Abstract
Abstract 4882
Thalidomide is an oral drug with anti myeloma activity. However, some patients fail to respond and serious side effects are common including thrombo-embolic disease and peripheral neuropathy. Identifying patients likely to respond has huge potential to better individualise treatment. Using proteomic methods, we have sought to identify a signature capable of distinguishing patients likely to respond to thalidomide-based therapy.
Serum samples from 36 consecutive newly diagnosed multiple myeloma patients were collected prior to initial treatment with thalidomide-based regimens. Samples were initially immunodepleted to enrich the low abundance proteins. This was followed by 2D-DIGE analysis, to identify differential expressed proteins. Identification of proteins found to be statistically significant different in expression between responders and non-responders to thalidomide was carried out using LC-MS/MS. These proteins Using commercially available ELISAs kits, the levels of candidate biomarker proteins was validated using the un-fractionated serum samples from the original cohort of patients. The expression levels of each of these proteins were used to generate a Receiver Operating Characteristic (ROC) curve. This permitted us to undertake a systematic diagnostic performance evaluation of each bio-marker alone and in combination..
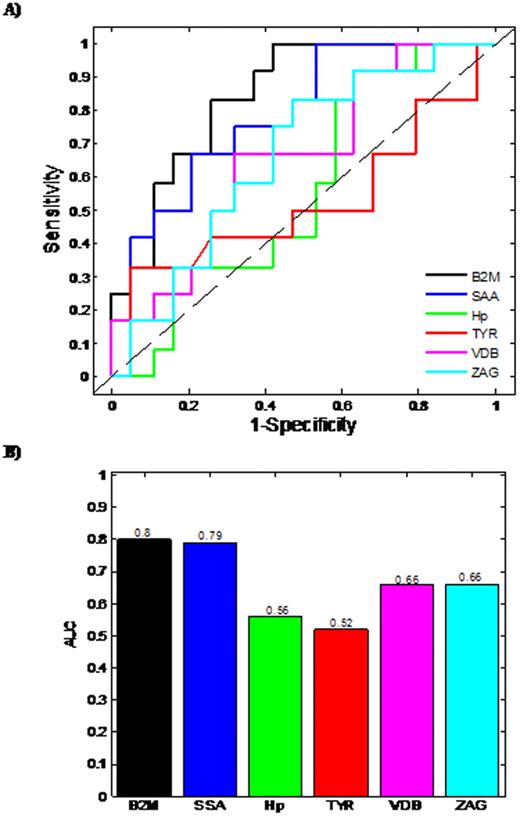
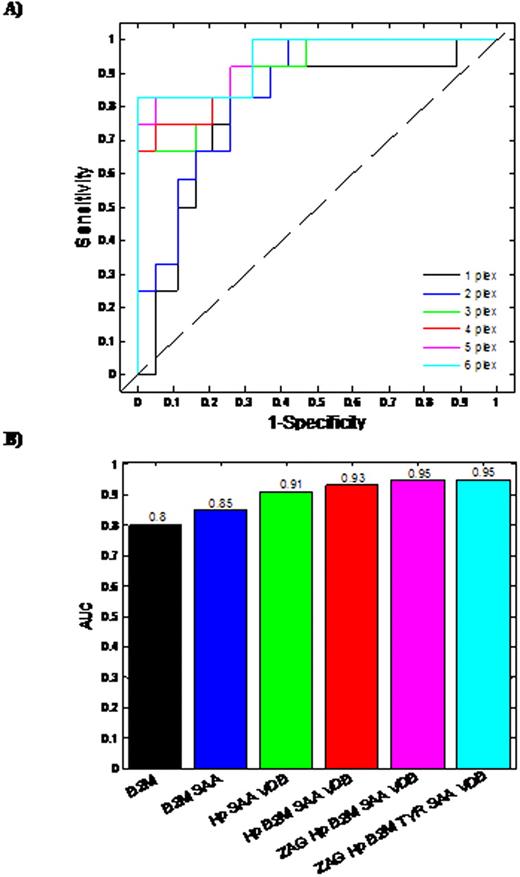
Based on Day 100 re-staging investigations and using International Myeloma Working Group uniform response criteria for multiple myeloma, 20 thalidomide responders and 16 non-responders were identified. The median patient age was 67 years (range 57-79 years), 18 male and 18 female. Six proteins were found to demonstrate a statistically different expression pattern between the responders/non-responders. Proteins found to have higher abundance level in the serum from thalidomide non-responders in comparison to responders, included Zinc alpha 2-glycoprotein (ZAG), Vitamin D binding Precursor (VDB), Transthyretin (TYR), Serum Amyloid A protein (SAA), beta-2-microglobulin (B2M), while Haptoglobin (Hp) had a lower abundance level in non-responders. Initially, Logistic regression (LR)was used to develop predictive models for each individual differentially expressed protein (Fig 1A, 1B). Using single protein LR models, B2M and SAA levels had the best predictive ability with Area Under Curve (AUC) values of 0.8 and 0.79, respectively, demonstrating acceptable performance. The remaining single protein models had AUC values less than 0.7, indicating minimal predictive ability. The predictive capability of models developed using combinations of proteins was also assessed. Logistic regression models were constructed and ROC analyses carried out on all possible permutations of the differentially expressed proteins. The most successful combinations of the bio-markers' models are shown (Fig 2A & 2B). The combination of three proteins (Hp+SAA+VDB) yields AUC values of 0.91. The best possible AUC resulted from the combination of Hp+SAA+VDB+ZAG+VDB, which yields an AUC of 95%, indicating an outstanding predictive capability.
Accurate prediction of an individual patient's drug response is an important prerequisite of personalized medicine. Using a panel of proteomic biomarkers, we have demonstrated the feasibility of predicting sensitivity and response to thalidomide in previously untreated myeloma. In the multiple of 3 or 4 protein combinations, these potential biomarkers can differentiate responders and non responders to thalidomide at diagnosis in up to 95 % of multiple myeloma patients.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal