Abstract
Neutrophils play a vital role in the immune defense, which is evident by the severity of neutropenia causing life-threatening infections. Granulocyte macrophage-colony stimulating factor (GM-CSF) controls homeostatic and emergency development of granulocytes. However, little is known about the contribution of the downstream mediating transcription factors signal transducer and activator of transcription 5A and 5B (STAT5A/B). To elucidate the function of this pathway, we generated mice with complete deletion of both Stat5a/b genes in hematopoietic cells. In homeostasis, peripheral neutrophils were markedly decreased in these animals. Moreover, during emergency situations, such as myelosuppression, Stat5a/b-mutant mice failed to produce enhanced levels of neutrophils and were unable to respond to GM-CSF. Both the GM-CSF–permitted survival of mature neutrophils and the generation of granulocytes from granulocyte-macrophage progenitors (GMPs) were markedly reduced in Stat5a/b mutants. GMPs showed impaired colony-formation ability with reduced number and size of colonies on GM-CSF stimulation. Moreover, continuous cell fate analyses by time-lapse microscopy and single cell tracking revealed that Stat5a/b-null GMPs showed both delayed cell-cycle progression and increased cell death. Finally, transcriptome analysis indicated that STAT5A/B directs GM-CSF signaling through the regulation of proliferation and survival genes.
Introduction
Neutrophils are the most abundant type of white blood cells in human peripheral blood and an integral part of the immune system. The essential role of neutrophils in infection is demonstrated by the severity of congenital, acquired, and therapy-induced neutropenia, which causes life-threatening infections.
Granulocyte-colony stimulating factor (G-CSF) and granulocyte macrophage-colony stimulating factor (GM-CSF) largely control the biology of neutrophils.1,2 GM-CSF, secreted by fibroblasts, endothelial cells, monocytes/macrophages, and activated T cells, regulates granulopoiesis and neutrophil mobilization to peripheral blood. The GM-CSF receptor is present on almost all types of myeloid progenitors and on mature monocytes, neutrophils, eosinophils, basophils, and dendritic cells. It consists of a ligand-specific subunit (GM-CSFRα) and the common beta subunit (βc),1 which is shared with the interleukin-3 (IL-3) and IL-5 receptors. GM-CSF is known to activate at least 3 signaling venues: the Janus kinase (Jak) 2/signal transducer and activator of transcription (STAT) pathway,3 the ras/mitogen-activated protein kinase (MAP kinase) pathway,4 and the phosphoinositide-3-kinase (PI3-kinase) pathway.5
Whereas G-CSF can activate 3 members of the STAT family (STATs 1, 3, and 5) with highest activation of STAT3, GM-CSF preferentially activates STAT5A and STAT5B (referred to as STAT5A/B throughout the text).3,6 STAT5A/B are transcription factors encoded by 2 juxtaposed genes7,8 that respond to diverse cytokines, including growth hormone, erythropoietin, interleukins, prolactin, and ligands of the epidermal growth factor family. STAT5A/B are key regulators in the development and function of various blood lineages, including regulatory T helper cells,9,10 erythroid precursors,11 and stem cells.12 STAT3 is also activated by an array of cytokines, interferons, and growth factors.13,14 Its role in granulopoiesis is ambivalent. On the one hand, it is a negative regulator of basal granulopoiesis, and mice lacking STAT3 in bone marrow progenitors display marked peripheral neutrophilia, similar and paradoxically, to mice lacking SOCS3 under resting conditions.15,16 SOCS3-mutant bone marrow cells and neutrophils show enhanced and prolonged STAT3 phosphorylation and unusual high activation of the MAP kinase pathway on G-CSF stimulation.15,17 On the other hand, STAT3 has been shown to be critical for G-CSF–mediated proliferation and mobilization of the granulocytic lineage.16,18 STAT1 does not appear to contribute substantially to the granulocyte lineage.19 A limited number of studies have addressed the role of STAT5A/B in granulopoiesis.3,20-23 Although these studies intended to analyze granulopoiesis in the absence of STAT5A/B, they had been performed with mice that were later shown to not be STAT5A/B deficient but express an N-terminally truncated hypomorphic STAT5A/B (Stat5ΔN) with remaining biologic activity.24 The conclusions of these studies therefore need to be viewed with caution and do not allow reliable conclusions without confirmation by further analysis of mouse lines with reliable full deletion of STAT5A/B activity in hematopoietic cells.
Strong activation of STAT5A/B has been observed in 30% of patients with acute myelogenous leukemia (AML), which is characterized by FMS-like tyrosine kinase 3 mutations (internal tandem duplications).25 Because the majority of AML cells are from the granulocyte lineage, we hypothesized that STAT5A/B contribute to the proliferation, differentiation, and/or survival of the granulocyte lineage. To elucidate the role of STAT5A/B in GM-CSF–mediated granulopoiesis and to understand the underlying mechanisms, we have analyzed granulopoiesis in mice with deletion of the Stat5a/b locus in all blood cells. In addition, purified granulocyte-macrophage progenitors (GMPs) were analyzed in vitro to define cell proliferation and survival in the presence and absence of STAT5A/B, also by time-lapse microscopy and long-term single cell tracking.
Methods
Generation of mice with a deletion of the Stat5a/b locus in hematopoietic lineages
Mice in which the locus encoding the Stat5a and Stat5b genes had been targeted with loxP sites (Stat5f/f mice)26 were crossed with transgenic mice expressing the Cre recombinase gene under the control of the Mx1 gene promoter.27 Stat5f/f:Mx1-Cre mice in the C57BL/6NCr congenic strain were used throughout this study. The Mx1-Cre transgene was activated through PolyIC (polyinosinic: polycytidylic acid; intraperitoneal injection) as described.27 Genotyping was performed by polymerase chain reaction (PCR) using genomic DNA and the following primers: (a) 5′-agcagcaaccagaggactac-3′; (b) 5′-cccattatcaccttc tttacag-3′; (c) 5′-gaaagcatgaaagggttggag-3′; (d) 5′-tacccgcttccattgctcag-3′; (e) 5′-gcggagccagcactattta-3′; and (f) 5′-ccggcatcaacgttttctttt-3′.
Primers a and b were used to detect the Stat5a/b-null alleles (590 bp), a and d for the floxed allele (530 bp), and a and c for the wild-type allele (450 bp). For checking deletion efficiency, primers a, b, and d were used for multiplex PCR. Primers e and f were used to detect the Mx1-Cre transgene (500 bp).
All animal experiments were approved by the Animal Care and Use Committee at the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). Animals were maintained in accordance with National Instiutes of Health (NIH) guidelines with the approval of the NIH Institutional Animal Care and Use Committee.
Hematologic data
Peripheral blood from retro-orbital puncture was collected in heparinized capillary tubes (Drummond Scientific Co), and complete blood counts (CBCs) were determined (HEMAVET HV950FS; Drew Scientific). 5-Fluorouracil (5-FU; 250 mg/kg) was injected intraperitoneally, and the CBC was measured every 3 days. To observe the in vivo cytokine response, either G-CSF or GM-CSF (both 50 μg/body weight per kilograms/day) were injected subcutaneously for 5 days and CBC was measured every day. The data were shown as normalized ratio using the following formula: ratio = (neutrophil count Stat5f/f/neutrophil count Stat5f/f:Mx1-Cre)/(neutrophil count Stat5f/f at day 0/neutrophil count Stat5f/f:Mx1-Cre at day 0).
To measure serum G-CSF and GM-CSF level at steady state (14 weeks after last PolyIC injection), enzyme-linked immunosorbent assays (Quantikine; R&D Systems) were performed according to the manufacturer's instructions.
Analyzing and sorting of bone marrow progenitor cells
Bone marrow (BM) was flushed from both femora and tibiae of 12- to 14-week-old mice. Bone marrow mononuclear cells (BMMNCs) were purified by Ficoll gradient centrifugation (Histopaque 1083; Sigma-Aldrich) or ACK solution (Lonza Walkersville). For sorting of GMPs (CD117+, Sca-1−, lineage−, CD34+, and CD16/32+),28 BMMNCs were stained with biotinylated antibodies specific for the following lineage markers: CD4 (H129.19), CD8 (53-6.7), B220 (RA3-6B2), Gr-1 (RB6-8C5), Ter-119 (Ter-119), IL-7Rα chain (B12-1 or A7R34), and in some experiments additionally CD3 (145-2C11) and CD19 (1D3). Lineage+ cells were partially removed with goat anti–rat IgG-conjugated magnetic beads (BioMag; QIAGEN), and the remaining cells were stained with streptavidin-Pacific blue (Invitrogen). Cells were stained with antibodies against CD16/32-PE (2.4G2; BD Biosciences), CD34-Alexa647 (RAM34), Sca-1-PE-Cy5.5 (E13-161.7 or D7; BD Biosciences), and CD117-PE-Cy7 (2B8; BD Biosciences). Cells were sorted or analyzed using a MoFlo (Dako Denmark) or a FACSAria (BD Biosciences) instrument. All antibodies were from eBioscience, if not otherwise mentioned. Living cells after sorting were counted by trypan blue exclusion.
Detection of phosphorylated STATs by FACS
Neutrophils from BM were starved in RPMI with 2% fetal bovine serum for 3 hours and then stimulated with G-CSF or GM-CSF (both 50 ng/mL) for 15 minutes, before fixation (2% paraformaldehyde) and permeabilization (cold methanol/acetone, 1:1 vol/vol). Phosphorylated STATs were detected with antiphosphorylated-STAT1 (4A), -STAT3 (4/P-STAT3), and -STAT5A antibody (47; all BD Biosciences) by FACS.
For analyzing GMPs, Sca-1− lineage− BM cells were obtained by magnetic lineage depletion (Biotin Selection Kit; StemCell Technologies) and starved for 2.5 hours in serum-free expansion medium (StemCell Technologies) before stimulation with either G-CSF or GM-CSF (both 100 ng/mL; PeproTech) for 15 minutes. Cells were fixed with Cytofix (BD Biosciences) and permeabilized with 100% cold ethanol. Phosphorylation of STATs in GMPs was detected by staining against antiphosphorylated-STAT3, -STAT5A, and surface markers CD117, CD48, and CD16/32.
Colony-formation assay of GMPs
A total of 300 GMPs were cultured in MethoCult M3234 (StemCell Technologies), supplemented with either G-CSF or GM-CSF (both 10 ng/mL; PeproTech) at 37°C, 5% CO2 for 8 days and microscopically scored. Cell numbers of colonies after 7 and 8 days for G-CSF and GM-CSF culture, respectively, were determined by picking individual colonies and microscopically counting living cells with trypan blue exclusion.
Functional analysis of neutrophils
For migration assay, phosphate-buffered saline (control) or 4% thioglycollate (TGA; Sigma-Aldrich) was injected intraperitoneally. After 4 hours, the percentage of neutrophils in peritoneal lavage was scored by May-Gruenwald-Giemsa staining. Neutrophils were isolated from BM using biotinylated anti–Gr-1 antibody, anti–rat IgG MicroBeads (Miltenyi Biotec), and LS Columns (Miltenyi Biotec). The purity was always more than 94%. Phagocytic activity of BM derived neutrophils was assayed by incubating 5 × 104 cells of neutrophils with 5 × 106 of Texas Red–conjugated opsonizing zymosan (Invitrogen) for 30 minutes and assessed by FACS. To measure superoxide production, BM-derived neutrophils were preincubated at 37°C for 15 minutes in the presence of 10μM hydroethidine (Fluka), and cell suspension was stimulated with 10μM of phorbol myristate acetate (Sigma-Aldrich) at 37°C for 10 minutes, before immediate FACS. Granulocytes were defined by successive gating on forward/side scatter and anti–Gr-1 staining. Neutrophil counts after in vitro culture were determined by trypan blue exclusion of dead cells.
Images were captured with a Nikon DXM 1200 digital camera on a Nikon ECLIPSE TE300 microscope (original magnification 10×/0.30 numeric aperture objective) with AxioVision Rel 4.7 software (Carl Zeiss). Captured images were processed by Microsoft Office PowerPoint 2003 and Adobe Photoshop Version 9 software.
Time-lapse imaging
GMPs from Stat5f/f orStat5f/f:Mx1-Cre mice were cultured in serum-free expansion medium, 10% fetal bovine serum, and 20 ng/mL GM-CSF in one 24-well dish applied with a Culture-Insert (IBIDI). Time-lapse imaging was performed with a CellObserver system (Carl Zeiss) at constant 37°C and 5% CO2. Phase-contrast images of each position were taken every 2.5 minutes with a 10× Neofluar objective (Carl Zeiss) and an AxioCamHRm camera (at 1388 × 1040 pixel resolution) with Carl Zeiss AxioVision 4.5 software.
Single-cell tracking
Affymetrix microarray
After starvation, GMPs were stimulated for 3 hours with GM-CSF (100 ng/mL) or left unstimulated. Total RNA from 4 different samples (Stat5f/f-unstimulated, Stat5f/f-stimulated and Stat5f/f:Mx1-Cre-unstimulated, Stat5f/f:Mx1-Cre-stimulated) was prepared by RNeasy Plus Mini Kit (QIAGEN) and processed/analyzed by NIDDK Core Facility at the NIH. Gene Chip expression 3′ amplification reagents in 2-cycle cDNA synthesis kit (Affymetrix) was used. Affymetrix gene expression analysis array for the Mouse430_2 was used. The microarray signals were referred by the Affymetrix Robust Multichip Average algorithm. Up- and down-regulated genes were selected based on P values (< .05) and fold change (> 2.0 or < −2.0) as assessed by analysis of variance with Partek Pro software (Partek). Multiple testing was covered by a false discovery rate correction (supplemental Table 3, available on the Blood website; see the Supplemental Materials link at the top of the online article). Potential STAT5A/B-regulated genes were identified by comparing genes that were up- or down-regulated in control GMPs on GM-CSF stimulation but were unchanged (within the 2-fold range) in STAT5a/b mutant GMPs. Hierarchical average linkage clustering of the statistical significant gene list (853 probes) was performed using the Euclidean distance by Partek Pro software. For the heat map, the mean values of each individual probe set were normalized according to the standard score calculation. The microarray analysis was performed with 3 independent biologic sample sets.
Statistical analyses
Statistical significance (P < .05) was determined by Student t test (2-tailed, unpaired/unequal variances), except for microarray analyses.
Results
Highly efficient conditional deletion of the Stat5a/b locus in hematopoietic cells
To explore the function of STAT5A/B in the development of the granulocyte lineage, the mouse Stat5a/b locus was deleted in all hematopoietic lineages using Cre-mediated recombination. Stat5f/f:Mx1-Cre mice carried a Mx1-Cre transgene27 and 2 Stat5a/b floxed alleles.26 The Mx1-Cre transgene was induced by PolyIC, and Stat5a/b deletion occurred in cells carrying interferon receptors. Importantly, and in contrast to approaches used in previous studies,3,22,23 this allows the reliable complete deletion of STAT5A/B without residual STAT5A/B activity. Cre-mediated deletion of the Stat5a/b locus was essentially complete in BMMNCs and granulocytes (supplemental Figure 1). As detailed in Table 1, the absolute number of peripheral neutrophils in Stat5f/f:Mx1-Cre mice was decreased by 50% (P < .001) in homeostasis. As expected, Stat5a/b-mutant mice also exhibited anemia and a decrease of white blood cell counts, in particular of lymphocytes (70% decrease) and monocytes (44% decrease). This demonstrates that STAT5A/B are required for the maintenance of normal steady-state granulopoiesis but not for the establishment of the granulocyte lineage.
Evaluation of steady-state hematopoiesis in Stat5f/f:Mx1-Cre mice
| . | Stat5f/f . | Stat5f/f:Mx1-Cre . | P . |
|---|---|---|---|
| Red blood cells, × 106/μL | 9.08 ± 0.48 | 5.94 ± 1.28 | < .001 |
| Hemoglobin, g/dL | 13.3 ± 0.6 | 8.0 ± 0.9 | < .001 |
| Hematocrit, percentage | 41.9 ± 2.0 | 25.9 ± 3.4 | < .001 |
| White blood cells, × 103/μL | 8.70 ± 1.61 | 3.05 ± 1.06 | < .001 |
| Neutrophils, × 103/μL | 1.54 ± 0.35 | 0.71 ± 0.35 | < .001 |
| Lymphocytes, × 103/μL | 6.80 ± 1.41 | 2.11 ± 0.78 | < .001 |
| Monocytes, × 103/μL/μL | 0.30 ± 0.09 | 0.17 ± 0.09 | < .001 |
| Eosinophils, × 103/μL | 0.05 ± 0.03 | 0.04 ± 0.03 | NS |
| Basophils, × 103/μL | 0.01 ± 0.01 | 0.01 ± 0.01 | NS |
| Platelets, × 103/μL | 850 ± 200 | 744 ± 164 | NS |
| . | Stat5f/f . | Stat5f/f:Mx1-Cre . | P . |
|---|---|---|---|
| Red blood cells, × 106/μL | 9.08 ± 0.48 | 5.94 ± 1.28 | < .001 |
| Hemoglobin, g/dL | 13.3 ± 0.6 | 8.0 ± 0.9 | < .001 |
| Hematocrit, percentage | 41.9 ± 2.0 | 25.9 ± 3.4 | < .001 |
| White blood cells, × 103/μL | 8.70 ± 1.61 | 3.05 ± 1.06 | < .001 |
| Neutrophils, × 103/μL | 1.54 ± 0.35 | 0.71 ± 0.35 | < .001 |
| Lymphocytes, × 103/μL | 6.80 ± 1.41 | 2.11 ± 0.78 | < .001 |
| Monocytes, × 103/μL/μL | 0.30 ± 0.09 | 0.17 ± 0.09 | < .001 |
| Eosinophils, × 103/μL | 0.05 ± 0.03 | 0.04 ± 0.03 | NS |
| Basophils, × 103/μL | 0.01 ± 0.01 | 0.01 ± 0.01 | NS |
| Platelets, × 103/μL | 850 ± 200 | 744 ± 164 | NS |
Data are mean ± SD of peripheral blood count analyses of Stat5f/f (n = 22) and Stat5f/f:Mx1-Cre mice (n = 23) at the age of 9 to 14 weeks. Peripheral blood was assayed at least 3 weeks after PolyIC injection.
NS indicates no significant difference.
Endogenous serum levels of the granulocytic lineage-supporting cytokines G-CSF and GM-CSF, measured by enzyme-linked immunosorbent assay, showed no significant difference between control and Stat5a/b-mutant mice under steady-state conditions at 14 weeks after PolyIC injection. However, the mean G-CSF level in blood was marginally increased in Stat5a/b-mutant mice: G-CSF (pg/mL), 130 (± 9; Stat5f/f), 189 (± 38; Stat5f/f:Mx1-Cre); GM-CSF (pg/mL), 26.6 (± 3.4; Stat5f/f), 26.2 (± 1.6; Stat5f/f:Mx1-Cre; mean of 3 mice per group ± SD, no significant difference). These results indicate that neither neutropenia observed in Stat5a/b-mutant mice is the result of suppressed G-CSF or GM-CSF productions, nor that aberrant cytokine levels might absorb further defects in granulopoiesis in these mice. This was further supported by the fact that both the total white blood cell count and the neutrophil count in peripheral blood of Stat5a/b-mutant mice were very constant over time, from 3 weeks after PolyIC injection to 15 weeks (Figure 1A-B). Thus, compensatory effects do not emerge in these mice, even after several months of hematopoietic Stat5a/b deletion.
Suppression of 5-FU and GM-CSF–induced granulopoiesis in STAT5f/f:Mx1-Cre mice. Persistent reduction of peripheral neutrophils in Stat5a/b-mutant mice in homeostasis without compensatory influences over time. (A) Similar peripheral white blood cell and (B) peripheral neutrophil counts of the same Stat5f/f:Mx1-Cre mice (n = 7) 3 weeks and 15 weeks after the last PolyIC injection. (C) Granulopoiesis after 5-FU injection. On injection of 5-FU, peripheral neutrophils from Stat5f/f and Stat5f/f:Mx1-Cre mice were counted every 3 days. Data are mean ± SD; n = 5 mice per group. (D) In vivo administration of G-CSF and GM-CSF. Stat5f/f and Stat5f/f:Mx1-Cre mice were injected subcutaneously with G-CSF or GM-CSF, and peripheral blood was collected daily 6 hours after injection. The ratio of neutrophils in Stat5f/f mice (n = 7) to the neutrophils in Stat5f/f:Mx1-Cre mice (n = 7) is shown. The ratio at day 0 was normalized to 1. *P < .05, **P < .01, fold increase of neutrophils in Stat5f/f versus Stat5f/f:Mx1-Cre mice on cytokine injection.
Suppression of 5-FU and GM-CSF–induced granulopoiesis in STAT5f/f:Mx1-Cre mice. Persistent reduction of peripheral neutrophils in Stat5a/b-mutant mice in homeostasis without compensatory influences over time. (A) Similar peripheral white blood cell and (B) peripheral neutrophil counts of the same Stat5f/f:Mx1-Cre mice (n = 7) 3 weeks and 15 weeks after the last PolyIC injection. (C) Granulopoiesis after 5-FU injection. On injection of 5-FU, peripheral neutrophils from Stat5f/f and Stat5f/f:Mx1-Cre mice were counted every 3 days. Data are mean ± SD; n = 5 mice per group. (D) In vivo administration of G-CSF and GM-CSF. Stat5f/f and Stat5f/f:Mx1-Cre mice were injected subcutaneously with G-CSF or GM-CSF, and peripheral blood was collected daily 6 hours after injection. The ratio of neutrophils in Stat5f/f mice (n = 7) to the neutrophils in Stat5f/f:Mx1-Cre mice (n = 7) is shown. The ratio at day 0 was normalized to 1. *P < .05, **P < .01, fold increase of neutrophils in Stat5f/f versus Stat5f/f:Mx1-Cre mice on cytokine injection.
Stat5a/b-mutant mice display aberrant granulopoiesis on myelosuppression or GM-CSF treatment
To investigate the role of STAT5A/B in emergency granulopoiesis, we induced myelosuppression in Stat5f/f:Mx1-Cre mice and Stat5f/f controls by 5-FU injection and measured peripheral blood neutrophils (Figure 1C). A more than 10-fold elevation of neutrophils was observed 18 days after injection in control but not in Stat5a/b-mutant mice (Figure 1C). This reaction is the response to the “cytokine storm” after the nadir stage induced by 5-FU. GM-CSF and G-CSF injections were performed for 5 days to establish to what extent Stat5f/f:Mx1-Cre mice respond to these typical granulocyte-related cytokines (Figure 1D). Whereas the response to G-CSF was virtually identical and increased peripheral blood neutrophils were observed in control and Stat5a/b-mutant mice to the same extent, the GM-CSF–induced increase of peripheral blood neutrophils was severely impaired in the absence of STAT5A/B (Figure 1D).
Stat5a/b-null mature neutrophils display normal function but lack GM-CSF–permitted survival
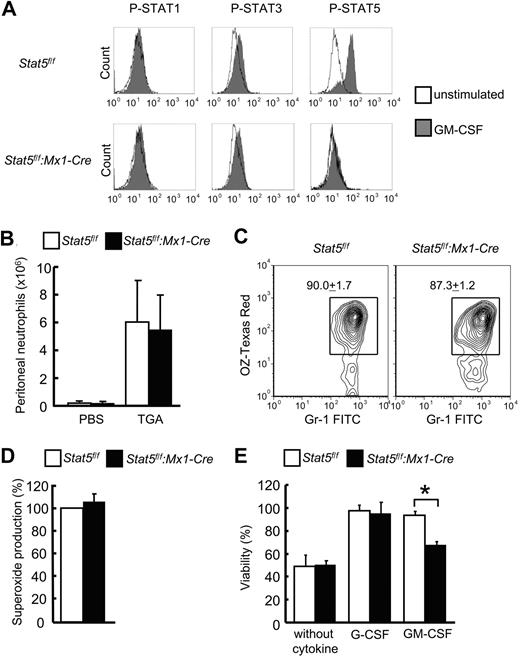
To determine which STAT-signaling pathway was stimulated by GM-CSF in mature neutrophils, phosphoflow cytometry was performed (Figure 2A). GM-CSF induced strong phosphorylation of STAT5A in control neutrophils of course not in Stat5a/b-null neutrophils. In contrast, STAT3 was only very weakly phosphorylated on GM-CSF stimulation. Importantly, no difference in STAT3 phosphorylation was observed between Stat5a/b-mutant mice and corresponding control mice. STAT1 was not phosphorylated on GM-CSF stimulation (Figure 2A).
Stat5a/b-null neutrophils have normal functions but are less responsive to GM-CSF–permitted survival. (A) Phosphoflow cytometry for STAT1, STAT3, and STAT5A in neutrophils derived from bone marrow of Stat5f/f and Stat5f/f:Mx1-Cre mice. Cells were stimulated with GM-CSF for 15 minutes after 3 hours of starvation. One representative experiment is displayed (n = 5 experiments). Control experiments are shown in supplemental Figure 2. (B) Migration assay by the induction of peritoneal exudates induced by the intraperitoneal injection of sodium TGA. At 4 hours after injection, peritoneal neutrophils from Stat5f/f and Stat5f/f:Mx1-Cre mice were counted. Data are mean ± SD (n = 18 mice per group for TGA, n = 4 mice per group for phosphate-buffered saline). (C) Phagocytosis assay with bone marrow neutrophils from Stat5f/f and Stat5f/f:Mx1-Cre mice. Flow cytometry determined the level of phagocytosis of Texas Red–conjugated opsonizing zymosan in Gr1+ neutrophils. Data are mean ± SD; n = 3 mice per group. One representative experiment is shown. (D) Oxidative burst assay. Superoxide production was determined in bone marrow mature neutrophils from Stat5f/f and Stat5f/f:Mx1-Cre mice. Cells were preincubated with hydroethidine and then activated by phorbol myristate acetate. Results (mean ± SD) are given as percentage to the control; n = 3 mice per group. (E) Survival assay. Bone marrow neutrophils from Stat5f/f and Stat5f/f:Mx1-Cre mice were cultured as indicated for 36 hours. Data are means ± SD (n = 3 mice per group). *P = .02.
Stat5a/b-null neutrophils have normal functions but are less responsive to GM-CSF–permitted survival. (A) Phosphoflow cytometry for STAT1, STAT3, and STAT5A in neutrophils derived from bone marrow of Stat5f/f and Stat5f/f:Mx1-Cre mice. Cells were stimulated with GM-CSF for 15 minutes after 3 hours of starvation. One representative experiment is displayed (n = 5 experiments). Control experiments are shown in supplemental Figure 2. (B) Migration assay by the induction of peritoneal exudates induced by the intraperitoneal injection of sodium TGA. At 4 hours after injection, peritoneal neutrophils from Stat5f/f and Stat5f/f:Mx1-Cre mice were counted. Data are mean ± SD (n = 18 mice per group for TGA, n = 4 mice per group for phosphate-buffered saline). (C) Phagocytosis assay with bone marrow neutrophils from Stat5f/f and Stat5f/f:Mx1-Cre mice. Flow cytometry determined the level of phagocytosis of Texas Red–conjugated opsonizing zymosan in Gr1+ neutrophils. Data are mean ± SD; n = 3 mice per group. One representative experiment is shown. (D) Oxidative burst assay. Superoxide production was determined in bone marrow mature neutrophils from Stat5f/f and Stat5f/f:Mx1-Cre mice. Cells were preincubated with hydroethidine and then activated by phorbol myristate acetate. Results (mean ± SD) are given as percentage to the control; n = 3 mice per group. (E) Survival assay. Bone marrow neutrophils from Stat5f/f and Stat5f/f:Mx1-Cre mice were cultured as indicated for 36 hours. Data are means ± SD (n = 3 mice per group). *P = .02.
The failure of Stat5f/f:Mx1-Cre mice to increase peripheral blood neutrophils after myelosuppression could be explained as a migratory defect or a survival defect of postmitotic mature neutrophils. To assess the possibility that STAT5A/B also regulate neutrophils, we performed several functional tests for mature neutrophils. The migratory ability of Stat5a/b-null neutrophils was tested in vivo on intraperitoneal injection of TGA, and in vitro assays were applied to gauge phagocytosis, oxidative burst, and cytokine-dependent survival of freshly isolated neutrophils. No significant differences in migration, phagocytic activity, or superoxide production were observed between control and Stat5a/b-null neutrophils (Figure 2B-D). In the absence of cytokines, the viability of both control and Stat5a/b-null neutrophils was reduced by 50% after 36 hours (Figure 2E). As expected, virtually all cells from control animals survived for 36 hours in the presence of G-CSF or GM-CSF. However, only 65% of Stat5a/b-null neutrophils survived in the presence of GM-CSF, whereas there was no increased cell death in mutant neutrophils in the presence of G-CSF (Figure 2E). Therefore, GM-CSF permits the survival of mature neutrophils via STAT5A/B signaling.
Impaired colony formation of GMPs in the absence of STAT5A/B
Additional explanations for the impaired GM-CSF–stimulated granulopoiesis in Stat5a/b-mutant mice could be defects in bone marrow granulocyte progenitor cells. On this account, we analyzed GMPs from Stat5f/f:Mx1-Cre mice and Stat5f/f controls. Although the percentage of GMPs in control and Stat5f/f:Mx1-Cre mice was similar, the absolute number of BMMNCs and therefore the absolute number of GMPs was decreased in mutant mice (BMMNCs from both femora and tibiae per mouse: Stat5f/f 3.5 ± 0.08 × 107 cells, Stat5f/f:Mx1-Cre 3.0 ± 0.26 × 107 cells, P = .024; percentage of GMPs in BMMNCs: Stat5f/f 0.8% ± 0.2%, Stat5f/f:Mx1-Cre 1.0% ± 0.04%, no significant difference).
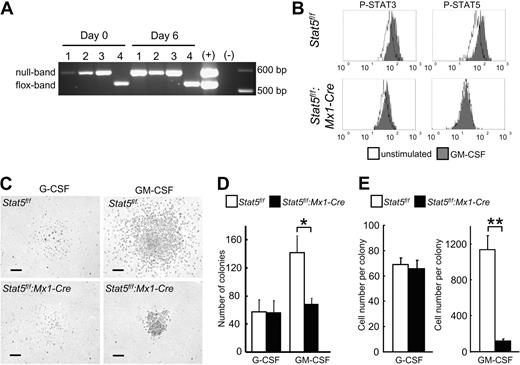
Complete Cre-mediated Stat5a/b deletion efficiency in the GMP population was confirmed by PCR. No residual Stat5f/f PCR product was observed in freshly purified GMPs from Stat5f/f:Mx1-Cre mice (Figure 3A, day 0). Moreover, no unrecombined escapees with potential growth advantage were detected after 6 days of in vitro culture (Figure 3A, day 6). These results demonstrated the complete deletion of the Stat5a/b locus in all progeny.
Reduced colony-forming potential of Stat5a/b-null GMPs. (A) Complete deletion of Stat5a/b in all GMPs from Stat5f/f:Mx1-Cre mice after PolyIC administration. No outgrowth of Stat5f/f cells after 6 days of culture. #1, #2, and #3 indicate independent experiments with cells from Stat5f/f:Mx1-Cre; #4, Stat5f/f. (B) Phosphoflow cytometry of GMPs revealed STAT5 activation by GM-CSF, and no aberrant STAT3 activation in GMPs from Stat5f/f:Mx1-Cre. One representative experiment of 2 is shown. (C-E) Colony formation assay with GMPs from Stat5f/f and Stat5f/f:Mx1-Cre mice cultured with either G-CSF or GM-CSF. Scale bars represent 100 μm. (D) The number of colonies from Stat5a/b-null GMPs cultured in GM-CSF was reduced. No difference in colony formation was observed with G-CSF. Data are mean ± SD; n = 3 independent experiments *P = .03. (E) Colonies derived from Stat5a/b-null GMPs cultured in GM-CSF were significantly smaller, consisting of much fewer cells; no difference was seen in G-CSF–derived colonies. Data are mean ± SEM. For G-CSF n = 40 colonies of each group from 2 experiments, for GM-CSF n = 45 colonies of Stat5f/f, for Stat5f/f:Mx1-Cre n = 37 colonies from 2 experiments. **P < .001.
Reduced colony-forming potential of Stat5a/b-null GMPs. (A) Complete deletion of Stat5a/b in all GMPs from Stat5f/f:Mx1-Cre mice after PolyIC administration. No outgrowth of Stat5f/f cells after 6 days of culture. #1, #2, and #3 indicate independent experiments with cells from Stat5f/f:Mx1-Cre; #4, Stat5f/f. (B) Phosphoflow cytometry of GMPs revealed STAT5 activation by GM-CSF, and no aberrant STAT3 activation in GMPs from Stat5f/f:Mx1-Cre. One representative experiment of 2 is shown. (C-E) Colony formation assay with GMPs from Stat5f/f and Stat5f/f:Mx1-Cre mice cultured with either G-CSF or GM-CSF. Scale bars represent 100 μm. (D) The number of colonies from Stat5a/b-null GMPs cultured in GM-CSF was reduced. No difference in colony formation was observed with G-CSF. Data are mean ± SD; n = 3 independent experiments *P = .03. (E) Colonies derived from Stat5a/b-null GMPs cultured in GM-CSF were significantly smaller, consisting of much fewer cells; no difference was seen in G-CSF–derived colonies. Data are mean ± SEM. For G-CSF n = 40 colonies of each group from 2 experiments, for GM-CSF n = 45 colonies of Stat5f/f, for Stat5f/f:Mx1-Cre n = 37 colonies from 2 experiments. **P < .001.
Cellular defects observed in the absence of STAT5A/B could be the result of the genuine loss of these transcription factors or an aberrant activation of other STAT members, as observed in other Stat5a/b-null tissues,31-33 or the deregulation of relevant receptors.
Gene expression profiling of control and Stat5a/b-null GMPs demonstrated almost equal levels of Stat1, Stat3, G-CSF receptor, and GM-CSF receptor mRNAs before and after GM-CSF stimulation in control and Stat5f/f:Mx1-Cre mice (supplemental Tables 1–2). Phosphoflow cytometry demonstrated GM-CSF–induced STAT5 phosphorylation in GMPs of control Stat5f/f mice. Although Stat3 mRNA levels were slightly increased in Stat5a/b-null GMPs with or without GM-CSF stimulation, increased phosphorylation of STAT3 was not detected in mature neutrophils or GMPs (Figures 2A, 3B), minimizing the possibility that the defects appeared because of inappropriate activity of other STATs.
Next we determined the functionality of Stat5a/b-null GMPs and to what extent STAT5A/B controlled GM-CSF–induced colony-forming potential of GMPs (Figure 3C-E). In the presence of GM-CSF, the numbers of Stat5a/b-null colonies were reduced by more than 50% (Figure 3D). Moreover, the average cell number of a Stat5a/b-null GMP-derived colony was only 11% of that from control GMPs (Figure 3E). In contrast, no difference in colony formation was seen between control and Stat5a/b-null GMPs in the presence of G-CSF (Figure 3D-E).
Impaired GM-CSF–induced proliferation and enhanced cell death of Stat5a/b-null GMPs
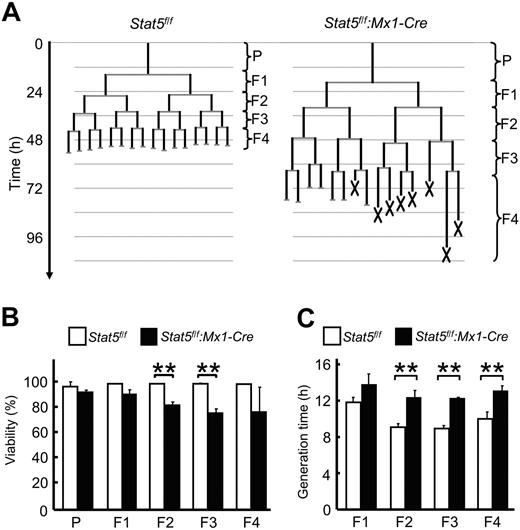
The reduction in GM-CSF–dependent number and size of colonies from GMPs could be explained by defects in cell survival, proliferation, or combinations of both during differentiation. To define the precise cell behavior of GMPs during their differentiation induced by GM-CSF through STAT5A/B, constant observation of the development at the single-cell level is required.34,35 Therefore, we performed time-lapse microscopy and single-cell tracking experiments (Figure 4; supplemental Video 1), a technology allowing the constant observation of individual GMPs and all their progeny throughout a developmental program.36 Analysis of several hundreds of GMPs and their progeny during culture with GM-CSF up to the fifth generation revealed increased cell death in the Stat5a/b-null GMPs, successively increasing from the F1 through the F4 generation (5%, 18%, 24%, and 24% in generation F1 through F4, respectively, Figure 4B, supplemental Video 2). Negligible cell death events occurred during development of control GMPs. In addition to increased cell death in Stat5a/b-null cells, their generation times were markedly longer in every generation compared with control cells (16%, 35%, 37%, and 31% longer in generation F1 through F4, respectively, Figure 4C). Whereas STAT5A/B are not essential for the establishment of the granulocyte lineage and the generation of GMPs, they are indispensable for GMPs and their progeny to respond to GM-CSF. The inability of Stat5a/b-null GMPs to form bona fide colonies is the result of 2 intrinsic defects, impaired cell proliferation, and cell survival.
Impaired proliferation and survival of Stat5a/b-null GMPs cultured with GM-CSF. Time-lapse microscopy and single-cell tracking were used to determine generation times and survival rates of GMPs and their progeny cultured in the presence of GM-CSF through 5 generations. (A) Typical GMP pedigrees generated by single-cell tracking. X represents cell death. (B) STAT5A/B-dependent survival. Initiating GMPs are referred to as parental cells (P). (C) Average generation time of all cells within 1 generation. For each experiment, more than 30 initiating GMPs and their progeny were tracked up to the fifth generation. Data are mean ± SD of 3 independent experiments (n > 30 pedigrees per experiment). **P < .01 in panels B and C.
Impaired proliferation and survival of Stat5a/b-null GMPs cultured with GM-CSF. Time-lapse microscopy and single-cell tracking were used to determine generation times and survival rates of GMPs and their progeny cultured in the presence of GM-CSF through 5 generations. (A) Typical GMP pedigrees generated by single-cell tracking. X represents cell death. (B) STAT5A/B-dependent survival. Initiating GMPs are referred to as parental cells (P). (C) Average generation time of all cells within 1 generation. For each experiment, more than 30 initiating GMPs and their progeny were tracked up to the fifth generation. Data are mean ± SD of 3 independent experiments (n > 30 pedigrees per experiment). **P < .01 in panels B and C.
STAT5A/B regulate genes for survival and proliferation in GMPs
To address potential mechanisms mediated by STAT5A/B to regulate proliferation and survival of the granulocyte lineage, we performed microarray-based gene expression profiling of GMPs from Stat5f/f and Stat5f/f:Mx1-Cre mice. Purified GMPs from both animal groups were treated either with or without GM-CSF after starvation, and total RNA was subjected for microarray analysis. The comparison of the expression levels of all mouse genes of these 4 sets of RNA samples allowed us to identify genes whose expression levels were influenced by GM-CSF treatment in control GMPs, but not in Stat5a/b-null GMPs. These genes most probably were directly regulated by GM-CSF stimulation via STAT5A/B engagement. A total of 389 known genes were induced and 211 known genes were suppressed by GM-CSF through STAT5A/B (cut-off P < .05, fold change is < −2.0 or > 2.0; supplemental Figure 3). Moreover, 67 predicted genes with unknown function were differentially expressed. As expected, typical STAT5A/B target genes, including Socs2 and Socs3, were identified. Our study demonstrates that both proliferation and survival of the granulocyte lineage were dependent on STAT5A/B. These 2 features are reflected in the gene expression profiles from GMPs: GM-CSF either induced the expression of genes (eg, Bcl2l1, Cflar, Dcun1d3, Eps8, Nek6, Nov/CCN3) or down-regulated the expression of genes (eg, Rgs2) linked to cell proliferation and survival in control, but not in Stat5a/b-null GMPs. The complete primary microarray datasets are available at the National Center for Biotechnology Information GEO database (accession no. GSE14672; http://www.ncbi.nlm.nih.gov/geo/).
Discussion
This study demonstrates the importance of STAT5A/B in the maintenance and expansion of the granulocytic lineage. In homeostasis as well as in cases of emergency, STAT5A/B is necessary to reach appropriate levels of neutrophils in the periphery. Reduced responsiveness to GM-CSF–mediated granulocytic cell lineage fates and functions at various cell stages of the granulocytic lineage are a reason for the diminished ability of Stat5a/b-mutant mice to keep normal neutrophil levels. Although mature Stat5a/b-null neutrophils display normal functions, their GM-CSF–mediated survival is reduced. GM-CSF–induced neutrophil generation in the bone marrow is severely affected by the loss of STAT5A/B. We could demonstrate at the single-cell level that GMPs have a reduced colony formation ability leading to both less and smaller colonies than control GMPs. The reduced number of progeny of individual GMPs was caused by a slower cell cycle and increased cell death during differentiation. Therefore, STAT5A/B controls survival and proliferation of granulocytic progenitors, which was reflected in a STAT5A/B target gene screen.
Although functional neutrophils were present in the absence of STAT5A/B, the number of peripheral blood neutrophils was reduced by 50%. Peripheral blood cell counts were stable over time after deletion of Stat5a/b, and no changes in G-CSF or GM-CSF serum levels were observed, indicating no obvious emerging compensatory mechanisms. Therefore, STAT5A/B are required for the maintenance of steady-state granulopoiesis but not for the establishment of the granulocyte lineage.
Previous studies could not reveal the role of STAT5A/B in homeostatic granulopoiesis. The deletion of only 1 of the 2 STAT5 genes, STAT5A or STAT5B, suffered from overlapping activities from the 2 STAT5 proteins, which are both expressed in hematopoietic cells, resulting in no obvious blood or blood progenitor phenotype.23 Nonconditional hypomorphic mice, which carry Stat5a/b genes encoding STAT5A/B lacking their respective N termini (referred as Stat5ΔN), have been used in numerous studies. However, because the truncated STAT5A/B in these mice has functional activity,24,37 results from these studies need to be viewed with caution. An increase of peripheral neutrophils was found in homeostasis in these mice.22 However, the neutrophil increase was not intrinsic to the loss of STAT5A/B in the granulocyte lineage, but was a secondary effect from an unexpected compensatory overproduction of G-CSF by liver endothelial cells.22 The mechanism of the enhanced G-CSF production was linked to an inhibitory STAT5B effect on these endothelial cells. As a consequence of the high G-CSF serum levels, both mature neutrophils and bone marrow-derived GMPs were increased, indicating that G-CSF–mediated granulopoiesis was largely unaffected by the Stat5ΔN mutation.22 However, only when Stat5ΔN bone marrow was transplanted into normal recipients, these mice developed neutropenia with diminished neutrophil survival indicating the true phenotype of STAT5A/B mutation for granulopoiesis.22
G-CSF and GM-CSF are the main regulators of neutrophil production and function, but other cytokines such as IL-3 and IL-6 also participate in granulopoiesis. Mice lacking G-CSF or the G-CSF receptor develop severe neutropenia with only 10% to 30% of peripheral neutrophil counts.38-40 G-CSF receptor mutant animals show hypogranulopoiesis but have normal distribution of uncommitted myeloid progenitors in the bone marrow. Moreover, neutrophil survival was diminished. Pathways other than STAT5A/B must contribute to granulopoiesis triggered by these cytokine receptors. STAT5A/B does not seem to play a founding role in G-CSF signaling. G-CSF activates mainly STAT3, and only weakly STAT5A/B, as well as several other pathways (MAP kinase, PI3 kinase, nuclear factor-κB).41 In our study, we observed a normal response to G-CSF in Stat5a/b-mutant mice at several stages of the granulocytic lineage, including neutrophil survival, neutrophil mobilization into the bloodstream, and granulopoiesis in the bone marrow. Our findings together with previous studies underline the largely independence of the G-CSF receptor from STAT5A/B signaling in various steps of granulopoiesis and shift the scientific interest to other G-CSF receptor signaling pathways, such as STAT3.
The role of STAT3 in granulopoiesis is controversial. The deletion of STAT3 results in an increase of peripheral neutrophils, which seems to be caused by enhanced survival of neutrophils.42 STAT3 activates SOCS3, a negative regulator of granulopoiesis,15 and studies postulate a negative feedback function of STAT3 in granulopoiesis. In Stat3-mutant mice, an enhanced ERK1/2 activation was detected in steady state and on G-CSF stimulation that seemed to be responsible, at least in part, for the enhanced neutrophil levels in these mice.42 On the other hand, mice with G-CSF receptor mutants that cannot signal via STAT3 or STAT5A/B show severe neutropenia, which can be rescued by constitutively active STAT3 overexpression, indicating a vital positive role of STAT3 in granulopoiesis.18 The contribution of alternative pathways other than the STAT pathway in granulopoiesis is very likely, but not well understood. A conditional double knockout mouse model leading to complete ablation of STAT5A/B and STAT3 would determine the overall contribution of STAT factors in granulopoiesis in relation to other pathways and might elucidate the level of redundancy of these crucial factors in granulopoiesis.
In contrast to the G-CSF receptor, the GM-CSF receptor preferentially signals via STAT5A/B and only leads to a weak phosphorylation of STAT3 in mature neutrophils and GMPs. Our study focused on the critical role of STAT5A/B in GM-CSF–mediated granulopoiesis. The generation and the survival of mature neutrophils were impaired in Stat5a/b-mutant mice, explaining the diminished neutrophil increase on GM-CSF injection, but were normal in response to G-CSF. Although GM-CSF receptor-null mice displayed only little disturbance in steady-state hematopoiesis,43 support from a defect in emergency granulopoiesis comes from studies with Listeria monocytogenes.44 However, the lacking neutrophil response in Stat5a/b-mutant mice on myelosuppression can hardly be explained by a sole role of missing GM-CSF responsiveness. More likely, a combination of several cytokines in the cytokine storm at the nadir stage, such as GM-CSF, IL-3, and IL-6, contributes to the vast generation of the granulocytic lineage. Many of these cytokines signal, at least in part, via STAT5A/B.
Stat5a/b-mutant mice have normal bone marrow progenitor cell distributions and only marginal lower total BMMNC numbers. Therefore, STAT5A/B is not necessary for the establishment of GMPs. In studies using unpurified bone marrow cells, reduced numbers of GM-CSF–induced colonies in Stat5a/b-mutant mice can be explained by either a reduced number of myeloid progenitors or by a reduced ability of progenitors to respond to GM-CSF.23 Both would lead to the same outcome with totally different biologic explanations. By knowing the exact cell number and identity of purified progenitors, we were able to draw conclusions of the clonogenicity of single GMPs. The 50% lower number of colonies derived from sorted Stat5a/b-null GMPs in response to GM-CSF can now be linked to a diminished colony-forming ability of preexisting GMPs. Moreover, we can show that GMPs from Stat5a/b-mutant mice have not per se a weakness in in vitro culture, as normal granulocytic colonies are formed in response to G-CSF. Not only is the number of colonies reduced on GM-CSF stimulation, but also the sizes were significantly smaller. Colonies can only arise when cells proliferate and survive during differentiation. By long-term single-cell tracking of individual differentiating GMPs, we could, for the first time, dissect survival and proliferation function of STAT5A/B in granulopoiesis. The cell cycle is markedly prolonged in every generation from GMPs onwards, and an increasing incidence of cell death appears, especially in later generations. Therefore, we postulate that STAT5A/B promotes both cell-cycle progression and survival. These 2 features are reflected in the gene expression profiles obtained from GMPs in the absence or presence of STAT5A/B. GM-CSF regulated the expression of a set of genes linked to cell proliferation and survival in Stat5f/f control GMPs compared with Stat5a/b-null GMPs. Among these genes was the gene for the antiapoptotic protein Bcl-X (Bcl2l1), which had been proposed as a STAT5A/B target.45 Epidermal growth factor pathway substrate number 8 (Eps8) had been linked to cell proliferation and survival46 and enhances epidermal growth factor-dependent mitogenic signals. NIMA-related expressed kinase 6 (Nek6) and other NEK family members contribute to the orchestration of mitotic progression and protect cells from chromosome instability.47 Nephroblastoma overexpressed (Nov)/CCN3 has been linked to self-renewal and maturation of hematopoietic cells.48 Down-regulated genes by GM-CSF stimulation in control but not in Stat5a/b-null GMPs included regulator of G-protein signaling 2 (Rgs2), which has been implicated as a potential proto-oncogene involved in leukemogenesis, and that is widely expressed in human AML, acute lymphoid leukemia, and acute-phase chronic myelogenous leukemia samples.49 Although these genes respond to GM-CSF through STAT5A/B, their specific contribution to proliferation and survival of GMPs has yet to be determined.
Taken together, our results demonstrate that GM-CSF not only controls the survival but also influences proliferation of the granulocyte lineage through the transcription factors STAT5A/B. The identification of genes that regulate these cell fates, survival, and proliferation might open new avenues for therapeutic interference in the future.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to B. Schauberger and K. Azadov for programming contributions, A. Kowarsch for statistical analyses, A. Roth and C. Raithel for technical assistance. We also thank G. Robinson for generating mouse-colony.
This work was supported by the German Research Council (DFG) to T.S. and the intramural program of the NIDDK/NIH and the Deutsche Forschungsgemeinschaft Mercator Visiting Professor program to L.H.
National Institutes of Health
Authorship
Contribution: A.K. designed and interpreted experiments, wrote the manuscript, and contributed to backcross and preparation of mice (experiments for Figures 1–2, 3C, supplemental Figures 1–2, Table 1) and microarray analysis; M.A.R. designed and interpreted experiments, wrote the manuscript, and performed experiments for Figures 1, 3, 4, and supplemental Figure 3, supplemental videos, and microarray analysis; J.M.S. conducted sorting for the GMP population; W.C. analyzed microarray data and supplemental Figure 3 and supplemental Table 3; M.C.W. prepared mice; B.-M.Z. contributed to backcross of mice and generated mouse colony; P.S.H. helped with experiments for Figure 4; J.J.O. organized sorting and conducted discussions; T.S. developed time-lapse microscopy and cell tracking technology, designed experiments, interpreted data, and wrote the manuscript; and L.H. designed experiments, interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lothar Hennighausen, NIDDK, NIH, Bldg 8, Rm 101, Bethesda, MD 20892; e-mail: lotharh@niddk.nih.gov.
References
Author notes
A.K. and M.A.R. contributed equally to this study.
T.S. and L.H. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal