Abstract
The factor H–related protein family (CFHR) is a group of minor plasma proteins genetically and structurally related to complement factor H (fH). Notably, deficiency of CFHR1/CFHR3 associates with protection against age-related macular degeneration and with the presence of anti-fH autoantibodies in atypical hemolytic uremic syndrome (aHUS). We have developed a proteomics strategy to analyze the CFHR proteins in plasma samples from controls, patients with aHUS, and patients with type II membranoproliferative glomerulonephritis. Here, we report on the identification of persons carrying novel deficiencies of CFHR1, CFHR3, and CFHR1/CFHR4A, resulting from point mutations in CFHR1 and CFHR3 or from a rearrangement involving CFHR1 and CFHR4. Remarkably, patients with aHUS lacking CFHR1, but not those lacking CFHR3, present anti-fH autoantibodies, suggesting that generation of these antibodies is specifically related to CFHR1 deficiency. We also report the characterization of a novel CFHR1 polymorphism, resulting from a gene conversion event between CFH and CFHR1, which strongly associates with aHUS. The risk allotype CFHR1*B, with greater sequence similarity to fH, may compete with fH, decreasing protection of cellular surfaces against complement damage. In summary, our comprehensive analyses of the CFHR proteins have improved our understanding of these proteins and provided further insights into aHUS pathogenesis.
Introduction
The complement system is a major component of the innate immune response to infection, and it is also involved in immunocomplex removal and the destruction of damaged or anomalous autologous cells. Improper complement activation is prevented by several plasma and membranes proteins, which perform a regulatory, inhibitory function. Complement factor H (fH) is the main inhibitor of the alternative pathway of the complement system.1 Deficiency or dysfunction of fH is associated with increased susceptibility to infections and to chronic diseases such as atypical hemolytic uremic syndrome (aHUS), type II membranoproliferative glomerulonephritis/dense deposits disease (MPGN2/DDD), and age-related macular degeneration (AMD).2,3
The fH gene, CFH, spans approximately 100 kb (kilobases) in the RCA (regulators of complement activation) gene cluster on human chromosome 1q32.4 fH is a single polypeptide chain glycoprotein of 155 kDa composed of 20 repetitive units of 60 amino acids named short consensus repeats (SCRs), which are arranged in a continuous fashion. Alternative splicing of CFH generates a shorter form of 43 kDa named fHlike-1 (CFHL1) consisting of SCR1 to SCR7 plus an additional hydrophobic tail of 4 amino acids.5,6 In close proximity to CFH there are 5 genes coding for proteins showing sequence and structural homology to fH that are known as fH-related proteins (CFHRs).7 CFH and the 5 CFHR genes (CFHR1-5) lie in a head-to-tail arrangement and are thought to have originated by successive duplication events within the RCA gene cluster.8
The CFHR proteins were first identified by Western blot analysis of human plasma with polyclonal anti-fH antibodies.9 They are all composed of SCR domains showing different degrees of identity with the homologous domains in fH. The concentrations of CFHR proteins in plasma are much lower than that of fH, and their biologic function is currently unknown. Binding to the physiologic fH ligand C3b has been reported for CFHR1, CFHR3, CFHR4, and CFHR5,10-12 and C3 convertase regulatory activity has been detected in CFHR5.13 Most recently, CFHR1 has been shown to inhibit the C5 convertase and the membrane attack complex formation,14 suggesting a sequential and complementary role for fH and CFHR1 in complement regulation.
The CFH-CFHR1-5 genomic region includes several large genomic duplications involving different exons of the CFH and CFHR1-5 genes, which have mediated genomic rearrangements through mechanisms of gene conversion and nonhomologous recombination. Several examples of gene conversion events between exon 23 of CFH and the homologous exon 6 of CFHR1 have been documented recently.8,15 Similarly, there is robust evidence of major rearrangements in the CFH-CFHR1-5 gene region that result in the deletion of the CFHR1 and CFHR3 genes (ΔCFHR1-CFHR3)16,17 and, occasionally, in the generation of CFH::CFHR1 hybrid genes.18 These CFH-CFHR1-5 rearrangements can be easily identified by MLPA (multiplex ligation-dependent probe amplification) technologies18 or by Western blot in the case of homozygote carriers.17 Chromosomes carrying both the deletion of the CFHR1 and CFHR3 genes and a CFH::CFHR1 hybrid gene are rare and are specifically associated with aHUS.18 In contrast, the deletion of the CFHR1 and CFHR3 genes (ΔCFHR1-CFHR3) without rearrangements in CFH is a common genetic polymorphism that is associated with a lower risk of AMD16,19 and with increased risk of aHUS because of the generation of anti-fH autoantibodies.17,20,21 The rearrangement that results in ΔCFHR1-CFHR3 is not a recurring phenomenon but a single event that became fixed in the human population a long time ago.19 Additional genetic alterations within the CFH-CFHR1-5 region are expected, and they probably have pathologic implications. In this context, the characterization of the products of CFH and CFHR1-CFHR5 genes would be most helpful.
We have characterized the CFHR proteins present in plasma samples from control persons, patients with aHUS, and patients with MPGN2/DDD by Western blot and have developed a strategy based on the analysis of partially purified CFHR proteins by 2-dimensional gel electrophoresis that increases resolution of these proteins and shows novel structural features. Our results improve our understanding of the genetic variability at the CFHR region. They suggest a specific relationship between the deficiency of CFHR1 and the generation of anti-fH autoantibodies and show a novel polymorphism in CFHR1 that confers increased risk to aHUS, thus providing further insights into the pathogenesis of aHUS.
Methods
Samples from patient and control persons
Blood samples were drawn from 151 patients with a clinical diagnosis of aHUS and from 13 patients with MPGN2/DDD. Serum and EDTA-plasma aliquots were stored immediately at −80°C until used. Peripheral blood leukocytes were used to prepare genomic DNA. All samples were obtained in the context of research studies on HUS and MPGN2/DDD that have the approval of the Ethics Committee from the University Hospital La Paz. Written informed consent was obtained from patients or their relatives in accordance with the Declaration of Helsinki. A previously described cohort of 141 patients with AMD22 was also included in some genetic studies as a disease control group.
Blood samples were also drawn from a total of 105 control persons (59 women and 46 men). Sixty-eight controls were healthy adult volunteers (aged 21-73 years; median, 34 years); 37 controls were anonymous children (aged 2-16 years; median, 6 years) who were about to undergo posttraumatic surgery in University Hospital La Paz but did not present signs of any infectious-related condition. Serum, EDTA-plasma, and DNA were obtained from these control samples, which were used for Western blot and genetic analyses. DNA samples from other 125 control persons22 were also available for some genetic studies.
Detection of fH autoantibodies in serum samples
Factor H autoantibodies in serum samples were detected with an enzyme-linked immunosorbent assay following the method previously described by Dragon-Durey et al.23 PBS (phosphate-buffered saline; pH 7.2) was used as the coupling buffer, PBS-Tween (0.1% Tween 20) was used for washing and blocking, and PBS-BSA (0.1% BSA) was used for preparing serum dilutions. Ninety-six–-well microtiter plates (Immuno Plates MaxiSorp; Nalge Nunc International) were coated overnight at 4°C with 0.2 μg of in-house purified fH24 in 100 μL of PBS. After washing and blocking, serial dilutions (1/100, 1/200, 1/400) of serum samples in PBS-BSA were added in duplicate and incubated at 4°C overnight. The plates were washed again, and 100 μL of HRP-goat anti–human IgGs (Nordic Immunology) diluted 1/2500 in PBS-Tween was added. The enzymatic reaction was developed with the peroxidase substrate ABTS (Roche Diagnostics GmbH). Absorbance values at 405 nm were compared with a reference positive sample, provided by Dr Marie Agnes Dragon-Durey (Hôpital Georges Pompidou, Paris, France), that had been given an arbitrary titer of 2000 AU/mL. Samples above 150 AU/mL (mean + 2 SDs obtained from 54 control samples) were considered positive.
Heparin-sepharose chromatography
Factor H, CFHL1, and CFHR proteins were partially purified by affinity chromatography on heparin-sepharose (GE Healthcare Bio-Sciences). Serum samples were dialyzed at 4°C against 20mM Tris-HCl pH 7.5, containing 50mM NaCl (equilibrating buffer). Dialyzed sera were incubated overnight at 4°C with heparin-sepharose beads. Beads were then centrifuged, and the supernatant was stored for further analysis. After several washes in equilibrating buffer, bound proteins were eluted with 20mM Tris-HCl pH 7.5, 500mM NaCl.
Two-dimensional electrophoresis
Isoelectrofocusing (IEF) was used in the first dimension, and standard SDS–polyacrylamide gel electrophoresis (PAGE) was used in the second dimension. For IEF, immobilized pH gradient (IPG) strips of 7 cm and different pH linear range were used (pH 3-10 or pH 4-7, ImmobilineDrystrips, GE Healthcare; pH 5-8, ReadyStrips, Bio-Rad). Samples were resuspended in IEF buffer (0.5% IPG buffer, 8 M urea, 2% Chaps, 0.002% bromophenol blue) and loaded onto the IPG strips manually. IPG strips were rehydrated at 20°C for 15 hours at 50 V, for 1 hour at 100 V, and for 1 hour at 1000 V in an IPGphor I horizontal apparatus (GE Healthcare). IEF was carried out at 5000 V for 5 hours 30 minutes. IPG strips were equilibrated in 50mM Tris-HCl, pH 6.8 buffer, containing 6M urea, 87% glycerol, 2% SDS, 0.002% bromophenol blue. For the second dimension, equilibrated IPG strips were placed on top of a 5% stacking–10% resolving polyacrylamide minigel, and electrophoresed in the following conditions: 50 mA/30 minutes plus 75 mA/30 minutes plus 100 mA/90 minutes.
After electrophoresis, gels were either stained with Coomassie Brilliant Blue G-250 (USB Corporation) for mass spectrometry (Proteomics Unit, Universidad Complutense, Madrid, Spain) or processed for Western blot analysis.
Western blot analysis
Proteins were transferred to nitrocellulose membranes at 100 V for 1 hour in a Mini Trans-Blot Cell apparatus (Bio-Rad). Membranes were blocked in ECL (electrochemiluminescence) Advance blocking agent (GE Healthcare) and incubated with primary antibodies for 2 hours. HRP-conjugated goat anti–rabbit or anti–mouse IgG (Bio-Source; Invitrogen) was used as secondary antibodies, and the membranes were developed with a chemiluminescent substrate (ECL Advance Kit; GE Healthcare) following the manufacturer's instructions.
Description of primary antibodies
Polyclonal antibodies were raised in rabbits. Antibody αCFHR1 was generated in the laboratory and recognizes fH, CFHL1, and the 2 isoforms of CFHR1: CFHR1α (37 kDa) and CFHR1β (43 kDa), containing 1 and 2 carbohydrate molecules, respectively.25 Antibody αCFHR3/4 was provided by Prof Peter Zipfel (Hans-Knöll Institute, Jena, Germany). This antibody recognizes fH, CFHL1, CFHR3, and the 2 products of the CFHR4 gene26 : CFHR4A (86 kDa) and CFHR4B (46 kDa). Antibody αCFHR5 recognizes CFHR5 (65 kDa) and, to a lesser extent, fH, CFHR1α, and CFHR1β; this was provided by Dr Jennifer McRae (St Vincent's Health, Melbourne, Australia). The monoclonal antibody 35H9 recognizes fH and CFHL1 and has been described previously.27 Working dilutions of the antibodies were 1:10 000 (MoAb 35H9), 1:50 000 (αCFHR1), 1:30 000 (αCFHR3/4), and 1:100000 (αCFHR5).
Genetic studies
Rearrangements in the CFH-CFHR1-5 genomic region were analyzed by MLPA. Copy number variations of CFHR1 and CFHR3 were determined by using the P236 A1 ARMD mix 1 from MRC-Holland. Because this mix does not include probes for the CFHR4 gene, another MLPA reaction was designed with probes for specific exons of CFH and the 5 CFHR genes. These probes were synthesized by Sigma-Aldrich and also included control probes for the BCAP31 gene, located on chromosome X, and for the MORF4L1 gene, located on chromosome 15. The hybridization sequences are shown in Table 1. Each probe pair hybridized to sequences immediately adjacent to the target region; the 3′ probes (R) were 5′-phosphorylated to provide a free phosphate for the ligation reaction. A nonspecific sequence between the hybridizing nucleotides and the nucleotides recognizing the primers was introduced to adjust the length of the fragments. The reagents for the MLPA reactions were purchased from MRC-Holland, and the reaction was carried out following the manufacturer's protocol. Two microliters of the final product plus 0.2 μL of internal standard (500LIZ) and 10 μL of deionized formamide (ABI no. 4311320) were analyzed in an ABI-3730 DNA Analyzer (Applied Biosystems).
Probes for MLPA analysis of the CFH-CFHR1-5 region
| Probe . | Hybridizing sequence (5′ to 3′) . |
|---|---|
| CFHex5L | GGGAATACCATTTTGGACAAGCAGTACG |
| CFHex5R | GTTTGTATGTAACTCAGGCTACAAGATTGAAGG |
| CFHex23L | TGGACAGCCAAACAGAAGCTTTATTC |
| CFHex23RCFHR1ex6R | GAGAACAGGTGAATCAGYTGAATTTG |
| CFHR1ex6L | GGACAGCCAAACAGAAGCTTTATTT |
| CFHR1ex2L | GTAATAGAAAACTTCCCCTGTAGGAACC |
| CFHR1ex2R | TGGGAAAATGGCTTATATTTTTCTTCATC |
| CFHR2ex5L | CCAACAAAATCTCATTCATTTCGAGCAAT |
| CFHR2ex5R | GTGTCAGAATGGGAAACTGGTATATCCCAG |
| CFHR3ex6L | AGCATTGTTACCCTAAATGTATGTCCAACTTC |
| CFHR3ex6R | CACTTTTCCACTTCTCACTCTTATGGTCTCA |
| CFHR4ex4L | GCACACACTGATTGGTAAATTTTATCCCTACAATG |
| CFHR4ex4R | GGACTTTCTTAGTTGAGTTGTGCATCGTATG |
| CFHR5ex5L | CGGGCCTAAGAAAATACAATGTGTGGATG |
| CFHR5ex5R | GAGAATGGACAACTTTACCCACTTGTGTTG |
| MORF4L1ex9L | GATGCTCCATAACTTAGAATGG |
| MORF4L1ex9R | GGTTACATCGCAATAAACCCAG |
| BCAP31ex6L | ACAGGAGCCTGAAGGCTGACCT |
| BCAP31ex6R | GCAGAAGCTAAAGGACGAGCTGG |
| Probe . | Hybridizing sequence (5′ to 3′) . |
|---|---|
| CFHex5L | GGGAATACCATTTTGGACAAGCAGTACG |
| CFHex5R | GTTTGTATGTAACTCAGGCTACAAGATTGAAGG |
| CFHex23L | TGGACAGCCAAACAGAAGCTTTATTC |
| CFHex23RCFHR1ex6R | GAGAACAGGTGAATCAGYTGAATTTG |
| CFHR1ex6L | GGACAGCCAAACAGAAGCTTTATTT |
| CFHR1ex2L | GTAATAGAAAACTTCCCCTGTAGGAACC |
| CFHR1ex2R | TGGGAAAATGGCTTATATTTTTCTTCATC |
| CFHR2ex5L | CCAACAAAATCTCATTCATTTCGAGCAAT |
| CFHR2ex5R | GTGTCAGAATGGGAAACTGGTATATCCCAG |
| CFHR3ex6L | AGCATTGTTACCCTAAATGTATGTCCAACTTC |
| CFHR3ex6R | CACTTTTCCACTTCTCACTCTTATGGTCTCA |
| CFHR4ex4L | GCACACACTGATTGGTAAATTTTATCCCTACAATG |
| CFHR4ex4R | GGACTTTCTTAGTTGAGTTGTGCATCGTATG |
| CFHR5ex5L | CGGGCCTAAGAAAATACAATGTGTGGATG |
| CFHR5ex5R | GAGAATGGACAACTTTACCCACTTGTGTTG |
| MORF4L1ex9L | GATGCTCCATAACTTAGAATGG |
| MORF4L1ex9R | GGTTACATCGCAATAAACCCAG |
| BCAP31ex6L | ACAGGAGCCTGAAGGCTGACCT |
| BCAP31ex6R | GCAGAAGCTAAAGGACGAGCTGG |
Reverse probes (R) were 5′-phosphorylated.
All CFHR1 and CFHR3 exons were amplified by polymerase chain reaction (PCR) with the use of specific oligonucleotides (Table 2). Direct sequencing of the PCR products was performed with the use of the BigDye Terminator Version 1.1 sequencing kit (Applied Biosystems) on an ABI Prism 3100-Avant Genetic Analyzer.
Oligonucleotides for direct sequencing of CFHR1 and CFHR3 genes
| Exon . | Domain . | Forward primer (5′ to 3′) . | Reverse primer (5′ to 3′) . | Size, bp . |
|---|---|---|---|---|
| CFHR1ex1 | SP | GGACTTTACTAAACTAGCTTCCAG | GCAACTTAGAGGATGGAGAG | 400 |
| CFHR1ex2 | SCR1 | TTATGTTATTTTCCCAGCAACAT | AATGACATCCATTTAATGAACAGA | 248 |
| CFHR1ex3 | SCR2 | AAGCGCAGAGATTACCAGAG | GATAACAGCATATGAGAGAACAG | 330 |
| CFHR1ex4 | SCR3 | CGTCTTGAAACATATTTGTAACTGTATT | GGTCGAATCTTTCTCAAAAATAATAC | 460 |
| CFHR1ex5 | SCR4 | TGTATTTTGATTTGCTCTCACAAT | GATTATTTTGTTACCAACAGCA | 314 |
| CFHR1ex6 | SCR5 | ATTTAAATCAATATGATGTTTTTACATAGTC | CAGAAATAAAGTCTGAAAAATTGCA | 703 |
| CFHR3ex 1 | SP | CTAGCTTCATGGTAGTGCAC | TTTAAGAGGGAAGCTGAGTG | 220 |
| CFHR3ex 2 | SCR1 | AAATGTTTGAGAGAAGGTGATATC | TTTTTCATATGATTTTCAAGAACTC | 472 |
| CFHR3ex 3 | SCR2 | CATTTACTTTATTTATTTATCATTGCTATG | TCTGAGACTGTCGTCCG | 282 |
| CFHR3ex 4 | SCR3 | CTTGTTCCCTCCTATAAAAGAAC | GCAACTAACATATTTTGCTGATAC | 547 |
| CFHR3ex 5 | SCR4 | TTGAAAATGCAGATGTCTTCC | GAACTCCTGACCTCATGG | 604 |
| CFHR3ex 6 | SCR5 | CATTTTATTTGCTCATGAAAGAG | TTCATTGATAAGAAGTCCAATATAAGA | 507 |
| Exon . | Domain . | Forward primer (5′ to 3′) . | Reverse primer (5′ to 3′) . | Size, bp . |
|---|---|---|---|---|
| CFHR1ex1 | SP | GGACTTTACTAAACTAGCTTCCAG | GCAACTTAGAGGATGGAGAG | 400 |
| CFHR1ex2 | SCR1 | TTATGTTATTTTCCCAGCAACAT | AATGACATCCATTTAATGAACAGA | 248 |
| CFHR1ex3 | SCR2 | AAGCGCAGAGATTACCAGAG | GATAACAGCATATGAGAGAACAG | 330 |
| CFHR1ex4 | SCR3 | CGTCTTGAAACATATTTGTAACTGTATT | GGTCGAATCTTTCTCAAAAATAATAC | 460 |
| CFHR1ex5 | SCR4 | TGTATTTTGATTTGCTCTCACAAT | GATTATTTTGTTACCAACAGCA | 314 |
| CFHR1ex6 | SCR5 | ATTTAAATCAATATGATGTTTTTACATAGTC | CAGAAATAAAGTCTGAAAAATTGCA | 703 |
| CFHR3ex 1 | SP | CTAGCTTCATGGTAGTGCAC | TTTAAGAGGGAAGCTGAGTG | 220 |
| CFHR3ex 2 | SCR1 | AAATGTTTGAGAGAAGGTGATATC | TTTTTCATATGATTTTCAAGAACTC | 472 |
| CFHR3ex 3 | SCR2 | CATTTACTTTATTTATTTATCATTGCTATG | TCTGAGACTGTCGTCCG | 282 |
| CFHR3ex 4 | SCR3 | CTTGTTCCCTCCTATAAAAGAAC | GCAACTAACATATTTTGCTGATAC | 547 |
| CFHR3ex 5 | SCR4 | TTGAAAATGCAGATGTCTTCC | GAACTCCTGACCTCATGG | 604 |
| CFHR3ex 6 | SCR5 | CATTTTATTTGCTCATGAAAGAG | TTCATTGATAAGAAGTCCAATATAAGA | 507 |
Results
Identification of CFHR protein deficiencies by Western blot
Serum samples from 115 control persons, 149 patients with aHUS, and 13 patients with MPGN2/DDD were analyzed by SDS-PAGE and Western blot with 3 polyclonal antibodies that recognize fH, its alternative splicing product, CFHL1, and the CFHR1-5 proteins. As illustrated in Figure 1, 4 banding patterns showing different CFHR proteins deficiencies were observed: deficiency of both CFHR1 and CFHR3 (DefR1R3), isolated deficiency of CFHR1 (DefR1), isolated deficiency of CFHR3 (DefR3), and deficiency of both CFHR1 and CFHR4A (DefR1R4A). Figure 1 also shows the normal, most common situation in which all CFHR proteins are present. The frequency of these complete CFHR deficiencies in control persons, patients with aHUS, and patients with MPGN2/DDD is shown in Table 3. In agreement with previous reports,17,20,21 we found that DefR1R3, present in approximately 2% of healthy controls, is associated with an increased risk of aHUS (12 of 149 vs 2 of 115; P = .026; OR = 4.95; 95% CI, 1.1-22.6) and is particularly elevated among patients with autoantibodies against fH (3 of 7 patients; see Table 7). Isolated deficiency of CFHR1 (DefR1) or CFHR3 (DefR3), as well as combined CFHR1/CFHR4A deficiency (DefR1R4A), were found only among patients with aHUS and patients with MPGN2/DDD.
Identification of CFHR proteins deficiencies in serum samples. Serum samples from control persons, patients with aHUS, and patients with GN were analyzed by SDS-PAGE and Western blot analysis by using 3 different antibody preparations (A-C) that recognize fH and CFHR proteins. Labels on the top of each gel refer to the 5 CFHR patterns observed: the most frequent pattern in which all the CFHR proteins analyzed are present (Normal), and 4 additional patterns showing different CFHR deficiencies (DefR1R3, DefR1, DefR1R4A, and DefR3). The serum samples chosen to illustrate the 5 patterns correspond to the control sample C1 and to the patients with aHUS H13, H119, H177, and H123, respectively. The position and size (kDa) of the molecular weight markers are shown to the left of gel A. The lane showing the DefR3 pattern in panel C has been repositioned.
Identification of CFHR proteins deficiencies in serum samples. Serum samples from control persons, patients with aHUS, and patients with GN were analyzed by SDS-PAGE and Western blot analysis by using 3 different antibody preparations (A-C) that recognize fH and CFHR proteins. Labels on the top of each gel refer to the 5 CFHR patterns observed: the most frequent pattern in which all the CFHR proteins analyzed are present (Normal), and 4 additional patterns showing different CFHR deficiencies (DefR1R3, DefR1, DefR1R4A, and DefR3). The serum samples chosen to illustrate the 5 patterns correspond to the control sample C1 and to the patients with aHUS H13, H119, H177, and H123, respectively. The position and size (kDa) of the molecular weight markers are shown to the left of gel A. The lane showing the DefR3 pattern in panel C has been repositioned.
Patterns of CFHR proteins as observed by Western blot analysis
| Pattern . | Controls (n = 115) . | aHUS (n = 149) . | MPGN2/DDD (n = 13) . |
|---|---|---|---|
| Normal | 113 | 133 | 9 |
| Def R1R3 | 2 | 12 | 2 |
| Def R1 | 0 | 1 | 1 |
| Def R3 | 0 | 2 | 1 |
| Def R1R4A | 0 | 1 | 0 |
| Pattern . | Controls (n = 115) . | aHUS (n = 149) . | MPGN2/DDD (n = 13) . |
|---|---|---|---|
| Normal | 113 | 133 | 9 |
| Def R1R3 | 2 | 12 | 2 |
| Def R1 | 0 | 1 | 1 |
| Def R3 | 0 | 2 | 1 |
| Def R1R4A | 0 | 1 | 0 |
Genetic analysis of CFHR deficiencies
Deficiency of CFHR1/CFHR3.
Genetic studies using specific MLPA probes showed that all persons presenting the DefR1R3 pattern were homozygous for the 84-kb long genomic deletion (ΔCFHR1-CFHR3) previously described by Zipfel et al.17 Analysis of this deletion was also carried out in additional control samples, as well as in a previously described cohort of patients with AMD.22 The results are shown in Table 4. In agreement with previous observations,16,19 we found that the ΔCFHR1-CFHR3 deletion is a protective factor for AMD. However, the ΔCFHR1-CFHR3 allele frequencies show no differences between control persons, patients with aHUS, or patients with MPGN2/DDD; nonetheless, there is an excess of ΔCFHR1-CFHR3 homozygotes in the aHUS cohort that significantly deviates from the expected value estimated from the ΔCFHR1-CFHR3 allele frequency. Thus, the aHUS population is not in Hardy-Weinberg equilibrium for the ΔCFHR1-CFHR3 polymorphism, most probably because of the presence of patients with fH autoantibodies (see “Discussion”).
Allelic frequency of ΔCFHR1-CFHR3 in patients and controls
| . | n . | ΔCFHR1-CFHR3 . | OR (95% CI) . | Homozygotes ΔCFHR1-CFHR3 observed vs expected . | Hardly-Weinberg equilibrium . |
|---|---|---|---|---|---|
| Controls | |||||
| Aged 0-30 y | 135 | 24.1 | NS | 7 vs 8 | NS |
| Aged 30-60 y | 43 | 25.58 | NS | 2 vs 3 | NS |
| Aged older than 60 y | 92 | 24.72 | NS | 6 vs 6 | NS |
| Totals | 269 | 24.53 | NS | 15 vs 16 | NS |
| AMD patients | 141 | 9,6 | 0.29 (0.181-0.475)* | 2 vs 1 | NS |
| aHUS patients | 137 | 21.12† | NS | 14 vs 6‡ | 0.00015 |
| MPGN2 patients | 11 | 18 | NS | 0 vs 0 | NS |
| . | n . | ΔCFHR1-CFHR3 . | OR (95% CI) . | Homozygotes ΔCFHR1-CFHR3 observed vs expected . | Hardly-Weinberg equilibrium . |
|---|---|---|---|---|---|
| Controls | |||||
| Aged 0-30 y | 135 | 24.1 | NS | 7 vs 8 | NS |
| Aged 30-60 y | 43 | 25.58 | NS | 2 vs 3 | NS |
| Aged older than 60 y | 92 | 24.72 | NS | 6 vs 6 | NS |
| Totals | 269 | 24.53 | NS | 15 vs 16 | NS |
| AMD patients | 141 | 9,6 | 0.29 (0.181-0.475)* | 2 vs 1 | NS |
| aHUS patients | 137 | 21.12† | NS | 14 vs 6‡ | 0.00015 |
| MPGN2 patients | 11 | 18 | NS | 0 vs 0 | NS |
NS indicates not significant.
P < .001.
The ΔCFHR1-CFHR3 allele frequency is not increased in aHUS compared with controls (21.12 vs 24.53).
The frequency of ΔCFHR1-CFHR3 homozygotes in aHUS is significantly higher than that expected from the ΔCFHR1-CFHR3 allele frequency. There is also an increase in the frequency of ΔCFHR1-CFHR3 homozygotes in aHUS compared with controls (14 of 137 compared with 15 of 269), which does not reach statistical significance probably because of the size of our cohorts.
Novel CFHR deficiencies.
Analysis of the ΔCFHR1-CFHR3 deletion was also carried out in the 6 patients presenting the novel CFHR deficiency patterns (DefR1, DefR3, and DefR1R4A). Additional MLPA analyses within the CFHR1-5 subregion of the RCA gene cluster and direct sequencing of CFHR1 and CFHR3 genes were also performed in these patients (see “Genetic studies” for details). These studies identified novel genetic alterations associated with the CFHR deficiencies that are summarized in Table 5. The 5 patients presenting CFHR1 or CFHR3 deficiency carry ΔCFHR1-CFHR3 in heterozygosis and therefore lack one CFHR1 allele and one CFHR3 allele. Sequencing analyses of the CFHR1 gene showed a heterozygous mutation in patient H119 that resulted in the substitution of a serine for a stop codon at position 160 in the SCR3 domain (c.479C > A; Ser160Stop; Figure 2A). This is a null mutation that generates a truncated CFHR1 protein that is probably not expressed. Similarly, sequencing analyses of all the exons and the promoter region of the CFHR3 gene identified a heterozygous mutation in patient H123 that results in the deletion of 2 nucleotides in exon 6 (c.839_840delTA). This deletion causes a frameshift mutation (p.I280KfsX6) that generates a stop codon 6 amino acids downstream of position 280, at the beginning of the SCR5 domain (Figure 2B). Mutation c.839_840delTA is therefore a null mutation that generates a truncated CFHR3 protein most probably not expressed. No mutations in CFHR3 were found in patients H150 and GN11.
Genetic alterations in patients with novel CFHR deficiencies
| Patient . | Deficiency . | ΔCFHR1-CFHR3 . | ΔCFHR1-CFHR4 . | CFHR1 mutation . | CFHR3 mutation . |
|---|---|---|---|---|---|
| H119* | DefR1 | Heterozygosis | No | Heterozygosis | No |
| GN9 | DefR1 | Heterozygosis | Heterozygosis | No | No |
| H177* | DefR1R4A | No | Homozygosis | No | No |
| H123 | DefR3 | Heterozygosis | No | No | Heterozygosis |
| H150 | DefR3 | Heterozygosis | No | No | No |
| GN11 | DefR3 | Heterozygosis | No | No | No |
| Patient . | Deficiency . | ΔCFHR1-CFHR3 . | ΔCFHR1-CFHR4 . | CFHR1 mutation . | CFHR3 mutation . |
|---|---|---|---|---|---|
| H119* | DefR1 | Heterozygosis | No | Heterozygosis | No |
| GN9 | DefR1 | Heterozygosis | Heterozygosis | No | No |
| H177* | DefR1R4A | No | Homozygosis | No | No |
| H123 | DefR3 | Heterozygosis | No | No | Heterozygosis |
| H150 | DefR3 | Heterozygosis | No | No | No |
| GN11 | DefR3 | Heterozygosis | No | No | No |
H119, H177, H123, H150 are patients with aHUS. GN9 and GN11 are patients with MPGN2/DDD.
Patient with aHUS with anti-fH autoantibodies.
Genetic analysis of CFHR1 and CFHR3 genes. Electropherograms showing the mutations identified in patients with aHUS (A) H119 (CFHR1 gene) and (B) H123 (CFHR3 gene). The 2 mutations were found in heterozygosis. The mutation in patient H123 could also be interpreted as c.838_839delAT (p.I280KfsX6). SwissProt accession numbers for protein sequences are Q03591 (CFHR1) and Q02985 (CFHR3). Nucleotide numbering is based on the translation start site; A in ATG is +1.
Genetic analysis of CFHR1 and CFHR3 genes. Electropherograms showing the mutations identified in patients with aHUS (A) H119 (CFHR1 gene) and (B) H123 (CFHR3 gene). The 2 mutations were found in heterozygosis. The mutation in patient H123 could also be interpreted as c.838_839delAT (p.I280KfsX6). SwissProt accession numbers for protein sequences are Q03591 (CFHR1) and Q02985 (CFHR3). Nucleotide numbering is based on the translation start site; A in ATG is +1.
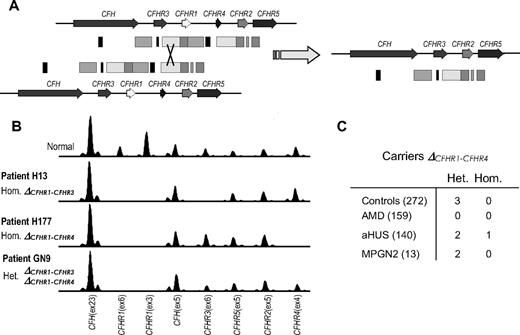
MLPA analysis in patients H177 (DefR1R4A) and GN9 (DefR1) resulted in the identification of a novel rearrangement that specifically removes the CFHR1 and CFHR4 genes (Figure 3A). We refer to this deletion as ΔCFHR1-CFHR4, and it probably involves a nonhomologous recombination event in the 3′ regions of the CFHR3 and CFHR4 genes. Patient H177, lacking both CFHR1 and CFHR4, is homozygous for this rearrangement, whereas patient GN9, lacking only CFHR1, is a compound heterozygote for ΔCFHR1-CFHR4 and ΔCFHR1-CFHR3 (Figure 3B). Further analysis of all the persons in our aHUS, MPGN2/DDD, and control cohorts identified the presence of this heterozygous rearrangement in some additional persons (Figure 3C), showing that ΔCFHR1-CFHR4 is a polymorphic trait in humans, although probably less frequent than ΔCFHR1-CFHR3.
Identification of ΔCFHR1-ΔCFHR4 deletion by MLPA. (A) Nonhomologous recombination event between homologous regions of the CFHR1 and CFHR4 genes that could have resulted in the ΔCFHR1-ΔCFHR4 deletion. (B) Electropherograms of the MLPA analyses showing heterozygous and homozygous deletions in patients with aHUS. (C) Frequency of the novel ΔCFHR1-ΔCFHR4 deletion in control persons and patients.
Identification of ΔCFHR1-ΔCFHR4 deletion by MLPA. (A) Nonhomologous recombination event between homologous regions of the CFHR1 and CFHR4 genes that could have resulted in the ΔCFHR1-ΔCFHR4 deletion. (B) Electropherograms of the MLPA analyses showing heterozygous and homozygous deletions in patients with aHUS. (C) Frequency of the novel ΔCFHR1-ΔCFHR4 deletion in control persons and patients.
CFHR protein deficiencies in patients with aHUS with anti-fH autoantibodies
Serum samples from control persons, patients with aHUS, and patients with MPGNII/DDD were screened for the presence of anti-fH autoantibodies basically as described.23 Autoantibodies were only detected in 7 pediatric patients with aHUS. The autoantibody titer and the main complement findings in the first serum sample available from these patients are shown in Table 6. Additional data on plasma treatment and renal function are also included in this table. Characterization of CFHR proteins by Western blot showed distinct CFHR deficiencies in 5 of these 7 patients (Table 7). Combined deficiency of both CFHR1 and CFHR3 was observed in only 3 patients (H87, H108, and H154), who were homozygous for the ΔCFHR1-CFHR3 deletion. Deficiency of CFHR1 and CFHR4A was observed in 1 patient (H177), presenting homozygous ΔCFHR1-CFHR4 deletion. Finally, a single deficiency of CFHR1 was detected in another patient (H119) who was a compound heterozygote for the ΔCFHR1-CFHR3 deletion and a missense mutation in the CFHR1 gene. Therefore, 5 of the 7 patients with fH autoantibodies showed homozygous CFHR1 deficiency, but the underlying genetic defects differ between the patients. Importantly, none of the patients presented mutations in the CFH, CFI, CFB, C3, or MCP genes.
Clinical and complement data in patients with aHUS with anti-fH autoantibodies
| Patient . | HUS onset . | First serum sample analyzed . | Current age and situation . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Age, y . | Plasma therapy . | Renal function . | Time after onset . | AutoAb titer, AU/mL* . | C3, mg/dL* . | C4, mg/dL* . | CFH, mg/dL* . | ||
| H87 | 5 | PE | Full recovery | Onset | 600 | 55.7† | 8.85† | 25 | 11 y; unknown |
| H108 | 4 | PE | Full recovery | 3 mo | 1600 | 86.2 | 28.4 | 22 | 8 y; ESRD |
| H119 | 4 | None | ESRD | 13 y | 600 | 117 | 16.6 | 28 | 20 y; RT since 1995 |
| H151 | 6 | PE + PI | Full recovery | 7 mo | 6000 | 66.7† | 16.7 | 31 | 8 y; stable with corticoids |
| H154 | 8 | PE | Full recovery | Onset | 18 000 | 58.5† | 19.6 | 26 | 10 y; stable with azathioprine |
| H171 | 3 | None | ESRD | 5 mo | 3600 | 81.3 | 40.6 | 25 | 4 y; RT since March 2009 |
| H177 | 7 | PF + 5% albumin | Death (myocarditis) | Onset | 16 000 | 58.5† | 19.6 | 22 | Died during first episode |
| Patient . | HUS onset . | First serum sample analyzed . | Current age and situation . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Age, y . | Plasma therapy . | Renal function . | Time after onset . | AutoAb titer, AU/mL* . | C3, mg/dL* . | C4, mg/dL* . | CFH, mg/dL* . | ||
| H87 | 5 | PE | Full recovery | Onset | 600 | 55.7† | 8.85† | 25 | 11 y; unknown |
| H108 | 4 | PE | Full recovery | 3 mo | 1600 | 86.2 | 28.4 | 22 | 8 y; ESRD |
| H119 | 4 | None | ESRD | 13 y | 600 | 117 | 16.6 | 28 | 20 y; RT since 1995 |
| H151 | 6 | PE + PI | Full recovery | 7 mo | 6000 | 66.7† | 16.7 | 31 | 8 y; stable with corticoids |
| H154 | 8 | PE | Full recovery | Onset | 18 000 | 58.5† | 19.6 | 26 | 10 y; stable with azathioprine |
| H171 | 3 | None | ESRD | 5 mo | 3600 | 81.3 | 40.6 | 25 | 4 y; RT since March 2009 |
| H177 | 7 | PF + 5% albumin | Death (myocarditis) | Onset | 16 000 | 58.5† | 19.6 | 22 | Died during first episode |
PE indicates plasma exchange; PF, plasmapheresis; PI, plasma infusion; ESRD, end-stage renal disease; and RT, renal transplant.
Normal values: AutoAb titer, < 150 AU/mL; C3, 77-135 mg/dL; C4, 14-47 mg/dL; CFH, 12-56 mg/dL.
Value out of normal range.
CFHR deficiencies and genetic alterations in patients with aHUS with anti-fH autoantibodies
| Patient . | CFHR1 deficiency . | CFHR3 deficiency . | CFHR4A deficiency . | ΔCFHR1-CFHR3 . | ΔCFHR1-CFHR4 . | CFHR1 mutation . | Other mutations* . |
|---|---|---|---|---|---|---|---|
| H87 | X | X | — | Homozygote | — | — | — |
| H108 | X | X | — | Homozygote | — | — | — |
| H154 | X | X | — | Homozygote | — | — | — |
| H177 | X | — | X | — | Homozygote | — | — |
| H119 | X | — | — | Heterozygote | — | Heterozygote | — |
| H151 | — | — | — | Heterozygote | — | — | — |
| H171 | — | — | — | — | — | — | — |
| Patient . | CFHR1 deficiency . | CFHR3 deficiency . | CFHR4A deficiency . | ΔCFHR1-CFHR3 . | ΔCFHR1-CFHR4 . | CFHR1 mutation . | Other mutations* . |
|---|---|---|---|---|---|---|---|
| H87 | X | X | — | Homozygote | — | — | — |
| H108 | X | X | — | Homozygote | — | — | — |
| H154 | X | X | — | Homozygote | — | — | — |
| H177 | X | — | X | — | Homozygote | — | — |
| H119 | X | — | — | Heterozygote | — | Heterozygote | — |
| H151 | — | — | — | Heterozygote | — | — | — |
| H171 | — | — | — | — | — | — | — |
X indicates homozygous deficiency; and —, no deficiency/no genetic alteration.
Genes analyzed were CFH, MCP, CFI, CFB, and C3.
Two-dimensional analysis of CFHR proteins
The analysis of serum samples by monodimensional electrophoresis and Western blot allowed detection of all products of CFH and CFHR genes thus far described. However, the bands corresponding to CFHR3 and CFHR4B overlap and could not be properly resolved. A similar situation was observed with CFHR1β and CFHL1. To solve these difficulties and to properly characterize these proteins, we performed 2-dimensional electrophoresis (2-DE), using IEF in the first dimension and SDS-PAGE in the second dimension.
Affinity chromatography on heparin-sepharose at low ionic strength (50mM NaCl) was used to remove major serum components (albumin and immunoglobulins) interfering with the 2D-gel analysis and to enrich the fH and CFHRs preparations.
The 2D-gel analysis of a normal control sample (C1) shows that fH and the CFHR proteins present complex patterns, including several isoforms (Figure 4A 2-DE). Isoform patterns corresponding to fH, CFHR4A, and CFHR5 were identified based on their molecular weights. Isoform patterns corresponding to CFHR1, CFHL1, CFHR3, and CFHR4B were assigned by comparison with the 2D-gels obtained from serum samples of patients with aHUS with complete deficiencies of CFHR1 or CFHR3 or both (Figure 4B). Differences between the theoretical and the observed isoelectric points were evident for most of the CFHR proteins (Figure 4A), suggesting the existence of posttranslational modifications that attached charged residues to the carbohydrate molecules of these proteins.
Analysis of CFHR proteins by 2-DE. (A) Western blots obtained after monodimensional (1DE) or 2-dimensional (2DE) electrophoresis of the heparin-sepharose eluate corresponding to control person C1, showing the Normal CFHR pattern. A single antibody solution containing equivalent amounts of the individual αCFHR1, αCFHR3/4, and αCFHR5 antibody preparations was used. The position and size (in kDa) of molecular weight markers is also indicated. (B) Two-dimensional analysis of CFHR1/CFHL1 and CFHR3/CFHR4B isoforms in control sample C1 and in 3 patients with aHUS showing the CFHR patterns DefR1R3, DefR1, or DefR3. CFHR4B isoforms are surrounded by dotted circles. Arrows indicate CFHL1 isoforms. The theoretical isoelectric points calculated with Compute pI/molecular weight program at ExPASy server (http://ca.expasy.org/tools/pi_tool.html) were CFH (6.12), CFHL1 (6.77), CFHR1 (7.53), CFHR3 (7.79), CFHR4A (4.85), CFHR4B (5.16), and CFHR5 (6.87).
Analysis of CFHR proteins by 2-DE. (A) Western blots obtained after monodimensional (1DE) or 2-dimensional (2DE) electrophoresis of the heparin-sepharose eluate corresponding to control person C1, showing the Normal CFHR pattern. A single antibody solution containing equivalent amounts of the individual αCFHR1, αCFHR3/4, and αCFHR5 antibody preparations was used. The position and size (in kDa) of molecular weight markers is also indicated. (B) Two-dimensional analysis of CFHR1/CFHL1 and CFHR3/CFHR4B isoforms in control sample C1 and in 3 patients with aHUS showing the CFHR patterns DefR1R3, DefR1, or DefR3. CFHR4B isoforms are surrounded by dotted circles. Arrows indicate CFHL1 isoforms. The theoretical isoelectric points calculated with Compute pI/molecular weight program at ExPASy server (http://ca.expasy.org/tools/pi_tool.html) were CFH (6.12), CFHL1 (6.77), CFHR1 (7.53), CFHR3 (7.79), CFHR4A (4.85), CFHR4B (5.16), and CFHR5 (6.87).
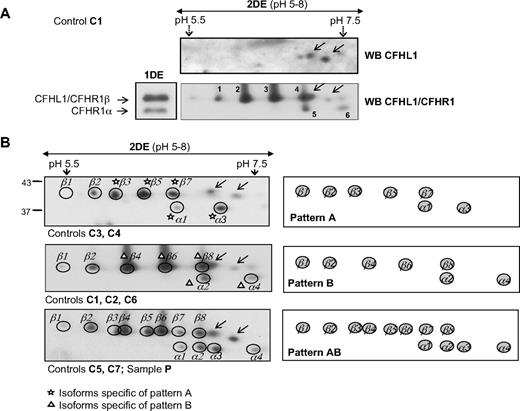
To determine whether the isoform patterns of the CFHR proteins remained constant in different persons, we analyzed 6 additional control persons (samples C2 to C7) and a “pooled” sample, including the 7 controls (sample P). All samples were analyzed by using IPG strips of 3 pH ranges to optimize isoform resolution of the different CFHR proteins.
Good separation of CFHR3 and CFHR4B by 2-DE was achieved by using IPG strips in the pH range of 3 to 10 (Figure 5A). Both proteins present a ladder pattern, but CFHR3 isoforms are more acidic, with an estimated pI range of 4.5 to 6, whereas CFHR4B isoforms are in the pI range of 6 to 7. Resolution of CFHR3 isoforms improves when using IPG strips in the pH range of 4 to 7. The patterns obtained in the 7 control persons and in the pooled control sample analyzed were very similar, with a total of 8 isoforms differing in both pI and molecular weight (Figure 5B). This isoform distribution explains the broad CFHR3 band observed by monodimensional analysis (Figure 4A lane 1DE). A single CFHR4B pattern was also apparent from the analysis of the same control samples (Figure 5B). A total of 5 isoforms with different pI and molecular weight were observed in all control samples analyzed, although the relative expression of the isoforms varied.
Characterization and variability of the CFHR3/CFHR4B region. (A) Separation of CFHR3/CFHR4B isoforms by 2DE. Western blot obtained with the heparin-sepharose eluate from control persons C1 and the αCFHR3/4 antibody preparation. (B) CFHR3 and (C) CFHR4B isoform patterns observed in individual samples (C2, C3) or in the pooled-control sample (P). The pH range used in the IEF step is shown at the top of the gels, and a schematic representation of each pattern is shown at the bottom.
Characterization and variability of the CFHR3/CFHR4B region. (A) Separation of CFHR3/CFHR4B isoforms by 2DE. Western blot obtained with the heparin-sepharose eluate from control persons C1 and the αCFHR3/4 antibody preparation. (B) CFHR3 and (C) CFHR4B isoform patterns observed in individual samples (C2, C3) or in the pooled-control sample (P). The pH range used in the IEF step is shown at the top of the gels, and a schematic representation of each pattern is shown at the bottom.
Resolution of the CFHR1/CFHL1 region was improved by using IPG strips with pH 5 to 8 in the IEF step, 10% SDS-PAGE in the second dimension, and Western blot with a monoclonal antibody that recognizes CFHL1 but not CFHR1. These conditions allowed proper identification of protein spots in the region, permitting discrimination between CFHL1 and CFHR1 isoforms. As shown in Figure 6A, 2 CFHL1 isoforms differing in molecular weight and pI were detected in the control sample analyzed. The minor isoform, partially overlapping with one of the CFHR1β isoforms, cannot be easily appreciated with the use of IPG strips in the pH range of 3 to 10 (Figure 4A). To confirm that the spots in the region assigned to CFHR1 isoforms indeed correspond to CFHR1, a replicate gel was stained with Coomassie Brilliant Blue G-250, and the spots matching those in the Western blot shown in Figure 6A were picked up manually and analyzed by peptide mass fingerprinting. All these spots were identified as CFHR1 (Swiss Protein Database entry Q03591).28
Characterization and variability of the CFHR1/CFHL1 region. (A) Two-dimensional electrophoretic analysis of the heparin-sepharose eluate obtained from control person C1, followed by Western blot with a polyclonal antibody preparation recognizing CFHR1 and CFHL1, or with a monoclonal antibody specific for CFHL1. Only the gel area corresponding to the CFHR1/CFHL1 region is shown. Spots numbered from 1 to 6 were picked up from a replicate gel stained with Coomassie and were identified as CFHR1 by matrix-assisted laser desorption-ionization time-of-flight spectrometry in a 4700 Proteomics Analyzer (PerSeptives Biosystems). (B) CFHR1 isoforms observed after 2DE and Western blot analysis of heparin-sepharose eluates from different control persons. C1 to C7 indicate individual samples; P, pooled-control sample. Isoforms characteristic of patterns A or B are labeled. Arrows indicate CFHL1 isoforms. A scheme of the 3 isoform patterns observed is shown on the right. The molecular weights (kDa) of the CFHR1 isoforms are indicated at the left.
Characterization and variability of the CFHR1/CFHL1 region. (A) Two-dimensional electrophoretic analysis of the heparin-sepharose eluate obtained from control person C1, followed by Western blot with a polyclonal antibody preparation recognizing CFHR1 and CFHL1, or with a monoclonal antibody specific for CFHL1. Only the gel area corresponding to the CFHR1/CFHL1 region is shown. Spots numbered from 1 to 6 were picked up from a replicate gel stained with Coomassie and were identified as CFHR1 by matrix-assisted laser desorption-ionization time-of-flight spectrometry in a 4700 Proteomics Analyzer (PerSeptives Biosystems). (B) CFHR1 isoforms observed after 2DE and Western blot analysis of heparin-sepharose eluates from different control persons. C1 to C7 indicate individual samples; P, pooled-control sample. Isoforms characteristic of patterns A or B are labeled. Arrows indicate CFHL1 isoforms. A scheme of the 3 isoform patterns observed is shown on the right. The molecular weights (kDa) of the CFHR1 isoforms are indicated at the left.
Interestingly, among the 7 control persons analyzed in this report, 3 distinct patterns of CFHR1 isoforms were identified (Figure 6B). Pattern A (Acidic), composed of 2 CFHR1α isoforms and 5 CFHRβ isoforms, was characteristic of control persons C3 and C4. Pattern B (Basic), also composed of 2 CFHR1α isoforms and 5 CFHRβ isoforms but slightly more basic than those in pattern A, was present in control samples C1, C2, and C6. Finally, pattern AB contains all isoforms observed in patterns A and B (4 α and 8 β); this pattern was observed in persons C5 and C7, and it was also obtained in the pooled sample P. The 2 CFHL1 isoforms were constant in the 7 control samples. The 3 CFHR1 isoform patterns closely resemble the 3 phenotypes observed by Susukida et al29 when analyzing samples from healthy volunteers in a study on fH polymorphism.
Genetic analysis of the CFHR1 A and B isoform patterns
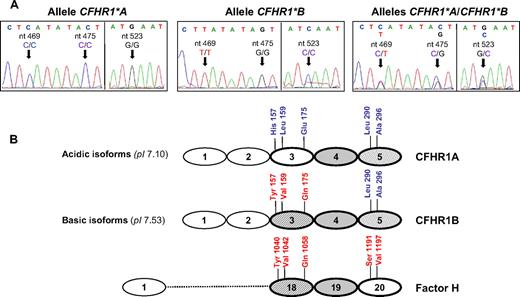
The CFHR1 2D isoform patterns A, B, and AB were suggestive of a polymorphism involving 2 allotypes, with patterns A and B as homozygotes or hemizygotes for each allele, and pattern AB corresponding to the heterozygote. To test this possibility, we sequenced all coding exons of the CFHR1 gene in the 7 control persons analyzed, and found that persons presenting pattern A and pattern B differed in their nucleotide sequences at different positions in exons 4, 5, and 6 (Figure 7A; supplemental figure, available on the Blood website; see the Supplemental Materials link at the top of the online article). Three nucleotide differences in CFHR1 exon 4 (c.469, c.475, and c.523) involve 3 amino acid changes in the SCR3 domain of CFHR1 (Figure 7A). Persons presenting pattern A were homozygotes or hemizygotes for a CFHR1 allele carrying His157-Leu159-Glu175 (CFHR1*A), and persons presenting pattern B were homozygotes or hemizygotes for a second CFHR1 allele carrying Tyr157-Val159-Gln175 (CFHR1*B) (Figure 7B). Interestingly, the nucleotide differences between the CFHR1*A and CFHR1*B alleles, including those involved in amino acid substitutions at positions p.157, p.159, and p.175, are differences that distinguish exon 4 of CFHR1 from exon 21 of CFH, respectively, suggesting that the CFHR1*B allele could have originated by a gene conversion event between CFHR1 and CFH.
Genetic basis for the 2D isoform patterns of CFHR1. (A) Electropherograms of CFHR1 exon 4 obtained from control persons presenting the CFHR1 2D isoform patterns A, B, and AB, respectively. The 3 nucleotide differences involving amino acid changes that distinguish the CFHR1*A and CFHR1*B alleles (c.469, c.475, and c.523) are shown. (B) Alignment of the homologous domains in CFHR1 (SCR3 to SCR5) and fH (SCR18 to SCR20). The amino acid differences between domains are indicated. SCR4 in CFHR1 and SCR19 in fH are identical. CFHR1A and CFHR1B isoforms differ in 3 amino acid residues located in the SCR3 domain (p.157, p.159, p.175); the theoretical pI is shown in brackets. The SCR3 domain in CFHR1B is identical to the SCR18 domain in fH. SwissProt accession numbers for protein sequences are Q03591 (CFHR1) and P08603 (fH). Nucleotide numbering is based on the translation start site: A in ATG is +1.
Genetic basis for the 2D isoform patterns of CFHR1. (A) Electropherograms of CFHR1 exon 4 obtained from control persons presenting the CFHR1 2D isoform patterns A, B, and AB, respectively. The 3 nucleotide differences involving amino acid changes that distinguish the CFHR1*A and CFHR1*B alleles (c.469, c.475, and c.523) are shown. (B) Alignment of the homologous domains in CFHR1 (SCR3 to SCR5) and fH (SCR18 to SCR20). The amino acid differences between domains are indicated. SCR4 in CFHR1 and SCR19 in fH are identical. CFHR1A and CFHR1B isoforms differ in 3 amino acid residues located in the SCR3 domain (p.157, p.159, p.175); the theoretical pI is shown in brackets. The SCR3 domain in CFHR1B is identical to the SCR18 domain in fH. SwissProt accession numbers for protein sequences are Q03591 (CFHR1) and P08603 (fH). Nucleotide numbering is based on the translation start site: A in ATG is +1.
Sequencing analyses of the CFHR1 and CFH genes in 129 controls and 151 patients with aHUS showed that the length of the gene conversion fragment is restricted to a short DNA sequence included within CFHR1 exon 4, which is internal to the primers used for PCR amplification of this exon. The data are consistent with the interpretation that this gene conversion event occurred a long time ago. This novel polymorphism increases the genetic variability at the CFHR1 gene, which should now be considered as a polymorphic locus, including 3 major alleles: CFHR1*A, CFHR1*B, and ΔCFHR1-CFHR3. The frequencies of these alleles in controls and patients with aHUS are shown in Table 8. Notably, the frequency of the CFHR1*B allele is significantly increased in the aHUS cohort, suggesting that carriers of this CFHR1 variant have an increased risk of developing aHUS. The analysis of the frequencies of the different CFHR1 genotypes in controls and patients with aHUS further showed that the association with aHUS is restricted to the condition of homozygosity for the CFHR1*B allele (carriers of 2 CFHR1*B copies) (Table 8). These data unravel a novel risk factor for aHUS and suggest that normal expression of the CFHR1*B allotype, more similar to fH, may compete or interfere with the normal function of fH, decreasing protection of cellular surfaces from damage by complement.
CFHR1 allele frequencies and genotype frequencies in aHUS association studies
| . | Frequencies* . | P . | OR (95% CI) . | |
|---|---|---|---|---|
| Controls (n = 129) . | aHUS (n = 151) . | |||
| Allele | ||||
| (A) CFHR1*A | 0.37 | 0.30 | ||
| (B) CFHR1*B | 0.39 | 0.48 | .033 | 1.46 (1.04-2.04) |
| (Del) ΔCFHR1-CFHR3 | 0.24 | 0.22 | ||
| Genotype | ||||
| A/A | 0.15 (19) | 0.13 (19) | ||
| B/B | 0.13 (17) | 0.28 (42) | .003 | 2.54 (1.36-4.73) |
| Del/Del | 0.07 (9) | 0.11 (16) | ||
| A/Del | 0.14 (18) | 0.09 (13) | ||
| B/Del | 0.20 (26) | 0.15 (22) | ||
| A/B | 0.31 (40) | 0.26 (39) | ||
| . | Frequencies* . | P . | OR (95% CI) . | |
|---|---|---|---|---|
| Controls (n = 129) . | aHUS (n = 151) . | |||
| Allele | ||||
| (A) CFHR1*A | 0.37 | 0.30 | ||
| (B) CFHR1*B | 0.39 | 0.48 | .033 | 1.46 (1.04-2.04) |
| (Del) ΔCFHR1-CFHR3 | 0.24 | 0.22 | ||
| Genotype | ||||
| A/A | 0.15 (19) | 0.13 (19) | ||
| B/B | 0.13 (17) | 0.28 (42) | .003 | 2.54 (1.36-4.73) |
| Del/Del | 0.07 (9) | 0.11 (16) | ||
| A/Del | 0.14 (18) | 0.09 (13) | ||
| B/Del | 0.20 (26) | 0.15 (22) | ||
| A/B | 0.31 (40) | 0.26 (39) | ||
Numbers in parentheses indicate number of participants in study.
Discussion
CFHR proteins are a group of minor plasma proteins that show structural and genetic homology to fH.7 Evolution of this family has resulted in genomic duplications that extend along the CFH-CFHRs subregion of the RCA gene cluster,8 and are the basis for the strong sequence similarity among fH and the CFHR proteins. The presence of these duplicated regions, however, favors gene conversion and nonhomologous recombination events that have been associated to aHUS.3
We report here the use of one-dimensional and 2D gel electrophoresis combined with Western blot analysis to analyze the CFHR proteins in plasma samples from patients with aHUS and patients with MPGN2/DDD. These analyses have resulted in the identification of persons carrying 3 novel CFHR protein deficiencies, DefR1, DefR3, and DefR1R4A, and have shown a previously unrecognized CFHR1 polymorphism. Genetic analysis in persons carrying these novel variations have shown that they have in part been generated as a consequence of the genomic instability in this region of the RCA gene cluster.
As expected from previous studies, DefR1R3 is the most frequent deficiency of CFHR proteins in our control population and in the patient cohorts. In agreement with previous results16 both the allelic frequency of ΔCFHR1-CFHR3 and the number of homozygotes are significantly associated with protection from AMD in the Spanish population (Table 4). In patients with aHUS, however, we could not replicate previous reports17 indicating that the ΔCFHR1-CFHR3 allele is associated with an increased predisposition to the disease; we found no differences between patients and controls for the frequency of the ΔCFHR1-CFHR3 allele. Importantly, our aHUS population is out of Hardy-Weinberg equilibrium for the CFHR1 polymorphism because of an excess of ΔCFHR1-CFHR3 homozygotes (pattern DefR1R3). Therefore, we conclude that the reported association of ΔCFHR1-CFHR3 with increased risk to aHUS only holds true for homozygous carriers.
A significant number of ΔCFHR1-CFHR3 homozygotes carry anti-fH autoantibodies,20 but whether a deficiency of CFHR1/CFHR3 is pathologically relevant or whether it is a marker for a different genetic defect is currently unknown. An important finding in our studies is the recognition that 2 patients with aHUS with anti-fH autoantibodies presented homozygous deficiency of CFHR1 but not of CFHR3 (Table 7). Moreover, the CFHR1 deficiency in these 2 patients was caused by a combination of different genetic defects. Patient H119 is a compound heterozygote for ΔCFHR1-CFHR3 and a nonsense mutation in CFHR1 (p.160S > Stop), whereas patient H177 is a homozygote for a novel genomic deletion involving the CFHR1 and CFHR4 genes (ΔCFHR1-CFHR4). Thus, among the 7 patients carrying anti-fH autoantibodies in our aHUS series, 5 of them presented a deficiency of CFHR1 as a consequence of 3 different genetic alterations. These data highlight a direct relationship between the lack of CFHR1 and the presence of anti-fH autoantibodies. They also suggest that the association of the autoantibodies with the genomic deletion encompassing CFHR1 and CFHR3 (ΔCFHR1-CFHR3) is secondary to this relationship. This conclusion is further supported by the fact that only 3 of the 12 homozygous ΔCFHR1-CFHR3 patients with aHUS develop anti-fH autoantibodies. In addition, the 2 patients with a single CFHR3 deficiency were negative for the anti-fH autoantibodies.
There is no clear explanation for the association between the deficiency of CFHR1 and the presence of anti-fH autoantibodies in patients with aHUS. These autoantibodies have been reported to recognize SCR19-SCR20 of fH,30 which are involved in binding to glycosaminoglycans and surface-bound C3b.31 As shown in Figure 7B, these SCRs are nearly identical to SCR4-SCR5 of CFHR1, so it is likely they can also recognize this protein. Unpublished data from our group suggest that anti-fH autoantibodies from CFHR1-deficient patients (patients H108, H154, and H177 in this report) crossreact with CFHR1, whereas this crossreactivity was not observed in the only patient with a normal CFHR1 expression that we could study (patient H151; C.A.-G. and P.S.-C., manuscript in preparation). Therefore, the autoantibodies in persons expressing CFHR1 may be different from those present in CFHR1-deficient persons. We would like to suggest that autoantibodies generated in the context of a CFHR1 deficiency are targeted to a region of the fH molecule that is critical for the developing of aHUS.
The physiologic function of CFHR1 is beginning to be understood. A recent report14 suggests CFHR1 has a complement regulatory activity at the level of the alternative pathway C5 convertase and the lytic pathway, a function mediated through the binding of the SCR1-SCR2 domains of CFHR1 to C5 and C5b6. Moreover, CFHR1 would also compete with fH for C3b and polyanionic binding on cell surfaces, thus interfering with the complement regulatory activity of fH. This potential fH-CFHR1 functional interference on cellular surfaces relies on the strong sequence similarity between SCR18 to SCR20 of fH and SCR3 to SCR5 of CFHR1 and is supported by genetic data from patients with aHUS.15,24
Characterization of CFHR1 by 2D gel electrophoresis and Western blot analysis showed acidic and basic isoforms of this protein in control persons (Figure 6). Genetic studies showed that the acidic and basic isoforms are indeed 2 CFHR1 allotypes (CFHR1*A and CFHR1*B) that differ in 3 amino acids within the SCR3 domain. Interestingly, these 3 amino acid differences make the SCR3 domain of the CFHR1*B allotype identical to the SCR18 domain of fH (Figure 7B). The 2 CFHR1 allotypes most probably correspond to the 2 human liver cDNA clones previously described,32 which were generated by a gene conversion event between the CFHR1 and the CFH genes. We present here the results of a case-control association study in the aHUS Spanish cohort showing that CFHR1*B, the allele presenting the highest sequence similarity to CFH, is strongly associated with increased risk of aHUS when it is present in homozygosity (2 copies; Table 8). These are novel and interesting data that may reflect a competition between CFHR1 and fH, decreasing the functional activity of fH on surfaces and thus predisposing to complement-mediated damage.
In conclusion, the analysis of CFHR proteins using 1- or 2-dimensional gel electrophoresis and Western blot analysis have allowed us to explore additional variability in the CFH-CFHR1-5 genomic region, providing new structural data on these proteins. For the first time, we have identified isolated deficiencies of CFHR1 and CFHR3, and established a specific relationship between the CFHR1 deficiency and the generation of anti-fH autoantibodies associated with aHUS. The discovery of a common CFHR1 allotype, which is associated with an increased risk of aHUS, is an exciting finding that may unravel a functional interference between CFHR1 and fH with consequences in pathology. These novel data warrant new studies that, in addition to increasing our understanding of the CFHR proteins, will probably improve our knowledge of the pathogenic mechanisms in aHUS.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Sheila Pinto for excellent technical assistance and Prof Peter F. Zipfel (Hans-Knöll Institute, Jena, Germany) and Dr Jennifer McRae (St Vincent's Health, Melbourne, Australia) for providing us with polyclonal antibodies recognizing CFHR proteins. We also thank Dr Marie Agnes Dragon-Durey (Hôpital Georges Pompidou, Paris, France) for the reference sample containing fH autoantibodies.
This work was supported by the Spanish Ministerio de Ciencia e Innovación (grants FIS 06/0625 to P.S.-C., SAF 2005-00913 and SAF 2008-00226 to S.R.d.C., SAF 2006-02948 to M.L.T.) and by grants from the CIBER de Enfermedades Raras (CIBERER) and the Fundación Renal Iñigo Álvarez de Toledo. C.A.G. was funded by the “Fundación para la Investigación Biomédica-Hospital Universitario La Paz” (FIBHULP).
Authorship
Contribution: C.A.-G. and R.M.-B. performed experiments, helped in the analysis of the data, and prepared the figures; M.L.-T. screened fH autoantibodies and analyzed data; and S.R.d.C., and P.S.-C. designed the research, analyzed results, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pilar Sánchez-Corral, Research Unit, Hospital Universitario La Paz, Paseo de la Castellana 261, 28046 Madrid, Spain; e-mail: psanchez.hulp@salud.madrid.org.
References
Author notes
*C.A.-G. and R.M.-B. contributed equally to this study.