to the GAPDH loading control using Multigage software and are indicated below each lane. (Bottom panels) Relative expression levels of HOXa9 mRNAs, normalized to U1A expression levels, using quantitative PCR in SEM cells (left) and RS4;11 (right) cells, which are overexpressing miR-128b, let7b, or miR-221 (n = 3). **P < .05. (E) Positions of the predicted (target scan) miR-221 target sequences are marked (X) in the 3′ UTRs of human CDKN1B mRNA. The solid arrows represent the position of primers used for cloning the 3′UTR segments that contains the predicted miR-221 binding sites. (F) Expression of the Lenilla luciferase reporter activity is significantly reduced when the CDKN1B-UTR-221 reporter vector containing the miR-221 binding sites of the 3′-UTR of CDKN1B is cotransfected together with miR-221. This reduction is not observed when the control and miR-221mut reporter vectors are used (n = 3). **P < .05. (G) Relative expression levels of CDKN1B mRNA is normalized to U1A mRNA expression levels using quantitative PCR in RS4;11 cells, which are overexpressing either miR-221 or a control vector. (H) Western blot analyses for p27 from total protein extracts of RS4;11 cells, which are overexpressing miR-221, or control vector. The relative intensities of each band were normalized to the β-actin loading control using Multigage software Version 2.2 (Fuji) and are indicated below each lane. (I) Cell-cycle analyses for SEM cells, which are overexpressing miR-221, miR-128b, let7b, or control vector (MDH). G1, S, and G2 populations are calculated using FlowJo software Version 7 (TreeStar). A representative experiment is shown (n = 3).
The target genes of miR-128b and miR-221. (A) Positions of the predicted (target scan) miR-128b target sequences are marked (X) in the 3′ UTRs of human MLL and AF4 mRNAs. The solid arrows represent the position of primers used for cloning the 3′UTR segments that contains the predicted miR-128b. (B) Expression of the Lenilla luciferase reporter is significantly reduced when MLL-UTR-128b and AF4-UTR-128b reporter vectors, containing the miR-128b binding sites of the 3′-UTR of MLL and AF4, respectively, are cotransfected together with miR-128b. This reduction is not observed when control or miR-128bmut expression vectors are used. The Renilla/firefly luciferase ratio is calculated and normalized against the control (MDH) (n = 3). **P < .05. (C) Relative expression levels of MLL-AF4, AF4-MLL, AF4-MLL+MLL, and MLL-AF4+AF4 mRNAs are normalized to U1A expression levels using quantitative PCR in RS4;11 cells, which are overexpressing miR-128b, control vector, or miR-221 (n = 3). **P < .05. (D) Western blot analyses for N320 (MLL N-terminal protein), MLL-AF4, and C180 (MLL C-terminal protein) from total protein extracts of RS4;11 cells, which are overexpressing miR-128b, control vector, or miR-221. The relative intensities of each band, N320 (top in top panel), MLL-AF4 (bottom in top panel), or C180 (middle panel), were normalized to the GAPDH loading control using Multigage software and are indicated below each lane. (Bottom panels) Relative expression levels of HOXa9 mRNAs, normalized to U1A expression levels, using quantitative PCR in SEM cells (left) and RS4;11 (right) cells, which are overexpressing miR-128b, let7b, or miR-221 (n = 3). **P < .05. (E) Positions of the predicted (target scan) miR-221 target sequences are marked (X) in the 3′ UTRs of human CDKN1B mRNA. The solid arrows represent the position of primers used for cloning the 3′UTR segments that contains the predicted miR-221 binding sites. (F) Expression of the Lenilla luciferase reporter activity is significantly reduced when the CDKN1B-UTR-221 reporter vector containing the miR-221 binding sites of the 3′-UTR of CDKN1B is cotransfected together with miR-221. This reduction is not observed when the control and miR-221mut reporter vectors are used (n = 3). **P < .05. (G) Relative expression levels of CDKN1B mRNA is normalized to U1A mRNA expression levels using quantitative PCR in RS4;11 cells, which are overexpressing either miR-221 or a control vector. (H) Western blot analyses for p27 from total protein extracts of RS4;11 cells, which are overexpressing miR-221, or control vector. The relative intensities of each band were normalized to the β-actin loading control using Multigage software Version 2.2 (Fuji) and are indicated below each lane. (I) Cell-cycle analyses for SEM cells, which are overexpressing miR-221, miR-128b, let7b, or control vector (MDH). G1, S, and G2 populations are calculated using FlowJo software Version 7 (TreeStar). A representative experiment is shown (n = 3).
CDKN1B mRNA possesses several putative miR-221 binding sites that are responsible for down-regulation by miR-221
As shown before,30,31 overexpression of miR-221 significantly reduced luciferase expression of reporter vectors containing the segments of the CDKN1B-UTR that have the putative miR-221 binding sites, but not expression from a control vector (Figure 5F). A Western blot extended this result and showed reduced levels of endogenous p27, the cell-cycle inhibitory protein that is the product of the CDKN1B gene, in miR221 overexpressing RS4;11 cells; the reduction in protein level was greater than that of the corresponding CDKN1B mRNA (Figure 5G-H). Moreover, cell-cycle analysis showed that more SEM (left) cells overexpressing miR-221 or miR-128b accumulated in S phase and fewer in G1 than did control SEM cells or cells expressing let7b (Figure 5I). This result is consistent with down-regulation of p27 by miR-221 because p27 inhibits G0/G1 entry; down-regulation of p27 caused by miR-221 overexpression thus decreases the fraction of G0/G1 cells and increases those in S phase.
These results clearly show that expression both of chimeric genes (AF4-MLL and MLL-AF4) and nonchimeric genes (MLL, AF4, and CDKN1B) are normally down-regulated by miR-128b and miR-221.
Discussion
We showed that several miRNAs, including miR-128b and miR-221, are down-regulated in primary cell samples of MLL-rearranged ALL relative to other types of ALL, and that reexpression of miR-128b and miR-221 cooperatively sensitizes 2 cultured lines of MLL-AF4 ALL cells, SEM and RS4;11, to killing by the glucocorticoid DEX. Glucocorticoids, DEX in particular, are among the most effective agents used in the treatment of lymphoid malignancies, including childhood ALL, resulting from their ability to induce apoptosis of lymphoid cells. Resistance to glucocorticoid-induced apoptosis is associated with the notoriously poor prognosis of MLL-AF4 ALL.19,20 Despite the central role of glucocorticoids in almost all protocols for ALL treatment, the mechanism by which glucocorticoids act on their target cells, and the molecular mechanisms underlying glucocorticoid resistance, remain poorly understood.32,33 Therefore, restoration of steroid sensitivity by miR-128b and miR-221 may lead to a better understanding of the mechanism of glucocorticoid function. As one example, Locked-nucleic-acid-modified oligonucleotides effectively antagonized liver-expressed miR-122 in nonhuman primates and is being tested in a clinical trial for hepatitis C virus34 (http://www.santaris.com/NewsReleases/santaris-pharma-begins-human-clinical-testing-of/Default.aspx). When attached to a carrier, such as a targeted liposome,35 delivery of miR-128b and miR-221 to MLL-AF4 ALL leukemic cells might be a promising adjunct to standard chemotherapy.
Most importantly, these 2 miRNAs have cooperative effect in inducing drug resistance. Two important targets of miR-128b are the 2 chimeric mRNAs, MLL-AF4 and AF4-MLL, which are generated by the chromosomal translocation t(4;11) that initiates the disease. Reduction of MLL-AF4 expression with small interfering RNAs induces apoptosis in the SEM MLL-AF4 ALL cell line.36 Overexpression of MLL-AF4, rather counterintuitively, reduces cell growth but also induces resistance to etoposide-mediated cytotoxicity.21,22 AF4-MLL, the reciprocal fusion protein, has oncogenic activity and may be important for leukemogenesis.23,24 Overexpression of both MLL-AF4 and AF4-MLL in mouse fibroblasts generates resistance to apoptosis.23 Thus, the down-regulation of miR-128b could lead to reduced steroid sensitivity through increasing expression levels of both oncogenic fusion proteins, although the mechanism by which these proteins might induce resistance to glucocorticoids is not known. One of the target genes of miR-221 is CDKN1B, which is transcriptionally activated by both the fusion MLL-AF4 protein and the wild type MLL protein.21,25 CDKN1B encodes p27, which is an important cell-cycle checkpoint regulator that causes cells to enter the G0 state from G1.37 Up-regulation of CDKN1B by MLL-AF4 and MLL is suggested to induce resistance to certain anticancer drugs by inducing quiescence.21,25 Therefore, 2 of the critical pathways that probably induce resistance to corticosteroids in MLL-AF4 ALL are normally down-regulated by miR-128b and miR-221. The regulation of this pathway would explain the cooperative effect of miR-221 and miR-128b, but other unknown proteins and pathways critically involved in glucocorticoid resistance may also be regulated by miR-128b and/or miR-221. Importantly, the proteins that directly act to induce steroid resistance of MLL-AF4 cells are unknown, and it is probable that miR-128 and miR-221 affect the stability or translation of some of these mRNAs as well. It will be interesting to determine the numbers of mRNAs down-regulated by miR-128 and miR-221 in lymphoid cells and how these may be involved in biologic functions related to the poor prognosis of MLL-AF4 ALL.
In conclusion, our results establish that miR-128b and miR-221, especially miR-128b, are functionally important in lymphoid cell biology. Most importantly, the effects of miR-128b and miR-221 on drug resistance are cooperative, and a combination of these miRNAs encompasses a promising target for disease therapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs R. Pieters and R. W. Stam for their contribution of providing MLL patient samples, Drs David Bartel, Beiyan Zhou, Christine Mayr, Michael Lam, Kazuaki Yokoyama, Seiichiro Kobayashi, and Arinobu Tojo for helpful discussion and comments, Dr Colin Sieff for gifting experimental material and critical readings of the manuscript, Mrs Takae Toyoshima for technical assistance, and Drs George Bell and Bingbing Yuan for help with the statistical analysis.
This work was supported by the Japan Society for the Promotion of Science (A.K.), US National Institutes of Health (grant R01 DK068348; H.F.L.), and the Dutch Cancer Society and the Netherlands Organization for Scientific Research (M.L.d.B.).
National Institutes of Health
Authorship
Contribution: A.K. designed, carried out, and analyzed all of the experiments and wrote the paper, all with the help and assistance of D.H.; D.S. and M.L.d.B. performed research; J.H. and P.K.R. contributed vital new reagents; S.A.A. contributed to collecting patient samples; and H.F.L. helped with the design and interpretation of the experiments and writing of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Harvey F. Lodish, Whitehead Institute for Biomedical Research, Nine Cambridge Center, Cambridge, MA 02142; e-mail: lodish@wi.mit.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal