Abstract
Cerebral malaria is a severe multifactorial condition associated with the interaction of high numbers of infected erythrocytes to human brain endothelium without invasion into the brain. The result is coma and seizures with death in more than 20% of cases. Because the brain endothelium is at the interface of these processes, we investigated the global gene responses of human brain endothelium after the interaction with Plasmodium falciparum–infected erythrocytes with either high- or low-binding phenotypes. The most significantly up-regulated transcripts were found in gene ontology groups comprising the immune response, apoptosis and antiapoptosis, inflammatory response, cell-cell signaling, and signal transduction and nuclear factor κB (NF-κB) activation cascade. The proinflammatory NF-κB pathway was central to the regulation of the P falciparum–modulated endothelium transcriptome. The proinflammatory molecules, for example, CCL20, CXCL1, CXCL2, IL-6, and IL-8, were increased more than 100-fold, suggesting an important role of blood-brain barrier (BBB) endothelium in the innate defense during P falciparum–infected erythrocyte (Pf-IRBC) sequestration. However, some of these diffusible molecules could have reversible effects on brain tissue and thus on neurologic function. The inflammatory pathways were validated by direct measurement of proteins in brain endothelial supernatants. This study delineates the strong inflammatory component of human brain endothelium contributing to cerebral malaria.
Introduction
Plasmodium falciparum remains one of the most deadly human pathogens causing a broad range of clinical manifestations ranging from asymptomatic or mild flulike symptoms to severe life-threatening anemia or cerebral malaria (CM).1 Pathological studies on postmortem samples from patients who died from CM have shown a correlation between sequestration of P falciparum–infected erythrocytes (Pf-IRBCs) in the brain microvessels and the malaria-related encephalopathy.2 The Pf-IRBC sequestration could compromise blood-brain barrier (BBB) integrity through multifactorial mechanisms, which are still under investigation.3 The underlying molecular mechanism remains unclear as to how intraerythrocytic P falciparum can lead to coma, seizures, and/or persistent cognitive defects, although not physically crossing the BBB and/or causing brain parenchymal leukocyte infiltrates.
In blood, circulating P falciparum parasites reach very high densities compared with other organisms. For example, bacteremia associated with sepsis has only 1 to 10 bacteria per milliliter of blood.4 Moderate HIV viremia is defined as approximately 10 to 100 000 virions per milliliter,5 whereas hepatitis C can reach 1 000 000 virions per milliliter of blood.6 A 10% P falciparum parasitemia, however, represents 400 000 000 parasites per milliliter of blood. In the brain microvasculature, only a fraction of the total parasite burden is sequestered in a heterogeneous distribution in parts of the endothelium that are covered by adherent Pf-IRBCs.2
The cerebral vasculature is highly specialized endothelium forming the actual site of the BBB. It is important to note that the BBB is a significant component of the innate immune system, and its activation is an important component in the initiation and subsequent cascade of inflammatory responses upon pathogen exposure.7 Vascular endothelia can also differ markedly depending on their location/organ. Endothelial cells derived from umbilical veins or aorta (macrovascular), or liver (microvascular with fenestrae), or lung and brain (microvascular and barrier forming) not only differ in morphology and site-specific gene expression but also respond differentially to pathogens.8,9 In accordance with this, our previous studies showed that Pf-IRBCs induce surface intercellular adhesion molecule-1 (ICAM-1) on human brain microvascular endothelial cells (HBMECs), whereas in contrast human umbilical vein endothelial cells (HUVECs) were nonresponsive.10 We previously showed that Pf-IRBCs compromise the electrical resistance of brain endothelium in a time- and dose-dependent manner.11 The Pf-IRBC–induced reduction of brain endothelium integrity was independent of its binding phenotype (high vs low adhesion) and a substantial role of soluble parasite factors in the activation of the BBB was implicated.10,11 To further understand the responses of the brain endothelium after interaction with Pf-IRBCs, we performed genome-wide transcriptional profiling of HBMECs after exposure with isogenic Pf-IRBCs, which were selected for high- or low-binding phenotypes.
Methods
Cell culture
P falciparum clone 3D7 was cultured in RPMI 1640 medium that contained 10% human serum.12 Parasite preparations were enriched for trophozoite stage by centrifugation of sorbitol-synchronized P falciparum culture on Percoll density gradients and examined by Giemsa staining for quality control. Selection (panning) of 3D7 for increased binding to brain endothelium was performed on nonactivated HBMECs in 3 cycles.11 Parasite cultures used during the studies were free from mycoplasma. Use of human erythrocyte for P falciparum culture was approved by the institutional review board of Johns Hopkins University Bloomberg School of Public Health.
HBMECs were isolated from human brain specimens and characterized as described previously.10,11 HBMECs (cells at passage < 6) from a single adult donor were used for the biologic replicate experiments. For microarray experiments, HBMECs were grown in collagen-coated 6-well plates until cells achieved about 80% to 90% confluence. The medium was then changed to RPMI 1640 with 5% fetal bovine serum for 24 hours before coincubation with Pf-IRBCs, at which time HBMECs are confluent. Endothelial cells were negative for mycoplasma infection (Vector Laboratories).
To approximate the conditions of brain sequestration, for all the studies, the Pf-IRBC to HBMEC ratio was kept at 50:1, which covers the whole monolayer surface of HBMECs by Pf-IRBCs. In addition, at this ratio we have observed maximal effects on electrical resistance of monolayer and increases in surface ICAM-1.10,11 The control uninfected red blood cells (RBCs) from the same donor as those used for parasite culture were maintained under similar conditions. For microarray studies, confluent HBMECs were incubated with Pf-IRBCs and uninfected RBCs at 37°C for 6 hours. After incubation, the culture supernatants were aspirated and stored at −70°C. Endothelial monolayers were subsequently washed twice with ice-cold phosphate-buffered saline and immediately resuspended into 1 mL Trizol reagent.
Microarray
Homogenization and lysis of cells was performed with Lysing Matrix D (Qbiogene) in Trizol reagent (Invitrogen) by rapid agitation in a FastPrep 120 Instrument (Qbiogene) for 15 seconds at speed setting 5. Subsequent to lysis, RNA extraction was performed with the Invitrogen PureLink Micro to Midi Total RNA Purification System, according to the manufacturer's recommended protocol. Quantification of RNA was performed using a NanoDrop spectrophotometer, and quality assessment was determined by RNA Nano LabChip analysis on an Agilent Bioanalyzer 2100.
Processing of templates for GeneChip Analysis followed the Affymetrix GeneChip Expression Analysis Technical Manual, Revision 5.13 Double-stranded cDNA was synthesized from 5 μg total RNA using the GeneChip Expression 3′ amplification reagents one-cycle cDNA synthesis kit (Affymetrix), and column-purified using the GeneChip Sample Cleanup Module. Biotinylated cRNA was synthesized using the GeneChip Expression 3′ amplification reagents for IVT labeling (Affymetrix). Resultant cRNAs were purified by column purification with the GeneChip Sample Cleanup Module (Affymetrix). Quality of biotinylated cRNA was assessed by RNA Nano LabChip analysis on an Agilent Bioanalyzer 2100. cRNA (15 μg) was fragmented by metal-induced hydrolysis in fragmentation buffer at 94°C for 35 minutes. Hybridization cocktails were prepared as recommended for arrays of “standard” format including incubation at 94°C for 5 minutes and 45°C for 5 minutes, and centrifugation at maximum speed for 5 minutes prior to pipetting into the GeneChips (Affymetrix Human Genome U133A 2.0). Hybridization was performed at 45°C for 16 hours at 60 rpm in the Affymetrix rotisserie hybridization oven. The signal amplification protocol for washing and staining of eukaryotic targets was performed in an automated fluidics station (Affymetrix FS450). The arrays were then transferred to the GCS3000 laser scanner (Affymetrix) and scanned at an emission wavelength of 570 nm at 2.5-μm resolution. Intensity of hybridization for each probe pair was computed by GCOS 1.4 software (Affymetrix). (For more detailed methods, please refer to the website of the Malaria Research Institute Gene Array Core Facility [MRI-GACF] at the Johns Hopkins Bloomberg School of Public Health: http://jhmmi.jhsph.edu.)
Primary analysis consisted of a quality assessment of the hybridization for each sample. Ratios of signal for probe sets at 5′ and 3′ regions of housekeeping genes were calculated and monitored as an indication of transcript quality for each sample. The hybridization intensity for each probe array was used in an expression algorithm, which designates each transcript as present (P), absent (A), or marginal (M). Microarray data analysis was limited to gene expression levels that were changed more than 2-fold. Primary statistical analysis of microarray data were done with Welch t test to identify significantly regulated genes. In the secondary analysis, the Benjamini and Hochberg procedure 18 of controlling false discovery rate was applied for correction of microarray data. All of the statistical analysis was done with Genesifter data analysis software (VizX Labs LLC, http://www.genesifter.net). A value of P less than .05 was accepted to be significant for all statistical analyses performed.
Cytokine and chemokine determinations
We performed both semiquantitative protein analysis using dot blot cytokine arrays (RayBio), and quantitative enzyme-linked immunosorbent assays (ELISAs) and BD Cytometric Bead Array on stored culture supernatants from microarray experiments as well as supernatants obtained from fresh coincubation studies done under similar conditions. The signals were detected by the enhanced chemiluminescence system on radiographic film, and then the National Institutes of Health (NIH) ImageJ program digitized the spots. The relative spot intensities were determined by dividing the values for each cytokine with the membrane-positive controls. The levels of tumor necrosis factor α (TNF-α), chemokine (C-C motif) ligand 20 (CCL20), and growth-regulated oncogene α (GRO-α) in endothelial culture supernatants were also determined with commercial ELISA kits (R&D Systems). Human interleukin 1β (IL-1β), IL-6, IL-8, regulated upon activation, normal T-cell expressed, and secreted (RANTES), and TNF-α levels in endothelial culture supernatants were measured using a Cytometric Bead Array–based immunoassay (Becton Dickinson) with standards ranging from 0 to 5000 pg/mL, with a 2-color flow cytometric analysis using FCAP Array software (Becton Dickinson). Concentrations of each cytokine in culture supernatants were determined from standard curves.
Kinetics of NF-κB nuclear translocation
To determine the kinetics of nuclear factor κB (NF-κB) nuclear translocation, nuclear and cytosolic extracts were prepared with slight modification of the previously described method of Dignam et al.14 HBMECs were incubated with or without Pf-IRBCs for indicated duration, ranging from 10 to 180 minutes. The cell pellet was resuspended in cell lysis buffer and allowed to swell on ice for 15 minutes, followed by centrifugation at 10 000g for 5 minutes. The nuclear pellet was resuspended in nuclear extraction buffer and incubated on ice for 15 minutes with occasional agitation. The extracted nuclei were pelleted at 10 000g for 15 minutes at 4°C, while the supernatants were collected as nuclear extracts and stored at −70°C. Equal amounts of proteins (Bradford) from each treatment were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis transferred to nitrocellulose membrane and the blots were probed with mouse anti-p65 monoclonal antibody (Santa Cruz Biotech Inc) followed by peroxidase-conjugated antimouse goat secondary antibody (Jackson ImmunoResearch).
Results
Pf-IRBCs with different binding phenotypes
The synchronized trophozoite-stage Pf-IRBCs were Percoll purified to greater than 90% for coincubations. At this stage, all parasites show knob formation on the infected erythrocyte surface (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Control/nonpanned Pf-IRBCs (clone 3D7) bound with approximately 25 Pf-IRBCs per 100 HBMECs. Pf-IRBCs were then sequentially panned in 3 consecutive rounds on HBMEC monolayer, resulting in an increase of the binding by more than 1 log.11 This adherent binding phenotype represented 10% of the panned and 1% of the nonpanned total Pf-IRBCs added in all experiments.
Both populations were then tested for their ability to activate HBMECs, increase ICAM-1 expression, and decrease monolayer resistance.10,11 We observed a 4-fold increase in HBMEC surface ICAM-1 at 6 hours for both panned and nonpanned Pf-IRBC coincubations compared with media and uninfected RBC controls. A maximum decrease in electrical resistance of the HBMEC monolayers observed at 6 hours was also similar for both the panned and nonpanned Pf-IRBCs. The same number of uninfected RBCs caused a slight increase in electrical resistance compared with media control from a monolayer effect.10,11 These Pf-IRBC populations were used in parallel to characterize the host HBMEC responses by microarray analysis.
Functional microarray analysis indicates immune response, inflammation, signal transduction, and apoptosis pathways
To determine the global gene expression profile of HBMECs in response to Pf-IRBC exposure, we performed 2 sets of independent experiments using both the panned and unpanned Pf-IRBCs. Initial data analysis using 4 groups (medium control, RBC control, Pf-IRBCs, and panned Pf-IRBCs [Pf-IRBC-Ps]) revealed no significant differences between 2 parasite lines as well as between medium and RBC controls of those experiments (supplemental Figure 2). We therefore grouped the panned and unpanned as well as medium and RBC controls of the independent experiments as biologic replicates for final statistical analysis.
Global transcriptional profiling of HBMECs after Pf-IRBC exposure for 6 hours revealed 372 probe sets representing 231 genes that were up-regulated and 216 probe sets representing 155 genes that were down-regulated. The average fold change was much higher for up-regulated genes compared with down-regulated genes. A complete listing of all the differentially regulated genes is provided as supplemental information in Tables 1–2.
Significantly up-regulated pathways
| Rank . | Pathway name . | No. of genes . | Gamma P value . |
|---|---|---|---|
| 1 | Cytokine-cytokine receptor interaction | 17 | .001 |
| 2 | Cell adhesion molecules (CAMs) | 8 | .001 |
| 3 | Epithelial cell signaling in Helicobacter pylori infection | 9 | .001 |
| 4 | Leukocyte transendothelial migration | 5 | .001 |
| 5 | Apoptosis | 12 | .001 |
| 6 | Toll-like receptor signaling pathway | 10 | .001 |
| 7 | Small cell lung cancer | 10 | .001 |
| 8 | B-cell receptor signaling pathway | 8 | .001 |
| 9 | MAPK signaling pathway | 16 | .001 |
| 10 | T-cell receptor signaling pathway | 9 | .001 |
| 11 | ErbB signaling pathway | 8 | .001 |
| 12 | Natural killer cell–mediated cytotoxicity | 9 | .002 |
| 13 | Jak-STAT signaling pathway | 9 | .003 |
| 14 | Adipocytokine signaling pathway | 6 | .006 |
| 15 | Complement and coagulation cascades | 6 | .008 |
| 16 | Chronic myeloid leukemia | 6 | .01 |
| 17 | TGF-beta signaling pathway | 6 | .01 |
| 18 | Pancreatic cancer | 5 | .039 |
| 19 | Antigen processing and presentation | 5 | .047 |
| Rank . | Pathway name . | No. of genes . | Gamma P value . |
|---|---|---|---|
| 1 | Cytokine-cytokine receptor interaction | 17 | .001 |
| 2 | Cell adhesion molecules (CAMs) | 8 | .001 |
| 3 | Epithelial cell signaling in Helicobacter pylori infection | 9 | .001 |
| 4 | Leukocyte transendothelial migration | 5 | .001 |
| 5 | Apoptosis | 12 | .001 |
| 6 | Toll-like receptor signaling pathway | 10 | .001 |
| 7 | Small cell lung cancer | 10 | .001 |
| 8 | B-cell receptor signaling pathway | 8 | .001 |
| 9 | MAPK signaling pathway | 16 | .001 |
| 10 | T-cell receptor signaling pathway | 9 | .001 |
| 11 | ErbB signaling pathway | 8 | .001 |
| 12 | Natural killer cell–mediated cytotoxicity | 9 | .002 |
| 13 | Jak-STAT signaling pathway | 9 | .003 |
| 14 | Adipocytokine signaling pathway | 6 | .006 |
| 15 | Complement and coagulation cascades | 6 | .008 |
| 16 | Chronic myeloid leukemia | 6 | .01 |
| 17 | TGF-beta signaling pathway | 6 | .01 |
| 18 | Pancreatic cancer | 5 | .039 |
| 19 | Antigen processing and presentation | 5 | .047 |
Selection of pathways was based upon number of genes more than 5 and significance (gamma P < .05). Pathway rankings are based on significance.
NF-κB signaling pathway (positive regulators of NF-κB activation cascade, NF-κB subunits, and cytosolic inhibitors of NF-κB)
| Gene name . | Gene symbol . | Ratio . | NFkB target . |
|---|---|---|---|
| NF-κB activation cascade | |||
| Tumor necrosis factor receptor superfamily, member 10b | TNFRSF10B | 3.62 | Yes |
| Receptor-interacting serine-threonine kinase 2 | RIPK2 | 4.12 | Yes |
| Baculoviral IAP repeat-containing 2 | BIRC2 | 5.75 | Yes |
| Tumor necrosis factor receptor superfamily, member 1A | TNFRSF1A | 2.16 | No |
| Tumor necrosis factor (TNF superfamily, member 2) | TNF | 8.81 | Yes |
| CD40 molecule, TNF receptor superfamily member 5 | CD40 | 6.35 | Yes |
| B-cell CLL/lymphoma 10 | BCL10 | 2.87 | Yes |
| TRAF 3 interacting protein 2 | TRAF3IP2 | 3.1 | No |
| CASP8 and FADD-like apoptosis regulator | CFLAR | 4.7 | Yes |
| Sequestosome 1 | SQSTM1 | 2.12 | Yes |
| NF-κB subunits | |||
| Nuclear factor of kappa light polypeptide gene enhancer in B cells 1 (p105) | NFKB1 | 8.28 | Yes |
| Nuclear factor of kappa light polypeptide gene enhancer in B cells 2 (p49/p100) | NFKB2 | 8.27 | Yes |
| V-rel reticuloendotheliosis viral oncogene homolog (avian) | cREL | 10.59 | Yes |
| V-rel reticuloendotheliosis viral oncogene homolog B | RELB | 11.56 | Yes |
| NF-κB inhibitory proteins | |||
| IKBα | NFKBIA | 21.61 | Yes |
| IKBϵ | NFKBIE | 3.62 | Yes |
| Gene name . | Gene symbol . | Ratio . | NFkB target . |
|---|---|---|---|
| NF-κB activation cascade | |||
| Tumor necrosis factor receptor superfamily, member 10b | TNFRSF10B | 3.62 | Yes |
| Receptor-interacting serine-threonine kinase 2 | RIPK2 | 4.12 | Yes |
| Baculoviral IAP repeat-containing 2 | BIRC2 | 5.75 | Yes |
| Tumor necrosis factor receptor superfamily, member 1A | TNFRSF1A | 2.16 | No |
| Tumor necrosis factor (TNF superfamily, member 2) | TNF | 8.81 | Yes |
| CD40 molecule, TNF receptor superfamily member 5 | CD40 | 6.35 | Yes |
| B-cell CLL/lymphoma 10 | BCL10 | 2.87 | Yes |
| TRAF 3 interacting protein 2 | TRAF3IP2 | 3.1 | No |
| CASP8 and FADD-like apoptosis regulator | CFLAR | 4.7 | Yes |
| Sequestosome 1 | SQSTM1 | 2.12 | Yes |
| NF-κB subunits | |||
| Nuclear factor of kappa light polypeptide gene enhancer in B cells 1 (p105) | NFKB1 | 8.28 | Yes |
| Nuclear factor of kappa light polypeptide gene enhancer in B cells 2 (p49/p100) | NFKB2 | 8.27 | Yes |
| V-rel reticuloendotheliosis viral oncogene homolog (avian) | cREL | 10.59 | Yes |
| V-rel reticuloendotheliosis viral oncogene homolog B | RELB | 11.56 | Yes |
| NF-κB inhibitory proteins | |||
| IKBα | NFKBIA | 21.61 | Yes |
| IKBϵ | NFKBIE | 3.62 | Yes |
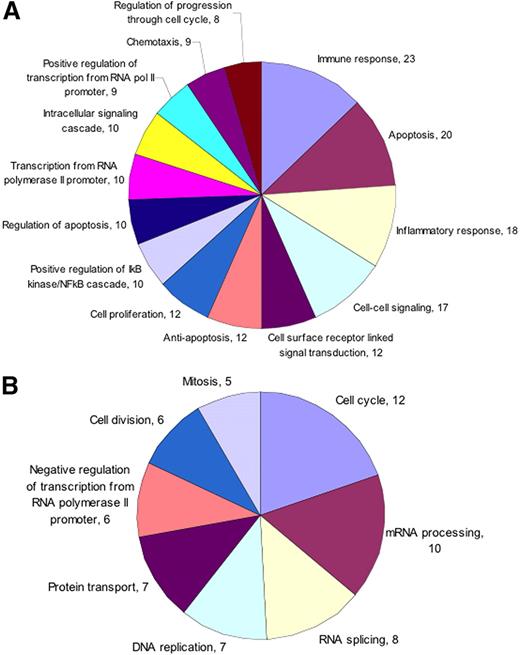
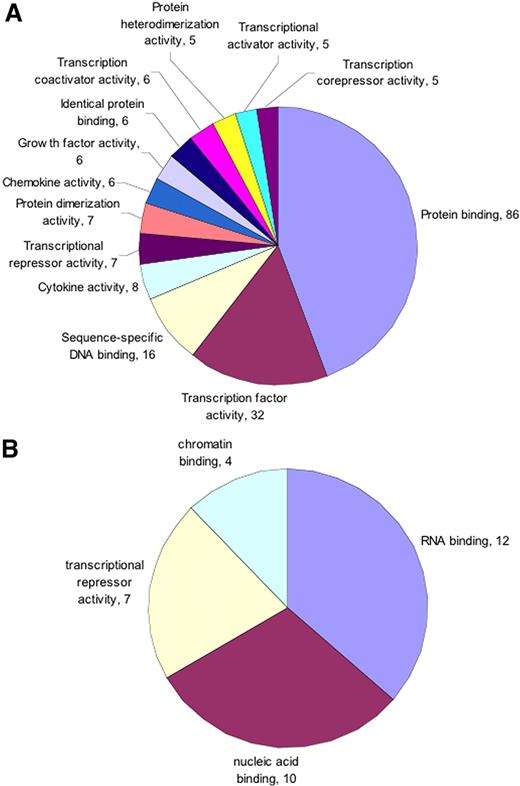
To functionally categorize differentially regulated genes, we performed Gene Ontology (GO) analysis on both up- and down-regulated genes separately.15 GO categorization was performed using the web-based tool Onto-Express (Wayne State University, http://vortex.cs.wayne.edu). The data were categorized based on biologic process, and molecular function categories with corrected P value less than .05 were plotted. We found that in the biologic process ontology of up-regulated genes, members belonging to immune response, apoptosis, inflammation, and signal transduction were more abundant (Figure 1). However, among down-regulated genes in the same category, genes mostly involved in cell cycle were in greater number (Figure 1). In the molecular function category of up-regulated genes, principal members belonged to protein binding, transcription, and cytokine and chemokine function (Figure 2). The majority of down-regulated genes were involved in RNA- or DNA-binding function (Figure 2).
Immune response and apoptosis are up-regulated by Pf-IRBC exposure. GO biologic process categories (A) up-regulated and (B) down-regulated were selected from total list obtained based on the number of genes and corrected P value. Only those categories were plotted that showed minimum 8 (up-regulated) or 5 (down-regulated) genes and have corrected P < .05. Numbers associated with each category represent number of genes.
Immune response and apoptosis are up-regulated by Pf-IRBC exposure. GO biologic process categories (A) up-regulated and (B) down-regulated were selected from total list obtained based on the number of genes and corrected P value. Only those categories were plotted that showed minimum 8 (up-regulated) or 5 (down-regulated) genes and have corrected P < .05. Numbers associated with each category represent number of genes.
Protein binding is up-regulated and RNA and DNA binding is down-regulated by Pf-IRBC exposure. GO molecular function categories (A) up-regulated and (B) down-regulated were selected from total list obtained based on the number of genes and corrected P value. Only those categories were plotted that showed minimum 5 (up-regulated) or 4 (down-regulated) genes and have corrected P value < .05. Numbers associated with each category represents number of genes.
Protein binding is up-regulated and RNA and DNA binding is down-regulated by Pf-IRBC exposure. GO molecular function categories (A) up-regulated and (B) down-regulated were selected from total list obtained based on the number of genes and corrected P value. Only those categories were plotted that showed minimum 5 (up-regulated) or 4 (down-regulated) genes and have corrected P value < .05. Numbers associated with each category represents number of genes.
The GO database does not cover every aspect of biology of different genes and many GO classes overlap with each other. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways are compiled from multiple sources, which integrate individual genes into a unified pathway. To illustrate differentially regulated genes into specific pathways, we performed pathway analysis using web-based software Pathway Express (Wayne State University, http://vortex.cs.wayne.edu). We identified 19 host response pathways that were significantly up-regulated in response to Pf-IRBC exposure (gamma P < .05 and at least 5 input genes). The most significantly affected pathways in terms of number of genes were the cytokine-cytokine receptor pathway (17 genes, gamma P = .001), followed by the mitogen-activated protein kinase signaling pathway (Table 1).
The NF-κB signaling pathway and its target genes were implicated in activated HBMECs
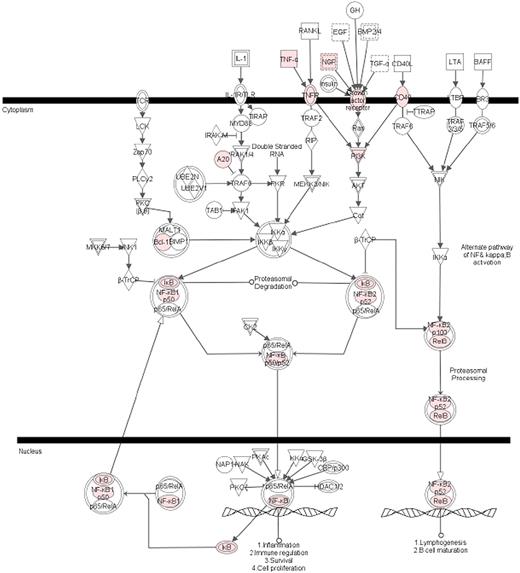
GO and pathway analysis revealed induction of several members of NF-κB activation cascade and 4 of 5 NF-κB subunits (REL-B, cREL, NF-κB1, and NF-κB2) (Table 2). Only one NF-κB subunit REL-A (p65) was not up-regulated, which is in accordance with its regulation at the protein level.1 We also observed up-regulation of 2 inhibitor proteins of NF-κB, IκBα and IκBϵ, which are responsible for NF-κB retention into the cytoplasm (Table 2). These observations led us to identify the up-regulated genes, which have earlier been shown to be the target of NF-κB. We compiled a list of NF-κB target genes from the literature,16-24 and upon comparison with our list we observed 72 NF-κB–regulated genes in our list of 221 up-regulated genes (Table 3). Interestingly, the average fold change for NF-κB target genes was significantly higher (> 30-fold) compared with non–NF-κB target genes (< 5-fold). Canonical pathway analysis by Ingenuity Pathways Analysis (IPA) software (Ingenuity Systems) identified NF-κB signaling pathway as the most significant among the up-regulated pathways (Figure 3).
List of IKK/NF-κB target genes in HBMECs, induced by Pf-IRBC or Pf-IRBC-P exposure
| Gene name . | Gene symbol . | Ratio . |
|---|---|---|
| Cytokine, chemokine, and receptors | ||
| Chemokine (C-C motif) ligand 20 | CCL20 | 779.33 |
| Chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) | CXCL1 | 174.23 |
| Chemokine (C-X-C motif) ligand 2 | CXCL2 | 286.98 |
| Chemokine (C-X-C motif) ligand 3 | CXCL3 | 54.69 |
| Chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) | CXCL6 | 5.99 |
| Interleukin 6 (interferon, beta 2) | IL-6 | 141.15 |
| Interleukin 8 | IL-8 | 132.04 |
| Interleukin 4 receptor | IL-4R | 3.18 |
| Intercellular adhesion molecule 1 (CD54), human rhinovirus receptor | ICAM1 | 54.61 |
| Tumor necrosis factor (ligand) superfamily, member 9 | TNFSF9 | 2.91 |
| Tumor necrosis factor receptor superfamily, member 5 | TNFRSF5 | 6.63 |
| Inhibin, beta A (activin A, activin AB alpha polypeptide) | INHBA | 17.46 |
| Leukemia inhibitory factor (cholinergic differentiation factor) | LIF | 7.44 |
| CD44 molecule (Indian blood group) | CD44 | 3.73 |
| Transcription factor and signaling molecules | ||
| Nuclear factor, interleukin 3 regulated | NFIL3 | 3.43 |
| Nuclear receptor subfamily 4, group A, member 1 | NR4A1 | 18.47 |
| Nuclear receptor subfamily 4, group A, member 2 | NR4A2 | 18.69 |
| V-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian) | MAFF | 10.12 |
| Jun oncogene | JUN | 7.94 |
| Activating transcription factor 3 | ATF3 | 11.02 |
| Interferon regulatory factor 2 | IRF2 | 2.17 |
| Early growth response 1 | EGR1 | 11.89 |
| Down syndrome critical region gene 1 | DSCR1 | 2.36 |
| Tissue factor pathway inhibitor 2 | TFPI2 | 2.49 |
| TNFAIP3 interacting protein 1 | TNIP1 | 5.89 |
| Tumor necrosis factor, alpha-induced protein 2 | TNFAIP2 | 29.74 |
| Tumor necrosis factor, alpha-induced protein 3 | TNFAIP3 | 26.54 |
| Tumor necrosis factor, alpha-induced protein 6 | TNFAIP6 | 9.01 |
| EH-domain containing 1 | EHD1 | 4.93 |
| Immediate early response 3 | IER3 | 12.01 |
| Immediate early response 5 | IER5 | 2.88 |
| Epstein-Barr virus–induced gene 3 | EBI3 | 6.68 |
| Jumonji, AT-rich interactive domain 1B | JARID1B | 2.23 |
| Baculoviral IAP repeat-containing 3 | BIRC3 | 46.22 |
| BCL2-like 1 | BCL2L1 | 2.97 |
| BCL2-related protein A1 | BCL2A1 | 11.11 |
| Kruppel-like factor 6 | KLF6 | 5.33 |
| Phorbol-12-myristate-13-acetate–induced protein 1 | PMAIP1 | 3.39 |
| Transporter activity | ||
| Solute carrier family 2 (facilitated glucose transporter), member 6 | SLC2A6 | 3.27 |
| Transporter 1, ATP-binding cassette, subfamily B (MDR/TAP) | TAP1 | 5.77 |
| Enzyme activity (eg, kinases, phosphatases) | ||
| Plasminogen activator, tissue | PLAT | 12.56 |
| Plasminogen activator, urokinase | PLAU | 6.24 |
| Mitogen-activated protein kinase kinase 3 | MAP2K3 | 3.12 |
| Protein phosphatase 1, regulatory (inhibitor) subunit 15A | PPP1R15A | 4.38 |
| GTP cyclohydrolase 1 (dopa-responsive dystonia) | GCH1 | 10.19 |
| Dual specificity phosphatase 1 | DUSP1 | 8.26 |
| Interferon-stimulated exonuclease gene 20 kDa | ISG20 | 6.88 |
| Superoxide dismutase 2, mitochondrial | SOD2 | 5.71 |
| Transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) | TGM2 | 3.49 |
| Others | ||
| Ferritin, heavy polypeptide 1 | FTH1 | 2.1 |
| Growth arrest and DNA-damage–inducible, alpha | GADD45A | 3.2 |
| Major histocompatibility complex, class I, B | HLA-B | 2.38 |
| Pentraxin-related gene, rapidly induced by IL-1 beta | PTX3 | 9.2 |
| Syndecan 4 | SDC4 | 4.89 |
| Zinc finger CCCH-type containing 12A | ZC3H12A | 50.3 |
| CD83 molecule | CD83 | 3.06 |
| Zinc finger protein 36, C3H type, homolog (mouse) | ZFP36 | 16.44 |
| Gene name . | Gene symbol . | Ratio . |
|---|---|---|
| Cytokine, chemokine, and receptors | ||
| Chemokine (C-C motif) ligand 20 | CCL20 | 779.33 |
| Chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) | CXCL1 | 174.23 |
| Chemokine (C-X-C motif) ligand 2 | CXCL2 | 286.98 |
| Chemokine (C-X-C motif) ligand 3 | CXCL3 | 54.69 |
| Chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) | CXCL6 | 5.99 |
| Interleukin 6 (interferon, beta 2) | IL-6 | 141.15 |
| Interleukin 8 | IL-8 | 132.04 |
| Interleukin 4 receptor | IL-4R | 3.18 |
| Intercellular adhesion molecule 1 (CD54), human rhinovirus receptor | ICAM1 | 54.61 |
| Tumor necrosis factor (ligand) superfamily, member 9 | TNFSF9 | 2.91 |
| Tumor necrosis factor receptor superfamily, member 5 | TNFRSF5 | 6.63 |
| Inhibin, beta A (activin A, activin AB alpha polypeptide) | INHBA | 17.46 |
| Leukemia inhibitory factor (cholinergic differentiation factor) | LIF | 7.44 |
| CD44 molecule (Indian blood group) | CD44 | 3.73 |
| Transcription factor and signaling molecules | ||
| Nuclear factor, interleukin 3 regulated | NFIL3 | 3.43 |
| Nuclear receptor subfamily 4, group A, member 1 | NR4A1 | 18.47 |
| Nuclear receptor subfamily 4, group A, member 2 | NR4A2 | 18.69 |
| V-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian) | MAFF | 10.12 |
| Jun oncogene | JUN | 7.94 |
| Activating transcription factor 3 | ATF3 | 11.02 |
| Interferon regulatory factor 2 | IRF2 | 2.17 |
| Early growth response 1 | EGR1 | 11.89 |
| Down syndrome critical region gene 1 | DSCR1 | 2.36 |
| Tissue factor pathway inhibitor 2 | TFPI2 | 2.49 |
| TNFAIP3 interacting protein 1 | TNIP1 | 5.89 |
| Tumor necrosis factor, alpha-induced protein 2 | TNFAIP2 | 29.74 |
| Tumor necrosis factor, alpha-induced protein 3 | TNFAIP3 | 26.54 |
| Tumor necrosis factor, alpha-induced protein 6 | TNFAIP6 | 9.01 |
| EH-domain containing 1 | EHD1 | 4.93 |
| Immediate early response 3 | IER3 | 12.01 |
| Immediate early response 5 | IER5 | 2.88 |
| Epstein-Barr virus–induced gene 3 | EBI3 | 6.68 |
| Jumonji, AT-rich interactive domain 1B | JARID1B | 2.23 |
| Baculoviral IAP repeat-containing 3 | BIRC3 | 46.22 |
| BCL2-like 1 | BCL2L1 | 2.97 |
| BCL2-related protein A1 | BCL2A1 | 11.11 |
| Kruppel-like factor 6 | KLF6 | 5.33 |
| Phorbol-12-myristate-13-acetate–induced protein 1 | PMAIP1 | 3.39 |
| Transporter activity | ||
| Solute carrier family 2 (facilitated glucose transporter), member 6 | SLC2A6 | 3.27 |
| Transporter 1, ATP-binding cassette, subfamily B (MDR/TAP) | TAP1 | 5.77 |
| Enzyme activity (eg, kinases, phosphatases) | ||
| Plasminogen activator, tissue | PLAT | 12.56 |
| Plasminogen activator, urokinase | PLAU | 6.24 |
| Mitogen-activated protein kinase kinase 3 | MAP2K3 | 3.12 |
| Protein phosphatase 1, regulatory (inhibitor) subunit 15A | PPP1R15A | 4.38 |
| GTP cyclohydrolase 1 (dopa-responsive dystonia) | GCH1 | 10.19 |
| Dual specificity phosphatase 1 | DUSP1 | 8.26 |
| Interferon-stimulated exonuclease gene 20 kDa | ISG20 | 6.88 |
| Superoxide dismutase 2, mitochondrial | SOD2 | 5.71 |
| Transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) | TGM2 | 3.49 |
| Others | ||
| Ferritin, heavy polypeptide 1 | FTH1 | 2.1 |
| Growth arrest and DNA-damage–inducible, alpha | GADD45A | 3.2 |
| Major histocompatibility complex, class I, B | HLA-B | 2.38 |
| Pentraxin-related gene, rapidly induced by IL-1 beta | PTX3 | 9.2 |
| Syndecan 4 | SDC4 | 4.89 |
| Zinc finger CCCH-type containing 12A | ZC3H12A | 50.3 |
| CD83 molecule | CD83 | 3.06 |
| Zinc finger protein 36, C3H type, homolog (mouse) | ZFP36 | 16.44 |
NF-κB signaling pathway was the most significant canonical pathway in HBMEC up-regulation. Genes, differentially regulated in HBMECs in response to Pf-IRBC coincubation, were clustered into canonical pathways using Ingenuity tool (Ingenuity Systems). The NF-κB pathway was statistically most significant (P = .001). Colored genes were up-regulated in the presence of Pf-IRBC.
NF-κB signaling pathway was the most significant canonical pathway in HBMEC up-regulation. Genes, differentially regulated in HBMECs in response to Pf-IRBC coincubation, were clustered into canonical pathways using Ingenuity tool (Ingenuity Systems). The NF-κB pathway was statistically most significant (P = .001). Colored genes were up-regulated in the presence of Pf-IRBC.
Protein quantification confirmed transcriptional response
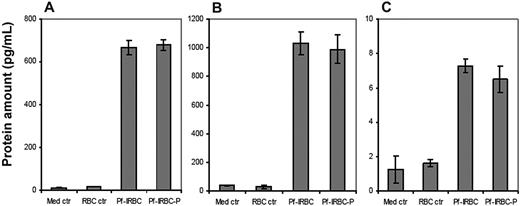
To confirm that the observed increase in transcription also translated into actual increases in release of proteins, the supernatants of HBMECs exposed to Pf-IRBCs, control RBCs, and media alone were analyzed for secreted proteins. The proinflammatory cytokines IL-6, IL-8, and TNF-α were increased in the Pf-IRBC supernatants as determined with a semiquantitative Western dot blot RayBio custom array (Table 4). Although there were discrepancies between the different methods, generally the increase in mRNAs in the Pf-IRBC–activated HBMECs was accompanied with an increase of the release of its protein/cytokine in the supernatants. GRO-α was not detected in the dot blot despite large change in transcription. Quantification of release of proteins as performed by ELISA for GRO-α, TNF-α, and CCL20 is depicted in Figure 4. GRO-α, CCL20, and TNF-α were apparent in Pf-IRBC–HBMEC coincubation supernatants, but not in control supernatants. The bead-based immunoassay quantitated IL-6, IL-8, RANTES (CCL5), TNF-α, and IL-1β. Release of CCL20, GRO-α, IL-6, and IL-8 was more than 100-fold increased, which correlated well with their transcriptional profile (Figures 4–5). TNF-α, which was associated with an 8-fold up-regulation at transcript level, showed a similar increase in protein expression (Figures 4–5). IL-1β, a gene that was neither up- nor down-regulated in microarray analysis, showed undetectable levels of protein expression by CBA in contrast to dot blot analysis. Although microarray data initially showed 5-fold up-regulation of RANTES when t test was used for statistical analysis, analysis by Welch t test and Benjamini and Hochberg multiple test correction yielded increase in RANTES as insignificant. However, we still observed increased RANTES in culture supernatants by both the CBA and dot blot analysis.
Comparison of array and dot blot cytokine results
| . | Array ratio . | Medium or RBCs, % of blot-positive control . | Pf-IRBCs, % of blot-positive control . |
|---|---|---|---|
| Eotaxin | A* | 0† | 0 |
| GCSF | Marginally up‡ | 0 | 0 |
| GM-CSF | A | 0 | 0 |
| Soluble ICAM-1 | 54 | 0 | 0 |
| IFNγ | A | 0 | 0 |
| IL-10 | A | 0 | 0 |
| IL-15 | No change§ | 0 | 0 |
| IL-1α | A | 0 | 0 |
| IL-1β | Marginally up | 0 | 29.4 |
| IL-3 | A | 0 | 0 |
| IL-4 | A | 0 | 0 |
| IL-5 | A | 0 | 0 |
| IL-6 | 141 | 58.9 | 109.3 |
| IL-7 | A | 0 | 0 |
| IL-8 | 132 | .6 | 99.3 |
| IP-10 | A | 0 | 0 |
| MCP-1 | No change | 70.6 | 69.8 |
| NAP-1A | Not in array | 0 | 0 |
| MIP-1α | A | 0 | 0 |
| GROα | 174 | 0 | 0 |
| PDGF-BB | No change | 0 | 19.6 |
| RANTES | Marginally up | 0 | 24.1 |
| SDF-1 | A | 0 | 0 |
| TGF-β | A | 0 | 0 |
| Fractalkine | A | 0 | 0 |
| TIMP-1 | No change | 0 | 0 |
| ENA-78 | A | 0 | 0 |
| TNF-α | 9 | 0 | 11.4 |
| Thrombopoeitin | A | 0 | 0 |
| GCP2/CXCL6 | 6 | 0 | 0 |
| . | Array ratio . | Medium or RBCs, % of blot-positive control . | Pf-IRBCs, % of blot-positive control . |
|---|---|---|---|
| Eotaxin | A* | 0† | 0 |
| GCSF | Marginally up‡ | 0 | 0 |
| GM-CSF | A | 0 | 0 |
| Soluble ICAM-1 | 54 | 0 | 0 |
| IFNγ | A | 0 | 0 |
| IL-10 | A | 0 | 0 |
| IL-15 | No change§ | 0 | 0 |
| IL-1α | A | 0 | 0 |
| IL-1β | Marginally up | 0 | 29.4 |
| IL-3 | A | 0 | 0 |
| IL-4 | A | 0 | 0 |
| IL-5 | A | 0 | 0 |
| IL-6 | 141 | 58.9 | 109.3 |
| IL-7 | A | 0 | 0 |
| IL-8 | 132 | .6 | 99.3 |
| IP-10 | A | 0 | 0 |
| MCP-1 | No change | 70.6 | 69.8 |
| NAP-1A | Not in array | 0 | 0 |
| MIP-1α | A | 0 | 0 |
| GROα | 174 | 0 | 0 |
| PDGF-BB | No change | 0 | 19.6 |
| RANTES | Marginally up | 0 | 24.1 |
| SDF-1 | A | 0 | 0 |
| TGF-β | A | 0 | 0 |
| Fractalkine | A | 0 | 0 |
| TIMP-1 | No change | 0 | 0 |
| ENA-78 | A | 0 | 0 |
| TNF-α | 9 | 0 | 11.4 |
| Thrombopoeitin | A | 0 | 0 |
| GCP2/CXCL6 | 6 | 0 | 0 |
A indicates absent called in more than 75% of gene probes.
0 indicates not detectable at less than 5% of positive control spots.
Marginally up mean fluorescence value of Pf-IRBC samples was nonstatistically higher (secondary analysis) than the control values.
No change mRNA levels remained unchanged between controls and Pf-IRBC samples.
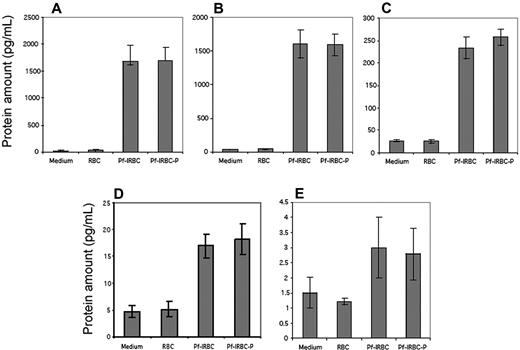
Quantification of inflammatory cytokines in HBMEC culture supernatants. Confluent monolayers of HBMECs were coincubated with Pf-IRBCs or Pf-IRBC-Ps for 6 hours and supernatants were assayed by ELISA. Protein levels of Groα (A) and CCL20 (B) were detected in high range, which is in accordance with their mRNA levels, in contrast to low levels of TNFα (C). Control HBMECs and HBMECs coincubated with RBCs showed very little to no target protein. Data are presented as mean ± SD of duplicate experiments with 2 biologic replicates (n = 4).
Quantification of inflammatory cytokines in HBMEC culture supernatants. Confluent monolayers of HBMECs were coincubated with Pf-IRBCs or Pf-IRBC-Ps for 6 hours and supernatants were assayed by ELISA. Protein levels of Groα (A) and CCL20 (B) were detected in high range, which is in accordance with their mRNA levels, in contrast to low levels of TNFα (C). Control HBMECs and HBMECs coincubated with RBCs showed very little to no target protein. Data are presented as mean ± SD of duplicate experiments with 2 biologic replicates (n = 4).
Quantification of protein expression in cell culture supernatants by BD Cytometric Bead Array (CBA). Confluent monolayers of HBMECs were coincubated with Pf-IRBCs or Pf-IRBC-Ps for 6 hours and supernatants were assayed by CBA (BD Cytometric Bead Array). Control HBMECs and HBMECs coincubated with RBCs showed very little to no protein. Protein levels of IL-6 (A) and IL-8 (B) were detected in high range, which is in accordance with their mRNA levels as detected by microarray experiments. Protein level of RANTES (C) was in the range of 250 pg/mL and again TNF-α (D) levels were in low range. IL-1β (E), which showed no up- or down-regulation in microarray experiments, showed insignificant levels of protein in HBMEC culture supernatants. Data are presented as mean ± SD of duplicate experiments with 2 biologic replicates (n = 4).
Quantification of protein expression in cell culture supernatants by BD Cytometric Bead Array (CBA). Confluent monolayers of HBMECs were coincubated with Pf-IRBCs or Pf-IRBC-Ps for 6 hours and supernatants were assayed by CBA (BD Cytometric Bead Array). Control HBMECs and HBMECs coincubated with RBCs showed very little to no protein. Protein levels of IL-6 (A) and IL-8 (B) were detected in high range, which is in accordance with their mRNA levels as detected by microarray experiments. Protein level of RANTES (C) was in the range of 250 pg/mL and again TNF-α (D) levels were in low range. IL-1β (E), which showed no up- or down-regulation in microarray experiments, showed insignificant levels of protein in HBMEC culture supernatants. Data are presented as mean ± SD of duplicate experiments with 2 biologic replicates (n = 4).
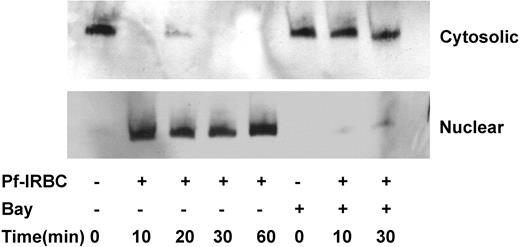
In quiescent HBMECs, p65 NF-κB is localized in cytosolic fraction; however, upon Pf-IRBC exposure it quickly translocates to the nucleus (Figure 6). Detection by Western blot suggests a translocation of more than 90% of cytosolic p65 to the nucleus within 10 minutes of Pf-IRBC exposure, suggesting direct and fast effect of parasite exposure. One-hour preincubation of HBMECs with IκB phosphorylation inhibitor Bay 11-7082 abolishes the Pf-IRBC–induced NF-κB activation.
The kinetics of p65 NF-κB nuclear translocation of Pf-IRBC–activated HBMECs and inhibition by IκB phosphorylation inhibitor Bay 11-7082. Confluent HBMECs were incubated in the absence or presence of Pf-IRBCs (5 × 108/100 × 20-mm culture dishes) for the times indicated. In addition, cells were also pretreated with Bay 11-7082 for 1 hour where indicated. Cytosolic and nuclear proteins (25 μg) were resolved on sodium dodecyl sulfate–polyacrylamide gel electrophoresis and Western blotting was performed.
The kinetics of p65 NF-κB nuclear translocation of Pf-IRBC–activated HBMECs and inhibition by IκB phosphorylation inhibitor Bay 11-7082. Confluent HBMECs were incubated in the absence or presence of Pf-IRBCs (5 × 108/100 × 20-mm culture dishes) for the times indicated. In addition, cells were also pretreated with Bay 11-7082 for 1 hour where indicated. Cytosolic and nuclear proteins (25 μg) were resolved on sodium dodecyl sulfate–polyacrylamide gel electrophoresis and Western blotting was performed.
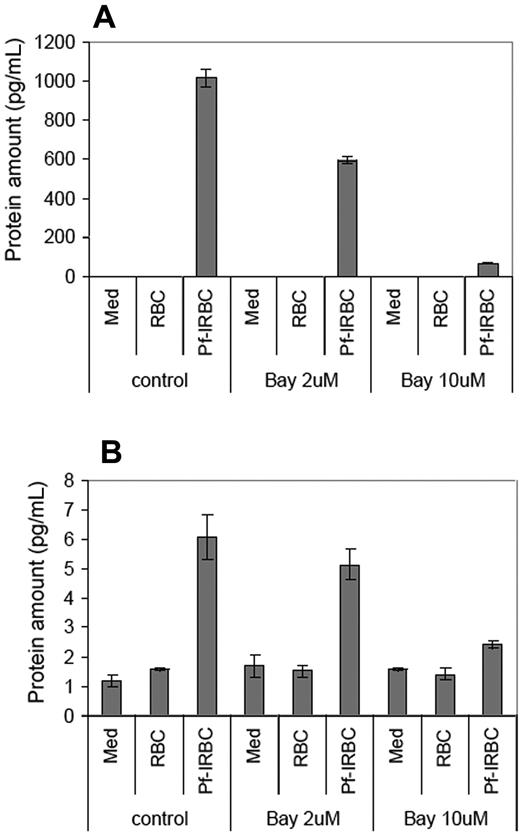
To confirm whether the up-regulation of target genes, especially cytokines, is controlled by NF-κB in HBMECs, we included BAY 11-7082 (2 and 10 μM), a specific inhibitor of IκB phosphorylation, during Pf-IRBC–HBMEC exposure experiments. As determined by ELISA, the levels of CCL20 and TNF-α in the culture supernatant decreased in the presence of NF-κB inhibitor, confirming NF-κB's role in its transcriptional regulation (Figure 7).
NF-κB inhibitor Bay 11-7082 reverses the expression of CCL20 and TNF-α. Confluent monolayers of HBMECs were pretreated with indicated concentrations of Bay 11-7082 for 1 hour, followed by 6 hours of coincubation with Pf-IRBCs. Protein levels of CCL20 (A) and TNF-α (B) were assayed in culture supernatants by ELISA. Data are presented as mean ± SD of duplicate experiments with 3 biologic replicates (n = 6).
NF-κB inhibitor Bay 11-7082 reverses the expression of CCL20 and TNF-α. Confluent monolayers of HBMECs were pretreated with indicated concentrations of Bay 11-7082 for 1 hour, followed by 6 hours of coincubation with Pf-IRBCs. Protein levels of CCL20 (A) and TNF-α (B) were assayed in culture supernatants by ELISA. Data are presented as mean ± SD of duplicate experiments with 3 biologic replicates (n = 6).
Discussion
Malaria-induced brain pathology is known to be associated with sequestration of Pf-IRBCs in microvasculature, resulting in complex cellular and immunomodulator interactions, involving coregulators such as cytokines and adhesion molecules. Our experimental design did not monitor the direct and immediate effects of binding of Pf-IRBCs to HBMECs, for example, through Pf-EMP-1 (P falciparum erythrocyte membrane protein 1), or monitor biomechanical changes, but instead analyzed the responses of the brain endothelium to the approximation of Pf-IRBCs. This was done at 6 hours of exposure because a maximum electrical resistance perturbation (barrier dysfunction) occurred at this time point.10,11 We observed significant up-regulation of more than 200 HBMEC genes and down-regulation of more than 150 genes in response to Pf-IRBC exposure. Functional categorization of differentially regulated genes showed effect on several important biologic pathways. Notable among the up-regulated genes were molecules belonging to the immune system, inflammatory response, signal transduction, cytokine-chemokines, and proapoptotic as well as antiapoptotic proteins. We have previously shown activation of NF-κB and subsequent increased expression of ICAM-1 on HBMEC surface in response to Pf-IRBC exposure.10 With this study, we confirmed the induction of ICAM-1 and up-regulation of NF-κB signaling pathway at the transcript level.
Compared with other reports on malaria-endothelial cell responses, we observed transcriptional changes in our study that appear to be specific for brain endothelium. In a recent study on HUVECs exposed to Pf-IRBCs, Chakravorty et al have observed no apparent induction of ICAM-1 or modulation of RNA transcription by Pf-IRBC alone.25 However, they did observe changes related to proadhesive and immune molecules at the transcriptional level when both low levels of TNF-α and Pf-IRBC were included during HUVEC coincubation experiments. Earlier we have also shown that HUVECs do not induce an increase in surface ICAM-1 expression in response to Pf-IRBCs.10 A previous transcriptional analysis by Kallmann et al of resting brain endothelium compared with resting HUVECs indicated 35 genes solely expressed in brain endothelium versus 20 solely in HUVECs.9 Notably, genes more specific to brain endothelium included those involved in immunoregulation (decorin and IL-6) or growth-supporting properties (TGF-β and brain-derived neurotropic factor), whereas IL-8 was present in both cell types. This disparity in responses with brain endothelium versus HUVECs emphasizes the heterogeneous nature of endothelial cells from different organs26 and the importance of focusing studies to the relevant organ endothelium.
As demonstrated by GO and pathway analysis, the dominant families of genes induced by Pf-IRBCs were the cytokine and chemokine families and genes related to their signaling pathways. In the CCL family, this included high levels of CCL20 and low levels of CCL5 (RANTES). In the CXC (growth-related oncogene) family, significantly elevated chemokines include CXCL1 (GRO-α, KC, MIP-2), CXCL2 (GRO-β, MIP-2β), CXCL3 (GRO-γ), and CXCL8 (IL-8, NAP-1, GCP-1). In addition, IL-6 (interferon beta2) is also largely up-regulated. The up-regulation of CCL20 was more than 700-fold, greatest among all the up-regulated genes. In culture supernatants, the increase in CCL20 protein was also detected. CCL20 is primarily expressed in ectodermal tissue and mucosal sites, to some extent in lymphoid tissues, and in peripheral blood mononuclear cells.27,28 TNF-α has been shown to induce CCL20 secretion and mRNA production in mouse endothelioma cell line bEnd3 and human dermal cells.28 Little is known about the presence and regulation of CCL20 in other tissues, such as brain. Recent studies indicate that CCL20 can mediate recruitment of dendritic cells to the central nervous system in various inflammatory, neurodegenerative conditions and also in parasitic infections.29 Our study showed a slight increase in mRNA for RANTES, whereas release of RANTES from HBMECs was clearly increased as measured by ELISA/multiplex and RayBio arrays. Studies in severe malaria have associated lower serum RANTES with thrombocytopenia.30,31 RANTES is often difficult to study epidemiologically in serum rather than plasma because release from platelets leads to overestimation with blood sample handling.32 A postmortem study on human cases showed increased RANTES and its receptors in the cerebrum and cerebellum by immunohistochemistry and RNA analysis.33 Our results implicate a small role for human brain endothelium in local production.
Raised levels of the inflammatory cytokines have been associated with severe malaria, particularly CM in African children. Cytokines can be derived from several sources, mainly immune cells. However with the brain capillary surface area approximately 20 m2, cytokines generated from activated endothelium can contribute considerably to plasma concentration.34 CM, compared with severe malaria, is associated with serum increases in IL-6,35 which is in accordance with findings in our data. In one study, increases in TNF-α distinguished severe malaria from healthy controls,35 whereas in a similar study, elevated TNF-α in serum distinguished CM from severe malaria.36 We observed moderate level of TNF-α up-regulation at transcript as well as protein level, suggesting brain endothelial cells also contribute to increase in TNF-α during CM. In a recent elegant study, Armah et al directly compared levels of 36 biomarkers in CSF to serum derived from Ghananese patients who died of CM, severe malaria, or no malaria. The authors concluded a general increase in the brain of proinflammatory and proapoptotic factors with a decrease in neuroprotective angiogenic factors.37 In P falciparum malaria, the proinflammatory cytokines may have direct systemic effects or adversely affect outcome by increasing the cytoadherence of infected erythrocytes to venular endothelium through up-regulation of adhesion molecules. Up-regulation of these chemokines and cytokines indeed highlights the active role of the endothelium in pathogenesis of CM.
Biologic process categorization and Ingenuity pathway analysis of differentially regulated genes led us to identify significant up-regulation of 4 NF-κB subunits and several members of its activation cascade. The mammalian NF-κB family consists of 5 members, REL-A (p65), REL-B, c-REL, p105/p50 (NF-κB1), and p100/p52 (NF-κB2).38 The NF-κB complex is composed of dimers formed from these subunits and in unstimulated cells is typically located in the cytoplasm bound to a member of the IκB family of proteins, α, β, and ϵ. All IκBs contain ankyrin repeat, protein-protein interaction domains, a feature shared with the p100 and p105 precursor proteins that can also function in an IκB-like manner and retain their NF-κB partner subunits in the cytoplasm.38 In our studies, we observed that 2 NF-κB inhibitory proteins, IκBα (> 20 fold) and IκBϵ (3-fold) were also up-regulated. Earlier NF-κB pathway has been implicated in malaria host responses including nitric oxide production39 or stimulation by Plasmodium glycosylphosphatidylinositols40 or Plasmodium hemozoin.41 Our laboratory has also demonstrated that induction of endothelial ICAM-1 was linked with nuclear translocation of p65 (REL-A) in response to Pf-IRBC exposure.10 In view of these findings, we screened all the up-regulated genes to identify NF-κB target genes. Including genes associated with the NF-κB pathways, its inhibitors, and activators, we identified more than 70 genes, which were shown earlier to be regulated by NF-κB (Tables 2–3). Interestingly, the average up-regulation for NF-κB target genes was significantly higher (> 30-fold) compared with nontarget genes (< 5 fold). The difference in average fold increases suggests that most of the nontarget genes might be regulated as secondary consequence of NF-κB activation. We also determined the kinetics of NF-κB activation by exposing the confluent HBMECs with Pf-IRBCs for different time points, which was similar for adherent and nonadherent Pf-IRBCs. We observed almost complete nuclear translocation of p65 within 10 minutes of Pf-IRBC exposure, suggesting a direct role for parasite factors in NF-κB activation (Figure 6) Bay 11-7082, a specific inhibitor of IκB phosphorylation, abolished the nuclear translocation of NF-κB and also reduced the secretion of CCL20 and TNF-α in culture supernatant.
How parasite exposure is activating NF-κB is not clear. It has been shown earlier that Plasmodium glycosylphosphatidylinositol and hemozoin can induce NF-κB activation in various cell types. However, purified hemozoin has no effect on expression of ICAM-1 and permeability properties of HBMEC monolayer.10 Our results also rule out the role of Pf-EMP-1–mediated interaction in endothelial NF-κB activation, as trypsinized, low-binding and high-binding Pf-IRBC evoked similar responses from HBMECs.11 Our observations indicate a role for soluble factors released or shed by Pf-IRBCs during coculture in endothelial activation. In recent years, ours as well as other laboratories have reported the role of parasite soluble factors in BBB alteration,11 HBVEC (human brain vascular endothelial cell) apoptosis,42 and activation of HUVECs.43 In view of these observations, further investigation to identify putative parasite soluble factors could logically be a next step forward.
NF-κB target genes could be classified in different functional groups. Those with a primarily inflammatory function were predominant. Genes for monocyte, macrophage, neutrophil, and T-cell chemoattractant chemokines and cytokines (CCL20, RANTES, CXCL1, CXCL2, CXCL3, CXCL6, IL8, IL-6, and TNF-α) were up-regulated consistent with the critical role of the endothelium in promoting leukocyte infiltration. Numerous publications in the past assert that accumulation of Pf-IRBCs, but not leukocytes, is a typical feature of human CM. In contrast, leukocyte involvement alone is considered a hallmark of murine CM. However, the studies in past decade have altered this model, allowing for some overlap as there are several reports of focal sequestration of leukocytes in brain microvasculature of human adults44 as well as pediatric CM patients.45 Our results strongly demonstrate potential for leukocyte recruitment at the site of parasite sequestration and/or after sequestration. Although the consensus is that there is no monocyte transmigration into the brain in CM (we do not see increase in monocyte chemotactic protein 1), it is still remains to be seen whether other intravascular sequestered immune cells, for example, dendritic cells, can potentially transmigrate toward the brain side during CM in a function of immune surveillance. This needs to be addressed using dendritic cell–specific markers in CM. The entry of leukocytes into the sites of infection is a multistep process; selectins on the vessels capture them and ICAM-1 and vascular cell adhesion molecule 1 firmly attach them to the endothelium.46 In our experiments, we observed up-regulation of ICAM-1 but not of any other selectin molecule or vascular cell adhesion molecule 1, which might explain the absence of significant leukocyte infiltration during CM.
A significant portion of the endothelial response was found to be directed toward countering the damaging effects of the malaria parasite exposure. Gene expression of a number of metallothionein 1 (MT1) subtypes—namely MT1K, MT1X, MT1F, MT1G, and MT1P2 and one metallothionein 2 (MT2) subtype MT2A—was significantly up-regulated (supplemental Table 1). Metallothioneins (MTs) are a family of metal-binding proteins that have been proposed to participate in a cellular defense against metal toxicity and free radicals. MTs can also protect the central nervous system by reducing the immune cell activation and recruitment, thereby minimizing the production of inflammatory cytokines.47 We also observed the up-regulation of superoxide dismutase (SOD), which provides the first line of defense against oxidative stress by catalyzing the dismutation of superoxide anion (O2−·) into hydrogen peroxide.48 Earlier it was shown that transient supplementation of SOD1 protects endothelial cells against P falciparum–induced oxidative stress and apoptosis and decreases the Pf-IRBC cytoadherence through a down-regulation of ICAM-1 expression.49
Another set of genes implicated in CM pathogenesis that show vigorous host responses to Pf-IRBC exposure are apoptosis-related genes. We observed significant alterations in the expression of endothelial genes involved in regulation of both proapoptotic and antiapoptotic pathways. We did not observed the brain endothelium going into apoptosis (data not shown). Several genes involved in regulation of apoptosis were up-regulated (eg, CFLAR, TRAIL receptor, RIPK2, GADD34, PUMA/JFY1); however, we did not observe up-regulation of any of the caspase genes in contrast to other models of CM. The caspase family of cysteine proteases includes proteases that initiate apoptosis and proteases that can act as executioners. Many of the NF-κB target genes belonging to antiapoptotic genes (eg, BCL10, BCL2L1, BCL2A1, BIRC3, BIRC5) were up-regulated, suggesting that NF-κB, besides playing a role in proinflammatory response, is also involved in defense against apoptosis.
In conclusion, the present study demonstrates that Pf-IRBC exposure to HBMECs induces a predominantly proinflammatory response that is mediated by activation of NF-κB. Although many have postulated that the ligand receptor interaction, for example, through Pf-EMP-1, is the primary driver of CM pathology, our data suggest that parasites cause the endothelial effects regardless of the level of high-affinity binding. However, in vivo cytoadherence is important to create the circumstances of approximation of the parasite-infected erythrocyte to the endothelium. Another distinction highlighted here is a separation from potential role of soluble Plasmodium factors from that of cytoadherence. Activation of NF-κB may be playing a major role in cytoprotection as well, as significant alteration in expression of apoptotic genes does not translate in functional apoptosis, probably due to the balancing effect of NF-κB–regulated antiapoptotic pathway. From a therapeutic point of view, inhibition of NF-κB has earlier been suggested as a possible approach to prevent proinflammatory consequences of endothelial cell activation. To achieve this goal, a method to modulate NF-κB would be a researchable option.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Anne Jedlicka and Jason Bailey at the Malaria Research Institute Gene Array Core Facility for performing RNA extraction, processing, and Affymetrix Chip hybridization.
This publication was made possible by UL1 RR 025005 from the National Center for Research Resources (NCRR) for provision of human erythrocytes. The expertise, facilities, and instrumentation for Affymetrix GeneChip experimentation and analyses are provided and supported by the Johns Hopkins University Bloomberg School of Public Health Malaria Research Institute. The data discussed in this publication have been deposited in National Center for Biotechnology Information's (NCBI's) Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) and are accessible through GEO Series accession number GSE9861.50
Authorship
Contribution: A.K.T., M.F.S., and D.J.S. conceived and designed the experiments; A.K.T. performed the experiments; W.S. and V.S. focused on array data analysis; all authors analyzed the entire data set; A.K.T., M.F.S., and D.J.S. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David J. Sullivan Jr, Johns Hopkins Bloomberg School of Public Health, Molecular Microbiology & Immunology And also Infectious Diseases School of Medicine, 615 N Wolfe St, E5628, Baltimore, MD 21205; e-mail: dsulliva@jhsph.edu.