Abstract
At the cellular level, development progresses through successive regulatory states, each characterized by their specific gene expression profile. However, the molecular mechanisms regulating first the priming and then maintenance of gene expression within one developmental pathway are essentially unknown. The hematopoietic system represents a powerful experimental model to address these questions and here we have focused on a regulatory circuit playing a central role in myelopoiesis: the transcription factor PU.1, its target gene colony-stimulating-factor 1 receptor (Csf1r), and key upstream regulators such as RUNX1. We find that during ontogeny, chromatin unfolding precedes the establishment of active histone marks and the formation of stable transcription factor complexes at the Pu.1 locus and we show that chromatin remodeling is mediated by the transient binding of RUNX1 to Pu.1 cis-elements. By contrast, chromatin reorganization of Csf1r requires prior expression of PU.1 together with RUNX1 binding. Once the full hematopoietic program is established, stable transcription factor complexes and active chromatin can be maintained without RUNX1. Our experiments therefore demonstrate how individual transcription factors function in a differentiation stage–specific manner to differentially affect the initiation versus maintenance of a developmental program.
Introduction
During development, gene activation is regulated by specific transcription factors, which interact with chromatin remodeling and modification factors to establish active chromatin structures. This is also true for the hematopoietic system. All mature blood cell lineages originate from hematopoietic stem cells (HSCs), which have the capacity to self-renew and differentiate. Lineage specification is regulated by a shift in balance of antagonistic transcription factors, which drive the expression of lineage-appropriate genes and repress alternative lineage fates.1-3 To direct differentiation into the distinct blood cell lineages at appropriate frequency, genes encoding hematopoietic regulators need to be expressed in a strictly controlled fashion both in terms of hierarchy and precise level of expression. If this hierarchy is altered by expression at an inappropriate developmental stage, differentiation is deregulated4 and cells can be reprogrammed into alternate lineages5,6 or are subverted into leukemia
Many hematopoietic lineage–specific genes are first transcriptionally activated in HSCs whereby transcription factors bind to their recognition sequences in a stable fashion and heritably maintain an active chromatin structure.7,8 Little is known about the order of events establishing this active chromatin state in development and how it is maintained in a lineage-specific fashion. To address this question in molecular detail, we studied a regulatory circuit that occupies a central role in myelopoiesis: the gene encoding for the transcription factor PU.1 and one of its targets, the colony-stimulating-factor 1 receptor gene (Csf1r), together with upstream factors regulating their expression. The developmental regulation of both genes has been extensively studied. Most transcription factors regulating their expression have been identified and important cis-regulatory elements and their chromatin structure have been characterized.8-18 Pu.1 is essential for myelopoiesis.19,20 Its expression is switched on at the onset of hematopoietic development and is maintained during myelopoiesis and B lymphopoiesis, but is down-regulated in erythroid cells and T cells.21 Overexpression of PU.1 in transgenic mice leads to erythroleukemia,22 and reduced levels of PU.1 cause myeloid leukemia.15 PU.1 expression levels therefore need to be tightly regulated in development. Csf1r is absolutely required for macrophage differentiation23 and its expression is crucially dependent on the prior expression of PU.1.9,24 In addition, Csf1r expression is tightly regulated. Csf1r mRNA is expressed at low levels in HSCs, but high-level expression is observed only in committed macrophage precursor cells.9 Both genes are targets for the transcription factor RUNX1. RUNX1 is absolutely required for definitive hematopoiesis as well as for Pu.1 and Csf1r expression, and thus for the establishment of myelopoiesis at later developmental stages.10,25 Interestingly, RUNX1 appears to be largely dispensable for hematopoietic development once stem cells have formed in the adult organism,26,27 indicating a fundamental difference in the molecular requirements for the establishment and the maintenance of the hematopoietic gene expression program. The molecular basis for this difference is unknown.
Using the differentiation of mouse embryonic stem (ES) cells as a model, we addressed these issues and performed an in-depth analysis of events in chromatin leading up to the transcriptional activation of Pu.1 and Csf1r during blood cell development and the molecular mechanisms regulating these processes. We focused on the following fundamental questions of (1) which type of chromatin alterations were the first ones to mark Pu.1 and Csf1r for activation and (2) whether RUNX1 was required for this initial chromatin activation. We also asked (3) whether RUNX1 primed chromatin by forming stable transcription factor complexes, (4) whether there was a developmental window during which RUNX1 had to act, and finally (5) whether RUNX1 was still needed once stable transcription factor complexes had formed on critical hematopoietic regulator genes. Our study shows that RUNX1 orchestrates blood-cell lineage–specific chromatin priming at a much earlier developmental stage than previously thought. We also provide direct proof for the idea that once RUNX1 has initiated the expression of hematopoietic transcription factor genes, a stable transcription factor circuit is formed on these genes and active chromatin is maintained in a heritable fashion.
Methods
Cell isolation and culture
The Bry-GFP ES cell line carries a GFP knocked into the brachyury locus.28 These cells and all other lines described here were cultured and fractionated as described.28 For generation of CD41+ cells, day-3.5 EB cells were replated in methylcellulose blast colony assays as previously described.28 After 4 days colonies were harvested, and cells were trypsinized, stained for CD41 expression, and sorted by fluorescence-activated cell sorting (FACS).
The inducible RUNX1 cell line (iRUNX1) contains the full-length murine proximal Runx1 isoform cDNA as defined by Telfer and Rothenberg29 and was generated by first targeting both Runx1 alleles in the Ainv18 ES cell line30 and subsequently introducing the RUNX1 cDNA expressed under the control of a DOX-responsive promoter.31 At day 3.5 of differentiation, DOX (Sigma-Aldrich) was added at a final concentration of 0.1 μg/mL. After 4 hours, EBs were dissociated. Flk1+ cells were isolated by FACS or by magnetic cell sorting (autoMACS; Miltenyi Biotec) using a biotin-coupled Flk1+ antibody (eBioscience) according to the manufacturer's instructions. Continuous induction of iRUNX1 was done with 0.1 μg/mL DOX; transient induction was done with 1 μg/mL DOX.
The inducible Dam ES cell line (iDAM) was generated by amplifying a V5-Dam fragment from the pLgw V5-EcoDam (Bas Van Steensel; NKI) and introducing it into Ainv18. The inducible RUNX1-Dam ES cell line (iRUNX1-DAM) was generated the same way, but V5-Dam was amplified from the pLgw V5-EcoDam and fused to RUNX1. Differentiation and induction of Flk1+ cells were done as described for iRUNX1.
Colony assays
Flk1+ wild-type and iRUNX1 cells were plated into the blast colony media described previously.28 After 4 days of culture, cells were replated at 105 cells/mL in a methylcellulose media supporting the growth of myeloid hematopoietic progenitors (StemCell Technologies) and colonies were counted after 7 days.
DNA methylation analysis and DamId
Bisulfite treatment of genomic DNA was performed using EZ DNA Methylation Kit (Zymo Research). Polymerase chain reaction (PCR) primers were complementary to the upper DNA strand after modification, and 2 rounds of nested PCR were performed using primers listed in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Gel-purified products were ligated into pCR2.1 plasmid (Invitrogen) and 20 clones were sequenced.
Dam-methylation analysis of genomic DNA purified from induced iDam and iRUNX1-Dam cells was performed exactly as described,32 except that gene-specific primers were used to detect differential GATC methylation after the ligation-mediated (LM)–PCR step. Primers used for both assays are depicted in supplemental Table 1.
ChIP assays and real-time PCR analysis
After sorting, cells were incubated for 2 hours under embryoid body–forming conditions to allow recovery, which did not change the expression of GFP/Bry and Flk-1. Undifferentiated ES cells and sorted cell populations were then cross-linked using formaldehyde (1% final concentration), lysed, and sonicated to obtain 0.5- to 1-kb fragments. The chromatin was mixed with cross-linked and sonicated Drosophila DNA in a ratio of 1:4. Chromatin was immunoprecipitated using anti-H3K27me3 (no. 07-449; Upstate Biotechnology), anti-H3K9ac (4441; Abcam), anti-H3K4me3 (8580; Abcam), or anti-H3 (no. ab-1791-100; Abcam) antibodies as described.33 Chromatin immunoprecipitation (ChIP) experiments detecting transcription factors were performed as described in Krysinska et al,9 with the following antibodies: anti-C/EBPβ (SC-150X; Santa Cruz Biotechnology), anti–Fli-1 (SC-356X), and anti-HA (H6908; Sigma-Aldrich). Precipitated DNA was quantified by real-time quantitative PCR as described.34 Signals observed with the specific antibody were divided by the signals obtained from an input control and were additionally normalized against the signal obtained with the H3 antibody or against an internal control in case of transcription factor ChIPs. Primers are listed in supplemental Table 1.
Replication timing
For replication timing analysis, ES cells and embryoid bodies were pulse labeled (30 minutes) with BrdU (50 mM; Sigma-Aldrich) before harvesting. BrdU-labeled ES cells and their derivatives were fixed with 70% cold ethanol, stained with propidium iodide, and sorted according to DNA content into 6 consecutive cell cycle fractions (G1-S, S1, S2, S3, S4, G2-M), and newly replicated DNA was purified using an anti-BrdU antibody (Becton Dickinson) as previously described.35 Real-time PCR was performed for each fraction to determine the replication timing profile of specific loci. Spiking with BrdU-labeled Drosophila DNA was used to control equal recovery of BrdU-labeled DNA.
In vivo footprinting analysis
RNA expression analysis
Total RNA was extracted using Trizol (Invitrogen) and contaminating genomic DNA was removed by DNaseI treatment. Total RNA (2 μg) was used for cDNA synthesis, using oligo(dT) 15-mer primer or random primers and MLLV reverse transcriptase followed by real-time PCR with primers listed in supplemental Table 1. Relative expression was calculated as a ratio to the GAPDH signal.
Results
Pu.1 and Csf1r activation occurs in a distinct developmental order and Pu.1 is first activated by chromatin unfolding and selective DNA demethylation
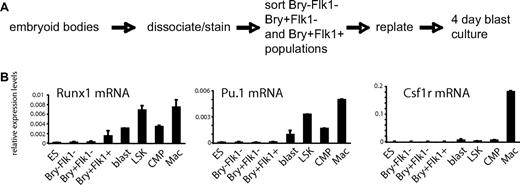
Embryonic hematopoiesis and the hierarchic control of gene expression are faithfully recapitulated in differentiating ES cells.25 The insertion of GFP into the brachyury locus provides a mesoderm-specific lineage tracer. During in vitro differentiation of such Bry-GFP ES cells, hematopoietic development proceeds from mesodermal cells to cells with both endothelial and hematopoietic potential (hemangioblasts) that express Bry-GFP and the VEGF-receptor 2 (Flk-1).28 Under appropriate culture conditions, this cell population rapidly develops into cells with hematopoietic potential expressing the CD41 surface marker via a hemogenic endothelium stage.31,38 From these differentiation cultures, we isolated precursor cell populations representing the sequential stages leading up to the formation of hematopoietic cells as outlined in Figure 1A. As shown in Figure 1B and supplemental Figure 1B, the ES cell culture system accurately recapitulated the hierarchic onset of expression of genes important for hematopoietic development. Genes encoding the transcription factors SCL, FLI-1, and RUNX1 were expressed first in Bry+Flk1+ cells. In contrast, the expression of genes required for the formation of specific blood cell lineages, such as those encoding C/EBPα, PU.1, and CSF-1R, was detected only in cultures containing cells committed to hematopoiesis (blast).
Hierarchy of transcription factor gene expression during ES cell differentiation. (A) Experimental strategy. Embryoid bodies were cultured for up to 3.75 days, and epiblast-like cells expressing neither marker (Bry−Flk1−), mesodermal cells expressing GFP only (Bry+Flk1−), and hemangioblast-enriched cell population expressing GFP and Flk-1 (Bry+Flk1+) were purified using fluorescence-activated cell sorting (FACS; supplemental Figure 1A). To obtain hematopoietic cells, we further cultured the (Bry+Flk1+) cells for 4 days under conditions supporting the generation of blast colonies and from these cultures isolated CD41+ cells, which have been shown to represent the first fully committed hematopoietic cells.39 (B) Timing of mRNA expression of hematopoietic regulator genes (Runx1, Pu.1, and Csf1r) in differentiating ES cells. ES indicates ES cells; blast, 4-day blast cell culture derived from +/+ cells; LSK, Lin−Sca-1+c-kithi cells; CMP, common myeloid precursor cell; and Mac, primary macrophages. Numbers in panel B represent mean values of 2 independent experiments analyzed in duplicate.
Hierarchy of transcription factor gene expression during ES cell differentiation. (A) Experimental strategy. Embryoid bodies were cultured for up to 3.75 days, and epiblast-like cells expressing neither marker (Bry−Flk1−), mesodermal cells expressing GFP only (Bry+Flk1−), and hemangioblast-enriched cell population expressing GFP and Flk-1 (Bry+Flk1+) were purified using fluorescence-activated cell sorting (FACS; supplemental Figure 1A). To obtain hematopoietic cells, we further cultured the (Bry+Flk1+) cells for 4 days under conditions supporting the generation of blast colonies and from these cultures isolated CD41+ cells, which have been shown to represent the first fully committed hematopoietic cells.39 (B) Timing of mRNA expression of hematopoietic regulator genes (Runx1, Pu.1, and Csf1r) in differentiating ES cells. ES indicates ES cells; blast, 4-day blast cell culture derived from +/+ cells; LSK, Lin−Sca-1+c-kithi cells; CMP, common myeloid precursor cell; and Mac, primary macrophages. Numbers in panel B represent mean values of 2 independent experiments analyzed in duplicate.
To analyze how and at which stage chromatin of Pu.1 and Csf1r was activated, we studied several different epigenetic features that have been shown to be associated with chromatin priming events. Global studies of histone modifications in mouse embryonic stem (ES) cells showed that many transcriptional regulator genes display “bivalent” chromatin marks at their promoter regions consisting of elevated levels of histone H3 lysine 4 trimethylation (H3K4me3), a mark associated with active transcription, and H3 lysine 27 trimethylation (H3K27me3), which is indicative of polycomb action and gene silencing.35,40,41 Neither Csf1r nor Pu.1 carries H3K27me3 or H3K4me3 in ES cells.42 To test whether the establishment of bivalent chromatin marks preceded the transcriptional activation of Pu.1 and Csf1r during ES cell differentiation, we performed ChIP assays for these modifications with purified ES cells, ES-derived cells, and controls. In addition, we examined possible changes in histone H3 lysine 9 acetylation (supplemental Figures 2-3). Elevated levels of H3K4me3 or H3K27me3 could readily be detected at the promoters of genes previously shown to harbor bivalent marks in ES cells or Flk1+ cells (data not shown), but were not found at any of the Pu.1 and Csf1r cis-elements. Histone acetylation at Pu.1 and Csf1r cis-elements was found only in cells actively expressing these genes, such as the RAW264 cell line or blast cell cultures (supplemental Figure 2).
Replication timing of certain lineage-specific genes has been shown to change during the differentiation of ES cells, and early replication is believed to be indicative of an open chromatin structure.35,43 Both Pu.1 and Csf1r replicated early in ES cells and their progeny (supplemental Figure 4); this feature was therefore not informative. We therefore examined chromatin structure directly using high-resolution in vivo footprinting assays using DNaseI or micrococcal nuclease (MNase). MNase is used to define nucleosome positioning and detects regions of nucleosome remodeling. DNaseI introduces single-strand cuts at the surface of nucleosomes generating cleavage patterns that are defined by the rotational positioning of nucleosomes, transcription factor binding, and chromatin folding. Under conditions of limited digestion, DNaseI can also be used to assay the general accessibility of chromatin in different cell types (which should not be confused with DNaseI hypersensitivity), because nucleosomal DNA can be masked by higher order chromatin structure.9,17 Another feature of DNaseI is that it can detect altered cleavage patterns caused by short-lived transcription factor DNA interactions.8
We compared ES cells, ES-cell derived cell populations, and Pu.1/Csf1r-expressing primary macrophages with 3T3 cells where both genes are epigenetically silenced.8,17 LM-PCR analysis of DNA digested by MNase into mainly monosomes and some dinucleosomes (supplemental Figure 5 and data not shown) revealed nucleosome loss over the transcription start site in macrophages as indicated by a loss of signal over this region. 3T3 cells show prominent cuts across the same region, indicating alternatively positioned nucleosomes over the promoter. The analysis of ES cells and hemangioblast cells revealed a similar digestion pattern across the promoter region as in 3T3 cells. Digestion with increased amounts of MNase led to a rapid loss of signal in ES cells both at the entire promoter region as well as at the URE (data not shown). This finding is in concordance with previous experiments demonstrating that ES cell chromatin is hyperplastic.44
The DNaseI accessibility analysis revealed striking differences in the degree of chromatin accessibility at the promoters of Pu.1 and Csf1r in the different cell types. As expected, DNaseI accessibility was low at both genes in 3T3 cells compared with macrophages, as indicated by weaker band intensities throughout the promoter regions (Figure 2A). For Csf1r, the same low accessibility was seen in ES cells, epiblast-like Bry−Flk1−, mesodermal Bry+Flk1−, and hemangioblast Bry+Flk1+ cells. High accessibility was seen only in purified CD41+ cells from blast cultures where the gene was transcriptionally activated. In contrast, at the Pu.1 promoter, DNaseI accessibility increased during ES cell differentiation reaching a level similar to that observed in macrophages in the Bry+Flk1+ population (Figure 2B). When examining the promoter from a greater distance using a different primer, we noted that chromatin unfolding was localized and was sometimes already apparent in Bry+Flk1− cells (Figure 2C). Furthermore, alterations in the digestion pattern in PU.1 nonexpressing Bry+Flk1+ cells were similar to those seen in PU.1-expressing ES cell–derived CD41+ cells and primary macrophages from mice. The same early increase in DNaseI accessibility was also seen at another important Pu.1 cis-regulatory element, the upstream regulatory element (URE; supplemental Figure 6A). This element carries enhancer elements that are absolutely required for correct Pu.1 expression.12,15 The Oct4 gene, which is progressively silenced in differentiating ES cells, showed a clear reduction in DNaseI accessibility that was characteristic for each differentiation stage45 (supplemental Figure 6B-C). Changes in relative DNaseI accessibility as compared to an internal control (rDNA genes) were confirmed by the quantification of band intensities as depicted in the figure. Altogether, these results indicated an early chromatin unfolding in the hemangioblast-containing cell population that is distinct from chromatin opening caused by loss or remodeling of nucleosomes.
Chromatin at Pu.1 cis-regulatory elements unfolds at the onset of hemangioblast formation and before the Csf1r promoter. Assay examining relative DNaseI accessibility of the Csf1r promoter (A) and the Pu.1 promoter (B,C) in ES cells, ES-derived cells, 3T3 fibroblast cells, CD41+ cells sorted from day-4 blast cell cultures, and macrophages (Mac). (C) LM-PCR looking at Pu.1 with a primer set hybridizing at greater distance from the transcription start. Naked DNA (DNA) served as control and sequences were annotated by an LM-PCR amplifying genomic DNA modified by a G-specific Maxam-Gilbert G reaction (G). For each experiment, equal DNaseI digestion of samples was confirmed by amplification with primers specific for rDNA genes. Regions of localized increases/decreases in DNaseI accessibility at Csf1r and Pu.1 are indicated as gray or white bars, respectively. Signal intensities were measured across the indicated regions and were calculated relative to the rDNA control, with ES cells set as 1. The nature of transcription factor binding sites and their position are indicated at the left. Note that chromatin opening was established in the absence of gene expression. This is further illustrated by the absence of increased DNaseI accessibility in the coding region compared with macrophages (C).
Chromatin at Pu.1 cis-regulatory elements unfolds at the onset of hemangioblast formation and before the Csf1r promoter. Assay examining relative DNaseI accessibility of the Csf1r promoter (A) and the Pu.1 promoter (B,C) in ES cells, ES-derived cells, 3T3 fibroblast cells, CD41+ cells sorted from day-4 blast cell cultures, and macrophages (Mac). (C) LM-PCR looking at Pu.1 with a primer set hybridizing at greater distance from the transcription start. Naked DNA (DNA) served as control and sequences were annotated by an LM-PCR amplifying genomic DNA modified by a G-specific Maxam-Gilbert G reaction (G). For each experiment, equal DNaseI digestion of samples was confirmed by amplification with primers specific for rDNA genes. Regions of localized increases/decreases in DNaseI accessibility at Csf1r and Pu.1 are indicated as gray or white bars, respectively. Signal intensities were measured across the indicated regions and were calculated relative to the rDNA control, with ES cells set as 1. The nature of transcription factor binding sites and their position are indicated at the left. Note that chromatin opening was established in the absence of gene expression. This is further illustrated by the absence of increased DNaseI accessibility in the coding region compared with macrophages (C).
Another indication for the establishment of a primed chromatin structure is the removal of methyl cytosine (mCpG) marks from the DNA.46 Several studies demonstrated that selective DNA demethylation accompanies the onset of chromatin reorganization during development.47,48 We therefore determined the developmental time point of demethylation at the Pu.1 promoter and also at the Pu.1 URE by bisulfite sequencing (Figure 3and data not shown). The majority of the CpGs at the Pu.1 promoter were methylated in ES cells, Bry−Flk1− cells, and mesodermal Bry+Flk1− cells and were unmethylated in a cell population enriched for HSCs (Lin−Sca1+c-kit+ cells [LSKs]), common myeloid precursor cells (CMPs), and macrophages. Demethylation of CpGs within the Pu.1 promoter started in the majority of cells in the hemangioblast Bry+Flk1+ cell population where methylation of specific CpGs was reduced up to 50% within 12 hours. Demethylation occurred at both DNA strands and coincided with the region of localized chromatin opening (Figure 2C and data not shown). This was also observed at the URE (data not shown). Csf1r regulatory elements remained methylated throughout ES cell differentiation and were demethylated only from the LSK stage onward47,49 (and data not shown).
Selective DNA demethylation at the Pu.1 promoter. DNA from the indicated cell types was subjected to bisulfite sequencing. (A) Map of the Pu.1 promoter indicating the position of CpGs and transcription factor binding sites. (B) Average DNA methylation levels of 20 independently sequenced clones.
Selective DNA demethylation at the Pu.1 promoter. DNA from the indicated cell types was subjected to bisulfite sequencing. (A) Map of the Pu.1 promoter indicating the position of CpGs and transcription factor binding sites. (B) Average DNA methylation levels of 20 independently sequenced clones.
In summary, our data show a clear hierarchy of chromatin priming events at Pu.1 and Csf1r. Although both genes are expressed only after commitment to the hematopoietic lineage, the reorganization of chromatin at Pu.1 starts significantly earlier than at Csf1r and occurs in the absence of elevated levels of active or bivalent chromatin marks.
Chromatin unfolding at Pu.1 is crucially dependent on RUNX1
To gain insight into the mechanism of Pu.1 chromatin unfolding, we investigated the role of specific transcription factors. Chromatin unfolding was not observed in ES cells and epiblast-like cells, therefore a central role of OCT1, ATF2, and Sp1/3, all of which have been shown to be involved in Pu.1 regulation and are functional in these cells, was ruled out14,17,50 (electrophoretic mobility shift assay [EMSA] data not shown). C/EBP family members that play an important role in Pu.1 regulation51 were not expressed in hemangioblasts (supplemental Figure 1 and data not shown) and could therefore be ruled out as well. RUNX1 has been shown to bind to several sites in the URE,13,17 and mice with a targeted mutation of the RUNX1 binding sites show a severe reduction in Pu.1 expression.10 In addition, the onset of intermediate-level Runx1 mRNA expression at the hemangioblast stage correlated with maximal levels of chromatin remodeling at Pu.1 (Figures 1, 2B). We therefore tested the idea that RUNX1 was crucial for chromatin unfolding at Pu.1 in these cells and measured Pu.1 and Csf1r expression as well as Pu.1 chromatin unfolding in cells derived from RUNX1−/− ES cell lines. RUNX1−/− cells can generate hemangioblasts that express Flk-1, SCL, and FLI-1, but do not produce cells of the definitive hematopoietic lineage25 (and data not shown). supplemental Figure 7, panels A through C, shows that RUNX1−/− blast culture cells do not express Pu.1, Csf1r, or Cebpa. Most importantly, chromatin unfolding at the Pu.1 cis-regulatory elements did not occur in the Flk1+ hemangioblast population (supplemental Figure 7D). This effect was specific for Pu.1 because the absence of RUNX1 had no effect on DNaseI accessibility at the Oct4 promoter (supplemental Figure 7E).
The experiments described so far suggested that RUNX1 was directly involved in directing chromatin remodeling at Pu.1 by binding to its cis-regulatory elements in hemangioblast cells. To test this idea, we performed dimethylsulfate (DMS) footprinting experiments on FACS-purified cells. DMS methylates the G(N7) position of guanines and the binding of transcription factors modifies this reaction. However, DNA-protein interactions need to be stable to be detected, because DMS has sufficient time to react at sites of dynamic interactions during the 5-minute incubation period. Short-lived interactions yield partial footprints or none at all.34 Stable transcription factor binding at Pu.1 cis-regulatory elements as indicated by alterations in DMS-reactivity including a clear protection of the RUNX1 binding site at the URE (supplemental Figures 8, 9A) was visible in cells expressing PU.1 (sorted CD41+ cells and macrophages). However, no alterations could be seen in hemangioblasts. This was in contrast to the promoter of the SCL gene in the same cells where 2 transcription factor binding sites were clearly protected from methylation (supplemental Figure 9B). To be able to efficiently bind to DNA, RUNX1 requires a partner, CBFβ, which is also indispensable for hematopoietic development.52 The lack of stable binding of RUNX1 in hemangioblasts did not correlate with the absence of expression of CBFβ mRNA, which was the same in the presence or absence of RUNX1 (data not shown).
These results indicated that the URE was either not a direct target of RUNX1 in ES cell–derived cells or that interaction of RUNX1 was too short lived to be detected by DMS in vivo footprinting. To evaluate the first possibility, we generated a RUNX1 knockout ES cell line (iRUNX1) that expressed a HA-tagged RUNX1 protein from a doxycycline (DOX)–inducible promoter.30 To test whether this protein was capable of binding to Pu.1, we first rescued hematopoietic development by continuous induction with DOX from the blast culture stage onward and examined RUNX1 binding to the URE by ChIP, using an antibody against the HA-tag of iRUNX1. In blast culture cell populations, we detected iRUNX1 binding to the URE in a complex that also contained C/EBPβ and FLI-1, as shown previously17 (supplemental Figure 10 and data not shown). Binding levels were comparable with those at Runx1 distal promoter, which is itself a target of RUNX1.53,54
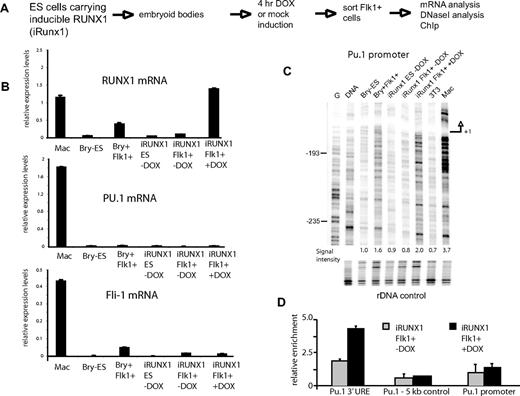
We next tested whether a short induction of iRUNX1 in Flk1+ cells in the absence of hematopoietic cell differentiation was sufficient to induce iRUNX1 binding and rescue chromatin remodeling. The induction of iRUNX1 by a 4-hour treatment with DOX in hemangioblasts at day 3.5 was not sufficient to switch on Pu.1 or Cebpa expression and did not alter expression of Fli1 (Figure 4 and data not shown), but chromatin remodeling was restored (Figure 4C). Also under these conditions no iRUNX1 binding to its target site was detected in hemangioblasts as measured by DMS in vivo footprinting (data not shown). However, using a large number of purified Flk1+ cells, we were able to detect the specific binding of iRUNX1 to the 3′ URE (Figure 4D) by ChIP. Binding was observed only in induced cells.
Induction of RUNX1 expression in RUNX1−/− ES cells carrying an inducible RUNX1 allele rapidly induces chromatin unfolding at Pu.1. (A) Experimental strategy. (B) Expression analyses of Runx1, Pu.1, and Fli1 mRNA in the indicated cell types and with/without doxycycline (DOX) induction (concentration 0.1 μg/mL). (C) DNaseI in vivo footprinting assays examining the Pu.1 promoter in the indicated cell types. Signal intensities were measured across the displayed regions and were calculated relative to the rDNA control, with ES cells set as 1. (D) iRUNX1 cells were differentiated into embryoid bodies and induced with 0.1 μg/mL DOX for 4 hours or left uninduced. Flk1+ cells were purified by magnetic cell sorting. ChIP assays were performed in duplicate with 107 cells for each assay and were normalized against the signal with an amplicon located on chromosome 2 (Chr2 control) representing a nonexpressed GAPDH pseudogene. Numbers in panels B and D are mean values between 2 measurements.
Induction of RUNX1 expression in RUNX1−/− ES cells carrying an inducible RUNX1 allele rapidly induces chromatin unfolding at Pu.1. (A) Experimental strategy. (B) Expression analyses of Runx1, Pu.1, and Fli1 mRNA in the indicated cell types and with/without doxycycline (DOX) induction (concentration 0.1 μg/mL). (C) DNaseI in vivo footprinting assays examining the Pu.1 promoter in the indicated cell types. Signal intensities were measured across the displayed regions and were calculated relative to the rDNA control, with ES cells set as 1. (D) iRUNX1 cells were differentiated into embryoid bodies and induced with 0.1 μg/mL DOX for 4 hours or left uninduced. Flk1+ cells were purified by magnetic cell sorting. ChIP assays were performed in duplicate with 107 cells for each assay and were normalized against the signal with an amplicon located on chromosome 2 (Chr2 control) representing a nonexpressed GAPDH pseudogene. Numbers in panels B and D are mean values between 2 measurements.
The weak binding of RUNX1 to its target suggested that RUNX1 only transiently interacted with Pu.1 in hemangioblasts. To test this idea by an alternative assay that also was more sensitive and required fewer cells, we constructed an inducible RUNX1−/− ES cell line (iRUNX-Dam) expressing a RUNX1-Dam-methylase fusion protein.55 Bacterial Dam-methylase targeted to specific binding sites methylates the adenines at nearby GATC (Dpn) sequences. This methyl mark stays behind after the factor has left.56 An ES cell line that expressed only Dam-methylase (iDam) served as control. The expression levels of iDam and iRUNX1-Dam were similar (data not shown). Figure 5 shows that a 4-hour induction of the RUNX1-Dam fusion protein in Flk1+ cells led to an increase of GATC methylation at the Pu.1 URE and to a lesser extent at the promoter compared with the iDam control. A similar increase was seen in the positive control looking at the distal Runx1 promoter, but not at the Chr2 control locus. Although Csf1r contains a RUNX1 binding site in the promoter,57 we were unable to see an increase in GATC methylation with iRUNX1-Dam at this position, substantiating the developmental stage–specific differences in the accessibility of chromatin between Csf1r and Pu.1 and demonstrating the specificity of our assay. The same results were obtained in a RUNX1 wt genetic background (data not shown), thereby confirming the results with a different ES cell clone.
RUNX1 binds transiently to Pu.1 in hemangioblasts. DamId assay analyzing GATC methylation levels at the indicated genes with hemangioblasts expressing Dam-methylase alone (iDam control) or RUNX1-Dam (iRUNX1-Dam) after a 4-hour induction with DOX. Differential methylation was detected by cleaving first with Dpn1, which cleaves only methylated DNA, and then with DpnII, which cleaves only unmethylated DNA.32 The Chr2 control represents a nonexpressed GAPDH pseudogene. The values represent the mean values of 3 independent experiments.
RUNX1 binds transiently to Pu.1 in hemangioblasts. DamId assay analyzing GATC methylation levels at the indicated genes with hemangioblasts expressing Dam-methylase alone (iDam control) or RUNX1-Dam (iRUNX1-Dam) after a 4-hour induction with DOX. Differential methylation was detected by cleaving first with Dpn1, which cleaves only methylated DNA, and then with DpnII, which cleaves only unmethylated DNA.32 The Chr2 control represents a nonexpressed GAPDH pseudogene. The values represent the mean values of 3 independent experiments.
Taken together, these experiments indicate that although RUNX1 is required for chromatin unfolding at Pu.1, it is unable to assemble stable transcription factor complexes on its target in hemangioblasts and interacts only transiently with its template.
RUNX1 is not required for the maintenance of active chromatin at Pu.1 and Csf1r
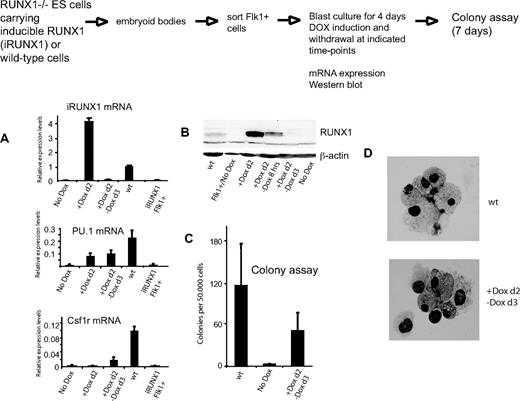
Using conditional gene targeting strategies, it was shown that RUNX1 is required to establish the hematopoietic system, but is not essential to maintain it.26,27,58 The molecular basis of this finding is unknown. To examine the role of RUNX1 in the maintenance of active chromatin at Pu.1, we made use of the fact that the DOX-responsive promoter expressing RUNX1 was rapidly switched off after DOX removal.59 To this end, we sorted hemangioblasts from iRUNX1 ES cells, plated them in blast mix, and induced them with DOX after 2 days of culture. Wild-type (wt) cells served as control. DOX was removed after 12 hours and cells were cultured for 4 days in total, then were harvested to measure Runx1, Pu.1, and Csf1r mRNA levels and to plate them into methylcellulose cultures to form hematopoietic colonies. Figure 6 shows that without DOX induction no Pu.1 or Csf1r mRNA was expressed and little or no definitive hematopoietic colonies were formed. DOX treatment alone did not influence colony formation in wt cells (data not shown). Twenty-four hours after DOX withdrawal, no Runx1 mRNA or RUNX1 protein could be detected (Figure 6A-B). RUNX1 induction was required to induce Pu.1 mRNA, but not for its sustenance after DOX withdrawal. This was also seen with C/EBP family members (supplemental Figure 11). Moreover, like in wt cultures (supplemental Figure 7B), Csf1r mRNA was induced with a delayed kinetics. Although colony numbers were reduced compared with wild-type cells where differentiation was continuous, myeloid cells had a normal morphology, demonstrating that a 12-hour pulse of DOX rescued definitive hematopoietic development, which was then sustained for at least 9 days (Figure 6C-D). DMS in vivo footprinting and ChIP experiments demonstrated that RUNX1 was not required to maintain the binding of all other factors at the 3′URE and the promoter after RUNX1 withdrawal (Figure 7 and data not shown). We noted that there was a weak, but reproducible, protection of the RUNX1 binding site at the URE. Inspection of the expression of other RUNX1 family members indicated the presence of RUNX2 and RUNX3 mRNA (supplemental Figure 11). When overexpressed, these factors can indeed rescue hematopoietic development in RUNX1 knockout ES cells.63 However, both were also present in the absence of DOX, indicating that these proteins may be able to cooperate with other factors at the URE once these have been induced, but that at the levels at which they are expressed in hemangioblasts are unable to replace RUNX1 at early stages of hematopoietic development.
A 12-hour pulse of RUNX1 expression is sufficient to rescue continuous Pu.1 mRNA expression, induce Csf1r mRNA expression, and rescue subsequent hematopoietic development. ES cells were differentiated as indicated. mRNA of the indicated cell populations (iRUNX1 and wt) was measured by real-time PCR (A) and RUNX1 protein was measured by Western blotting (B) using an anti-RUNX1 antibody after 4 days of blast culture (except for the +DOXd2 samples, which were measured before or after 8 hours of withdrawal, as indicated). (C) Number of hematopoietic colonies before and after DOX withdrawal; (D) Giemsa-stained cytospin of representative colonies. mRNA of iRUNX1 Flk1+ cells was prepared straight after cell sorting. Numbers in panels A and C represent mean values of 2 independent experiments analyzed in duplicate.
A 12-hour pulse of RUNX1 expression is sufficient to rescue continuous Pu.1 mRNA expression, induce Csf1r mRNA expression, and rescue subsequent hematopoietic development. ES cells were differentiated as indicated. mRNA of the indicated cell populations (iRUNX1 and wt) was measured by real-time PCR (A) and RUNX1 protein was measured by Western blotting (B) using an anti-RUNX1 antibody after 4 days of blast culture (except for the +DOXd2 samples, which were measured before or after 8 hours of withdrawal, as indicated). (C) Number of hematopoietic colonies before and after DOX withdrawal; (D) Giemsa-stained cytospin of representative colonies. mRNA of iRUNX1 Flk1+ cells was prepared straight after cell sorting. Numbers in panels A and C represent mean values of 2 independent experiments analyzed in duplicate.
Active chromatin is maintained after RUNX1 withdrawal. (A) In vivo DMS footprinting assay examining the Pu.1 3′ URE conducted with 3T3 cells, macrophages (Mac), and CD41+ cells sorted after 4 days of a blast culture that had been continuously induced with 1 μg/mL doxycycline at day 2 (+DOX) and where the inducer was removed after day 3 (−DOX). Transcription factor binding sites are indicated on the right. Guanines displaying hyperreactivity to DMS-methylation are marked by •; protections from methylation are indicated by white circles; the weak protection over the RUNX site is indicated by a gray circle. Only reproducible changes are marked. Remaining band intensity (with protected bands) or fold enhancement (with hyperreactive bands) is indicated next to the bands with the G reaction set as 1. (B) ChIP assay demonstrating binding of C/EBPβ to its binding sites at the 3′ URE and the Pu.1 promoter in blast cultures from wt cells (■) and induced iRUNX1 cells (iRUNX-DOX) where DOX has been withdrawn as described in panel A. (C) Network diagram illustrating the initiating role of RUNX1 in establishing open chromatin and the expression of hematopoietic master regulators such as PU.1 in pluripotent hematopoietic precursor cells as well as the maintenance of the active state in differentiating myeloid cells. Once Runx1 is activated in pluripotent hematopoietic cells,60,61 a regulatory circuit is established capable of maintaining open chromatin in the absence of RUNX1 and driving myelopoiesis. The induction of secondary factors driving macrophage-specific genes has been described in Krysinska et al9 and Laslo et al.62 The relative importance of the different transcription factors is illustrated by thin (less important) and bold (more important) lines. Numbers in panel B represent mean values of 2 independent experiments analyzed in duplicate.
Active chromatin is maintained after RUNX1 withdrawal. (A) In vivo DMS footprinting assay examining the Pu.1 3′ URE conducted with 3T3 cells, macrophages (Mac), and CD41+ cells sorted after 4 days of a blast culture that had been continuously induced with 1 μg/mL doxycycline at day 2 (+DOX) and where the inducer was removed after day 3 (−DOX). Transcription factor binding sites are indicated on the right. Guanines displaying hyperreactivity to DMS-methylation are marked by •; protections from methylation are indicated by white circles; the weak protection over the RUNX site is indicated by a gray circle. Only reproducible changes are marked. Remaining band intensity (with protected bands) or fold enhancement (with hyperreactive bands) is indicated next to the bands with the G reaction set as 1. (B) ChIP assay demonstrating binding of C/EBPβ to its binding sites at the 3′ URE and the Pu.1 promoter in blast cultures from wt cells (■) and induced iRUNX1 cells (iRUNX-DOX) where DOX has been withdrawn as described in panel A. (C) Network diagram illustrating the initiating role of RUNX1 in establishing open chromatin and the expression of hematopoietic master regulators such as PU.1 in pluripotent hematopoietic precursor cells as well as the maintenance of the active state in differentiating myeloid cells. Once Runx1 is activated in pluripotent hematopoietic cells,60,61 a regulatory circuit is established capable of maintaining open chromatin in the absence of RUNX1 and driving myelopoiesis. The induction of secondary factors driving macrophage-specific genes has been described in Krysinska et al9 and Laslo et al.62 The relative importance of the different transcription factors is illustrated by thin (less important) and bold (more important) lines. Numbers in panel B represent mean values of 2 independent experiments analyzed in duplicate.
In summary, our experiments suggest that (1) RUNX1 is absolutely required to unfold chromatin at Pu.1 in hemangioblasts, (2) that RUNX1 interacts with Pu.1 in a transient fashion, and (3) that RUNX1 is required for the activation of Pu.1, Csf1r, and Cebpa transcription, but it is not essential once hematopoietic precursors have formed, and (4) that this is based on the fact that once stable transcription factor complexes are assembled at Pu.1 cis-regulatory elements, they remain in the absence of RUNX1.
Discussion
In this study, we made several significant new observations. First, we show that chromatin activation at Pu.1 and Csf1r follows a clear hierarchy that is defined by a differential transcription factor dependency. Although RUNX1 binds to both loci,10,64 it is sufficient to initiate chromatin remodeling at Pu.1 at the onset of hematopoietic development, but not at Csf1r. In previous experiments examining Csf1r activation, we demonstrated that at this gene chromatin unfolding is crucially dependent on the expression of high levels of PU.1.9 We recently found that RUNX1 binding to Csf1r is also dependent on the presence of PU.1 (data not shown). This indicates that although RUNX1 is required to activate important hematopoietic regulator genes and thus generate hematopoietic cells in the first place, its priming activity is context dependent, ensuring that in the embryo Csf1r is not expressed outside the hematopoietic system.
Our second important result demonstrates that at Pu.1 chromatin unfolding and the selective demethylation of DNA precede the establishment of active histone marks. Neither Pu.1 nor Csf1r carries significant levels of bivalent chromatin marks in ES cells or their immediate progeny, but instead are organized in DNaseI inaccessible chromatin and DNA is methylated. It is therefore likely that both genes have to remain firmly silenced until needed to ensure that blood cell development proceeds normally.
Another important result from this study is our finding that RUNX1 directly interacts with its target sequences in hemangioblasts but that this interaction is transient. Experiments looking at the interaction of the unliganded estrogen receptor with chromatin demonstrated that incomplete factor complexes occupy their binding sites with a very short half-life and are often detectable only in synchronized cells.65 Using a high-affinity antibody, we could previously show that unstable protein-DNA interactions that could induce chromatin remodeling in a hit-and-run fashion do occur in precursor cell lines,34 and our experiments provide direct evidence that this is also the mechanism operating in the differentiation system described here. Hemangioblasts express several transcription factors capable to bind to Pu.1, but none of them is able to initiate chromatin unfolding in the absence of RUNX1. For example, FLI-1 can drive high-level Pu.1 expression in the absence of PU.1,17 but cannot bind to the URE without RUNX1 in hemangioblasts.
To rescue hematopoietic development in Runx1 knockout ES cells, both the DNA binding domain of RUNX1 as well as its transcription activation domain are necessary,63 indicating that RUNX1 is likely to cooperate with other transcription factors to form transient complexes on target DNA that initiate chromatin unfolding. In contrast, the DNA binding domain of SCL/TAL1, another TF critical in hematopoietic development, is not required to rescue blood cell development in SCL/TAL1 knockout ES cells, indicating that this factor collaborates with other proteins to perform its crucial function on its DNA targets.66 Taken together, our results indicate that transcription factors change chromatin gradually until a threshold is reached. This finding has important consequences with respect to our current thinking of how and when cell fate decisions are regulated, because it suggests that even subcomplexes of transcription factors that are unable to form stable and heritable complexes on their target genes can play a regulatory role.
Last, but not least, our study sheds light on the molecular details of how a stable transcriptionally active state is maintained. The actual transcriptional activation of Pu.1 occurs in a sequential fashion (Figure 7C). First, chromatin is unfolded and other lineage-specific transcription factors are induced. Thereafter, these factors form stable transcription factor complexes on Pu.1 cis-regulatory elements, which, in turn, instigate the establishment of active histone marks and drive the onset of gene expression. In this context, it is interesting to note that the Pu.1 URE contains 2 C/EBP binding sites crucial for enhancer function.16 Similar to PU.1, C/EBPα and C/EBPβ are expressed in hematopoietic cells, but not in hemangioblasts and their expression is also crucially dependent on RUNX1. Moreover, C/EBP binding is maintained in the absence of RUNX1. It is therefore likely that C/EBP proteins help to stabilize an assembled enhancer complex at the URE in hematopoietic cells that forms an activated chromatin structure.
RUNX1 is absolutely required for the commitment of hemangioblasts to the definitive hematopoietic lineage. Similar to what has recently been shown in the mouse,58 our study defines a brief developmental window after which RUNX1 is no longer essential for hematopoietic differentiation and the activation of myeloid-specific genes. As outlined in Figure 7C, we propose that this is because RUNX1 is crucial for the initial chromatin remodeling at transcription factor genes that serve as master regulators for specific blood cell lineages. Our data suggest that once these transcription factors are expressed, stable transcription factor complexes are formed on these genes and active chromatin is maintained, meaning that an epigenetic memory for active gene expression is established. Once this has occurred, RUNX1 becomes less important and is used only in certain genomic contexts as one of many other transcription factors that in combination are necessary to efficiently maintain the hematopoietic transcriptional network. This could explain why RUNX1 is not generally essential for adult hematopoiesis,26,27 but has been shown to be necessary for maintaining normal blood cell development over time,67 suggesting that its deletion impacts on the long-term stability of transcription factor complexes in a continuously replicating precursor cell population. Although our results are currently based on the detailed analysis of only a few genes, we believe that similar principles operate in the entire hematopoietic system and at many different developmental pathways.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Bas van Steensel (Amsterdam, The Netherlands) for the Dam methylase expression plasmids. We also thank Bertie Göttgens (Cambridge, United Kingdom), Peter Cockerill, Gina Doody, and Reuben Tooze (all in Leeds, United Kingdom) for helpful comments on the paper and Liz Straszcinski and Adam Davidson for expert help with cell sorting.
Work in C.B.'s laboratory is funded by grants from the Biotechnology and Biological Sciences Research Council (BBSRC, Swindon, United Kingdom), the European Community (Integrated Project EuTRACC, grant no. LSHG-CT-2007-037445), and Leukemia Research (London, United Kingdom). A.F.'s laboratory is supported by the MRC, and D.G.T.'s laboratory is supported by National Institutes of Health (NIH, Bethesda, MD) grant CA41456. G.L.'s and V.K.'s laboratories are supported by Cancer Research UK (CRUK, London, United Kingdom) and the BBSRC.
National Institutes of Health
Authorship
Contribution: M.H., M.L., H.K., C.L., and L.M. designed and performed experiments and analyzed data; D.C., A.W., and R.I. performed experiments; H.J. designed experiments and analyzed data; A.F. and D.G.T. helped write the paper; V.K. and G.L. designed and performed experiments and helped write the paper; and C.B. conceived the study, designed experiments, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Constanze Bonifer, University of Leeds, St James University Hospital, The Wellcome Trust Brenner Bldg, Leeds LS9 7TF, United Kingdom; e-mail: c.bonifer@leeds.ac.uk.
References
Author notes
*M.H., M.L., H.K., and C.L. contributed equally to this study.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal