Abstract
Caspases play a critical role in regulation of apoptosis, cell differentiation, inflammation, and innate immunity, and several are mutated or have altered expression in non-Hodgkin lymphoma (NHL). To study the impact of genetic variation in caspases on NHL risk, we analyzed tag single nucleotide polymorphisms (SNPs) in 12 caspase and related genes in 3 population-based case-control studies (1946 cases and 1808 controls). Gene-based analysis, adjusting for the number of tagSNPs genotyped in each gene, showed significant associations for CASP8, CASP9, and CASP1. SNP-based analysis showed that CASP8 rs6736233 (odds ratio (OR) CG = 1.21; ORCC = 2.13; P trend = .011); CASP9 rs4661636 (ORCT = 0.89; ORTT = 0.77; P trend = .011); and CASP1 rs1785882 (ORAT = 1.12; ORAA = 1.30; P trend = .0054) were significantly associated with NHL risk and consistent across studies. It is noteworthy that genetic variants in CASP8 were associated with risk of all major NHL subtypes. Our findings suggest that genetic variation in caspases may play an important role in lymphomagenesis.
Introduction
Caspases are highly conserved intracellular cysteine proteases that mediate apoptosis and are categorized as initiator caspases (1, 2, 4, 5, 8, 9, 10, 11, and 12) or effector caspases (3, 6, 7, and 14).1 Initiator caspases are the first to be activated in apoptosis and in turn activate effector caspases, orchestrating programmed cell death.1 Caspases contribute to biologic processes important in lymphomagenesis, including cytokine maturation (eg, caspases 1, 5, and 11), nuclear factor kappa beta (NF-κB) activation (eg, caspases 1, 2, and 8), and B-cell maturation/proliferation (eg, caspases 3 and 8).1
There is growing evidence that caspase genes are altered in non-Hodgkin lymphoma (NHL).2,3 Somatic mutations in caspases 3 and 10 have been reported,3,4 and NHL tumor expression array analyses have shown that caspases 1, 2, 9, and 10 were differentially expressed by NHL subtypes.2,5,6 There is also preliminary evidence from association studies that single nucleotide polymorphisms (SNPs) in several caspase and caspase-related genes may be associated with risk of NHL or its subtypes.7,8 To investigate whether genetic variation in caspase genes plays in lymphomagenesis, we genotyped tagSNPs in 12 caspase or caspase-related genes in 1946 NHL cases and 1808 controls pooled from 3 independent population-based case-control studies conducted in the United States and Australia.
Methods
Three population-based case-control studies of NHL participated in this pooled analysis: the National Cancer Institute–Surveillance, Epidemiology and End Results (NCI-SEER) NHL study, conducted within the SEER Iowa, Detroit, Los Angeles, and Seattle registries9 ; the Connecticut NHL study, conducted among female residents of Connecticut10 ; and the New South Wales study, conducted among residents of New South Wales and the Australian Capital Territory, Australia11 (supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). The protocols for each study were approved by the Institutional Review Boards of the NCI, each SEER center for the NCI-SEER study, Yale University, the Connecticut Department of Public Health, and the NCI for the Connecticut study, and all participating institutions for the New South Wales study. All study participants provided informed consent, in accordance with the Declaration of Helsinki. NHL subtypes were grouped according to the World Health Organization (WHO) classification using the International Lymphoma Epidemiology Consortium (InterLymph) guidelines.12 DNA was extracted from blood10-12 or buccal cells,10 and tagSNPs were genotyped at the NCI Core Genotyping Facility (supplemental data). In total, 79 tagSNPs in 12 caspase or caspase-related genes and 23 additional SNPs within 9 regions adjacent to these genes used to expand genomic coverage were selected (supplemental Table 2); the latter 23 SNPs did not show any noteworthy evidence of association (supplemental Table 3). SNPs in caspase genes showing association in a previous report from the Connecticut study8 were completely tagged (supplemental Table 2), and results for those SNPs in the pooled analyses can found in supplemental Tables 3 to 8. However, stronger effects were found for novel SNPs genotyped in the same caspase genes for the pooled analysis reported here and are featured in “Results and discussion.”
Genotype-specific risks of NHL for each SNP were estimated as odds ratios and 95% confidence intervals for the heterozygote and less common homozygote genotypes, with the more common homozygote as the reference, using unconditional logistic regression. Our approach for defining the referent group follows genetic convention for tagSNPs, but we note that the directionality of effects is to be determined by follow-up studies to establish the causal variants. Models were adjusted for age, race, sex, and study center. Polytomous multivariate unconditional logistic regression models were used to evaluate the effect among different NHL subtypes.
To obtain a gene-level summary of association and adjust for the number of tag SNPs in each gene, taking into account the underlying linkage disequilibrium pattern, we computed the minimum P value (“minP test”), which assesses the statistical significance of the smallest P trend within each gene by permutation-based resampling methods (10 000 permutations).13 A significance level of P < .05 was interpreted as evidence of association. To account for multiple comparisons across the 12 caspase genes tested, we applied the false discovery rate (FDR)14 to the minP test for all NHLs. An FDR value less than 0.2 was considered evidence that an association had a relatively low probability of being a false discovery. We emphasize SNPs with associations for NHL risk overall, as well as with histologic subtypes, in contrast to SNPs, which show association with only specific histologies, because the latter analyses have reduced power and are probably false-positive associations.15
Results and discussion
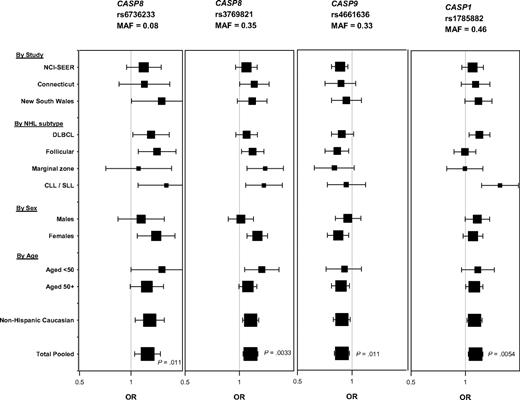
Cases and controls showed comparable distributions by age and race within each study (supplemental Table 1). We found significant evidence of association at the gene level for CASP8, CASP9, and CASP1 with NHL and one or more subtypes (Table 113 ; Figure 1). FDR values for the associations with NHL were 0.15. There was also evidence for association at the gene level for CASP8AP2 and CASP14 with diffuse large B-cell lymphoma (DLBCL), CASP4 with chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL), and CASP2 with marginal zone lymphoma (Table 1). Within genes showing an association with NHL, CASP8 rs6736233 and rs3769821 (r2 = 0.13, D′ = 0.98) and CASP1 rs1785882 were associated with increased risk and CASP9 rs4661636 was associated with decreased risk of NHL overall and with one or more of its subtypes (Figure 1; supplemental Tables 3-4). These associations were consistent across study, age, sex, and among non-Hispanic whites (Figure 1; supplemental Tables 3, 5-9). It is also noteworthy that in the aggregate genetic variants in CASP8 were significantly associated with risk of all 4 major B-cell subtypes (Figure 1; supplemental Table 4). In addition, CASP1 rs1785882 was associated with risk of NHL overall and 2 of its subtypes (DLBCL and CLL/SLL; Figure 1; supplemental Tables 3 and 4).
Summary of permutation test (minP test) for P trends for gene-based CASP SNPs for NHL overall and by subtype
| Gene . | SNPs/gene no. . | NHL, P . | DLBCL, P . | FL, P . | MZL, P . | CLL/SLL, P . |
|---|---|---|---|---|---|---|
| CASP1* | 4 | .019** | .032** | .48 | .74 | .001** |
| CASP2† | 4 | .18 | .34 | .34 | .023** | .76 |
| CASP3 | 6 | .16 | .46 | .62 | .74 | .43 |
| CASP4‡ | 6 | .49 | .94 | .37 | .42 | .01** |
| CASP5 | 10 | .50 | .18 | .61 | .86 | .08 |
| CASP6 | 5 | .37 | .25 | .71 | .37 | .23 |
| CASP7 | 13 | .73 | .82 | .66 | .37 | .87 |
| CASP8§ | 9 | .027** | .16 | .050** | .033** | .042** |
| CASP9‖ | 4 | .036** | .22 | .043** | .23 | .45 |
| CASP10 | 3 | .05 | .35 | .21 | .13 | .06 |
| CASP8AP2¶ | 9 | .60 | .040** | .83 | .21 | .27 |
| CASP14# | 6 | .31 | .007** | .57 | .18 | .16 |
| Gene . | SNPs/gene no. . | NHL, P . | DLBCL, P . | FL, P . | MZL, P . | CLL/SLL, P . |
|---|---|---|---|---|---|---|
| CASP1* | 4 | .019** | .032** | .48 | .74 | .001** |
| CASP2† | 4 | .18 | .34 | .34 | .023** | .76 |
| CASP3 | 6 | .16 | .46 | .62 | .74 | .43 |
| CASP4‡ | 6 | .49 | .94 | .37 | .42 | .01** |
| CASP5 | 10 | .50 | .18 | .61 | .86 | .08 |
| CASP6 | 5 | .37 | .25 | .71 | .37 | .23 |
| CASP7 | 13 | .73 | .82 | .66 | .37 | .87 |
| CASP8§ | 9 | .027** | .16 | .050** | .033** | .042** |
| CASP9‖ | 4 | .036** | .22 | .043** | .23 | .45 |
| CASP10 | 3 | .05 | .35 | .21 | .13 | .06 |
| CASP8AP2¶ | 9 | .60 | .040** | .83 | .21 | .27 |
| CASP14# | 6 | .31 | .007** | .57 | .18 | .16 |
The minP test assesses the true statistical significance of the smallest P trend within each gene (determined by dichotomous logistic regression, comparing NHL or NHL subtypes with controls; SNPs listed in Table S2) by permutation-based resampling methods (10 000 permutations) that automatically adjust for the number of tag SNPs tested within that gene and the underlying linkage disequilibrium pattern.13
NHL indicates non-Hodgkin lymphoma; DLBCL, diffuse large B-cell lymphoma; FL, follicular lymphoma; MZL, marginal zone lymphoma; and CLL/SLL chronic lymphocytic leukemia/small lymphocytic lymphoma.
CASP1: significant SNPs for NHL, rs1785882; DLBCL, rs1785882; CLL/SLL, rs1785882, rs501626.
CASP2: significant SNPs for MZL, rs7810486.
CASP4: significant SNPs for CLL/SLL, rs10791740, rs11226565, rs7123277.
CASP8: significant SNPs for NHL, rs6736233, rs3769821; Follicular, rs6736233, rs3769821, rs2293554; MZL, rs3769825, rs3769821, rs700636; CLL/SLL, rs3769825, rs6736233, rs3769821.
CASP9: significant SNPs for NHL, rs4661636, rs4646047.
CASP8AP2: significant SNPs for DLBCL, rs12661230.
CASP14: significant SNPs for DLBCL, rs714920.
P < .05.
Association between the most noteworthy SNPs in CASP8, CASP9, and CASP1 and risk of NHL by study, NHL subtype, sex, age, and ethnicity, based on the additive model. Square symbols represent odds ratios; symbol size is proportional to number of cases. Horizontal lines represent 95% confidence intervals. The x-axis ranges from an odds ratio of 0.5 to 2.0. Number of cases and controls by study (NCI-SEER: 990 cases, 828 controls; Connecticut: 436 cases, 515 controls; New South Wales: 520 cases, 465 controls); by NHL subtype (DLBCL, n = 600; follicular, n = 540; marginal zone, n = 160; and CLL/SLL, n = 161); by sex (males: 840 cases, 711 controls; females: 1106 cases, 1097 controls); by age (< 50 years: 484 cases, 408 controls; age ≥ 50 years: 1462 cases, 1400 controls); and by ethnicity (non-Hispanic whites: 1751 cases, 1578 controls; all ethnicities combined: 1946 cases, 1808 controls). P values are from additive (ie, trend) model. MAF indicates minor allele frequency.
Association between the most noteworthy SNPs in CASP8, CASP9, and CASP1 and risk of NHL by study, NHL subtype, sex, age, and ethnicity, based on the additive model. Square symbols represent odds ratios; symbol size is proportional to number of cases. Horizontal lines represent 95% confidence intervals. The x-axis ranges from an odds ratio of 0.5 to 2.0. Number of cases and controls by study (NCI-SEER: 990 cases, 828 controls; Connecticut: 436 cases, 515 controls; New South Wales: 520 cases, 465 controls); by NHL subtype (DLBCL, n = 600; follicular, n = 540; marginal zone, n = 160; and CLL/SLL, n = 161); by sex (males: 840 cases, 711 controls; females: 1106 cases, 1097 controls); by age (< 50 years: 484 cases, 408 controls; age ≥ 50 years: 1462 cases, 1400 controls); and by ethnicity (non-Hispanic whites: 1751 cases, 1578 controls; all ethnicities combined: 1946 cases, 1808 controls). P values are from additive (ie, trend) model. MAF indicates minor allele frequency.
This is the first comprehensive evaluation of genetic variation in caspase genes and risk of NHL. Our results suggest that SNPs in initiator caspases (ie, CASP8, CASP9, and CASP1) affect lymphomagenesis.
The main initiator caspases in mammals are caspase 8 (located in the death receptor-mediated apoptosis pathway) and caspase 9 (located in the intrinsic mitochondrial apoptosis pathway).1 Lan et al19 recently reported that higher mitochondrial DNA copy number was associated with risk of NHL, which is consistent with impaired mitochondrial apoptosis. We acknowledge, however, that the observed associations could reflect other biologic functions mediated by these genes in addition to apoptosis. In particular, caspase 8 plays a broad role in regulating lymphocyte homeostasis, NF-κB activation, and differentiation of monocytes into macrophages,1 all of potential relevance for NHL etiology.20 One report found that persons with CASP8 mutations had decreased T-, B-, and NK-cell activation and decreased lymphocyte apoptosis.21 Although CASP8 rs1045485 has been associated with risk of breast cancer, melanoma, and glioma,22-24 we did not detect an association with NHL.
Caspases 1, 4, and 5 play a key role in maturation of proinflammatory cytokines in cells infected by certain pathogens, and caspase 1 is the most efficient caspase in the process.25 For example, caspase 1, initially known as interleukin-1B–converting enzyme, is critical for maturation of interleukin-1B1 and also regulates interferon-γ production.25 Further, caspase 1 plays a role in NF-κB activation,1 so the underlying biologic basis of the CASP1 association we report here could be the result of one or more of these functions.
In conclusion, our study provides evidence that common genetic variants in CASP8, CASP9, and CASP1 are associated with risk of NHL and one or more subtypes. If replicated in larger studies, a comprehensive strategy of fine mapping followed by functional analyses should be carried out and gene-environment interactions should be explored.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Peter Hui (Information Management Services Inc, Silver Spring, MD) for programming support.
DNA extraction, genotyping, and statistical analysis for this project were supported by the Intramural Research Program of NIH (NCI). The NCI-SEER study was also supported by the Intramural Research Program of NIH (NCI) and by the Public Health Service (contracts N01-PC-65064, N01-PC-67008, N01-PC-67009, N01-PC-67010, and N02-PC-71105). The Connecticut study was supported by NIH (grant CA62006; T.Z.) from the NCI. The New South Wales study was supported by the National Health and Medical Research Council of Australia (project grant 990920; B.A.), the Cancer Council New South Wales, and the University of Sydney Medical Foundation.
National Institutes of Health
Authorship
Contribution: Q.L., L.M.M., P.H., M.P.P., M.Y., S.J.C., N.R., and S.S.W. conceived and led the project; Q.L., B.A., P.H., T.Z., M.P.P., J.R.C., Y.Z., A.G., W.C., T.R.H., C.M.V., S.D., B.L., A.K., M.S., N.R., and S.S.W. obtained and provided data and DNA samples; M.Y. and S.J.C. carried out bioinformatics and genotyping; Q.L., L.M.M., P.H., I.M., M.P.P., M.Y., N.C., S.J.C., N.R., and S.S.W. performed statistical analysis; Q.L., L.M.M., N.R., and S.S.W. drafted and revised the manuscript; and B.A., P.H., I.M., T.Z., M.P.P., J.R.C., Y.Z., A.G., W.C., M.Y., T.R.H., C.M.V., S.D., B.L., A.K., M.S., S.H.Z., N.C., and S.J.C. provided input on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Qing Lan, Occupational & Environmental Epidemiology Branch, Division of Cancer Epidemiology and Genetics, NCI, NIH, DHHS, MSC 7240, 6120 Executive Blvd, EPS 8109, Bethesda, MD 20892-7240; e-mail: qingl@mail.nih.gov.