Abstract
Clonal evolution and outgrowth of cellular variants with additional chromosomal abnormalities are major causes of disease progression in chronic lymphocytic leukemia (CLL). Because new DNA lesions occur during S phase, proliferating cells are at the core of this problem. In this study, we used in vivo deuterium (2H) labeling of CLL cells to better understand the phenotype of proliferating cells in 13 leukemic clones. In each case, there was heterogeneity in cellular proliferation, with a higher fraction of newly produced CD38+ cells compared with CD38− counterparts. On average, there were 2-fold higher percentages of newly born cells in the CD38+ fraction than in CD38− cells; when analyzed on an individual patient basis, CD38+2H-labeled cells ranged from 6.6% to 73%. Based on distinct kinetic patterns, interclonal heterogeneity was also observed. Specifically, 4 patients exhibited a delayed appearance of newly produced CD38+ cells in the blood, higher leukemic cell CXC chemokine receptor 4 (CXCR4) levels, and increased risk for lymphoid organ infiltration and poor outcome. Our data refine the proliferative compartment in CLL based on CD38 expression and suggest a relationship between in vivo kinetics, expression of a protein involved in CLL cell retention and trafficking to solid tissues, and clinical outcome.
Introduction
B-cell chronic lymphocytic leukemia (CLL) is a relatively common and incurable adult disease of unknown etiology. Although the majority of circulating CLL cells are not proliferating,1,2 a small proliferative compartment does exist,3 and lymph nodes and bone marrow contain aggregates of activated, dividing cells.4 In addition, accessory signals delivered in the microenvironment of these solid tissues are essential for neoplastic cell survival and expansion,5 suggesting that clonal accumulation involves rescue of leukemic cells from death. Thus, a dynamic balance between birth and death is characteristic of CLL clones.3
The disease has a strikingly variable clinical course, and different biologic variables help predict clinical outcome,6,7 including immunoglobulin (Ig) heavy variable (IGHV) gene mutation status8 and the percentage of cells expressing zeta-chain–associated protein kinase 70 (ZAP-70)9,10 and CD38.6,11 CD38 is a surface membrane bound ectoenzyme that functions as a receptor.12-15 Expression of CD38 on normal B cells depends on cell maturation and can be induced upon stimulation.16 The prognostic value of CD38 is linked to its capacity to promote leukemic cell survival.17-20 Moreover, within each CLL clone, cells expressing CD38 are enriched in expression of Ki-67,21 suggesting that CD38+ cells represent a cycling subset.
Deuterium (2H) incorporation into newly synthesized DNA can be precisely quantified by gas chromatography/mass spectrometry (GC/MS) providing an established measure of DNA replication.22,23 This approach documented that in vivo CLL cells proliferate at rates ranging from 0.08% to 1.7% of the clone per day,3,24 and patients with higher birth rates appear at risk of more active disease.3
In this study, we used in vivo 2H labeling to better understand the phenotype of the most actively proliferating cells within each CLL clone. 2H enrichment in genomic DNA was compared between CD38+ and CD38− cells of 13 patients, revealing intraclonal heterogeneity in cellular proliferation, with CD38+ cells proliferating more rapidly. We also identified interclonal heterogeneity with 2 patient groups of distinct kinetic patterns and CXC chemokine receptor 4 (CXCR4) levels. The group with higher CXCR4 levels showed a delayed appearance of 2H-labeled CD38+ cells in blood and seemed at a higher risk for lymphoid organ infiltration and inferior outcome. Our data suggest relationships between CLL kinetics, expression of a molecule involved in CLL cell retention and trafficking to solid tissues,25,26 and clinical course.
Methods
Patients and 2H2O protocols
The study was approved by the North Shore–LIJ Health System's Institutional Review Board and was conducted according to the principles of the World Medical Association Declaration of Helsinki. Thirteen CLL patients (Table 1), diagnosed by established criteria,27 were selected from a larger cohort who participated in 2H2O protocols. All subjects provided written informed consent prior to enrollment. Exclusion criteria for patient participation can be found in supplemental Methods (available on the Blood website; see the Supplemental Materials link at the top of the online article). Every patient received 2H, in the form of 2H2O, during a labeling period (6-12 weeks) and was followed until the end of washout period (week 24) in accordance to protocols reported in supplemental Methods. CLL cells were studied at 3 time points (Figure 1A). Because previously published data showed delayed appearance of labeled cells in the blood of some patients,3 time points were selected (1) during the labeling period (3 or 4 weeks), (2) at or near the end of the labeling period (6 or 8 weeks), and (3) after 2H2O washout (usually week 24).
Clinical and laboratory features of the CLL patients involved in this study
| CLL patient no. . | % CD38+ . | % ZAP-70+ . | Cytogenetic abnormalities, % cells . | IGHV mutation status* . | Lymph node/spleen/liver enlargement† . | Rai stage at diagnosis and when 2H2O study began . | Status . | Time to first treatment, mo‡ . | Mean WBC (beginning-end of study)§ . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Δ11q22 . | Tri12 . | Δ13q14 . | Δ17p23 . | |||||||||
| 189 | 2 | 7 | < 5 | < 5 | 70 | < 5 | M/UM | +/−/− | 0-IV | Expired | 53 | 107 (76-175) |
| 280 | 1 | 29 | < 5 | < 5 | 99 | < 5 | M | −/+/+ | 0-II | Alive | 108 | 58 (61-75) |
| 321 | 10 | 95 | < 5 | 59 | < 5 | < 5 | UM | −/−/− | 0-0 | Alive | None | 73 (102-90) |
| 332 | 66 | 88 | na | < 5 | 93.5 | < 5 | UM | +/−/− | I-I | Alive | 28 | 74 (na-87) |
| 355 | 25 | 18 | 68 | < 5 | 25 | < 5 | UM | +/−/− | I-I | Expired | 88 | 228 (71-310) |
| 452 | 36 | 48 | na | na | na | na | M/UM | +/−/− | 0-I | Alive | 36 | 79 (63-105) |
| 546 | 7 | 6 | 0 | 0 | 78 | 0 | M | −/−/− | 0-0 | Alive | None | 32 (27-35) |
| 569 | 98 | 40 | 0 | 0 | 0 | 0 | UM | −/+/− | II-II | Alive | None | 73 (52-95) |
| 606 | 8 | 5 | 0 | 0 | 68 | 0 | M | −/−/− | 0-0 | Alive | None | 29 (30-26) |
| 625 | 54 | 57 | 80 | 0 | 0 | 0 | UM | ++/−/− | II-II | Alive | 17 | 297 (245-261) |
| 822 | 22 | 56 | 0 | 0 | na | 0 | UM | −/+/− | 0-III | Alive | 108 | 43 (30-75) |
| 875 | 14 | 18 | 0 | 0 | 0 | 0 | M | −/−/− | 0-0 | Alive | None | 15 (15-15) |
| 931 | 63 | 49 | na | na | na | na | UM | −/−/− | 0-0 | Expired | None | 20 (17-28) |
| CLL patient no. . | % CD38+ . | % ZAP-70+ . | Cytogenetic abnormalities, % cells . | IGHV mutation status* . | Lymph node/spleen/liver enlargement† . | Rai stage at diagnosis and when 2H2O study began . | Status . | Time to first treatment, mo‡ . | Mean WBC (beginning-end of study)§ . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Δ11q22 . | Tri12 . | Δ13q14 . | Δ17p23 . | |||||||||
| 189 | 2 | 7 | < 5 | < 5 | 70 | < 5 | M/UM | +/−/− | 0-IV | Expired | 53 | 107 (76-175) |
| 280 | 1 | 29 | < 5 | < 5 | 99 | < 5 | M | −/+/+ | 0-II | Alive | 108 | 58 (61-75) |
| 321 | 10 | 95 | < 5 | 59 | < 5 | < 5 | UM | −/−/− | 0-0 | Alive | None | 73 (102-90) |
| 332 | 66 | 88 | na | < 5 | 93.5 | < 5 | UM | +/−/− | I-I | Alive | 28 | 74 (na-87) |
| 355 | 25 | 18 | 68 | < 5 | 25 | < 5 | UM | +/−/− | I-I | Expired | 88 | 228 (71-310) |
| 452 | 36 | 48 | na | na | na | na | M/UM | +/−/− | 0-I | Alive | 36 | 79 (63-105) |
| 546 | 7 | 6 | 0 | 0 | 78 | 0 | M | −/−/− | 0-0 | Alive | None | 32 (27-35) |
| 569 | 98 | 40 | 0 | 0 | 0 | 0 | UM | −/+/− | II-II | Alive | None | 73 (52-95) |
| 606 | 8 | 5 | 0 | 0 | 68 | 0 | M | −/−/− | 0-0 | Alive | None | 29 (30-26) |
| 625 | 54 | 57 | 80 | 0 | 0 | 0 | UM | ++/−/− | II-II | Alive | 17 | 297 (245-261) |
| 822 | 22 | 56 | 0 | 0 | na | 0 | UM | −/+/− | 0-III | Alive | 108 | 43 (30-75) |
| 875 | 14 | 18 | 0 | 0 | 0 | 0 | M | −/−/− | 0-0 | Alive | None | 15 (15-15) |
| 931 | 63 | 49 | na | na | na | na | UM | −/−/− | 0-0 | Expired | None | 20 (17-28) |
Note that WBC data for CLL 569 were incomplete (only beginning and end values were available) and WBC count of CLL 332 at the beginning of the study was not available.
na indicates data not available.
Two percent or less difference from the germline gene defines a patient as IGHV unmutated (UM); more than 2% difference defines a patient as IGHV mutated (M).
Lymph nodes: + indicates 1-1.5 cm; ++, 1.5-3 cm. + for spleen and liver defines them as palpable by physical examination or enlarged on imaging (ultrasound or computed tomography scan). Only data 6 months before or after the study are included.
Time from initial diagnosis to first treatment for clinical progression of CLL.
Mean WBC count recorded during 2H2O protocol period; in parentheses are values at the beginning and end of the protocol.
Isolation of cell fractions based on expression of CD38 and Ki-67
CD38
Previously cryopreserved peripheral blood mononuclear cells (PBMCs) from CLL patients were thawed and incubated with murine monoclonal IgG1 anti–human CD5–fluorescein isothiocyanate (FITC), CD38–phycoerythrin (PE), CD3–peridinin-chlorophyll-protein complex (PerCP), and CD19–allophycocyanin (APC; all from BD Biosciences). After sorting into CD19+CD5+CD38+ and CD19+CD5+CD38− fractions with a BD FACSAria (Becton Dickinson Immunocytometry Systems; Figure 1B), cells were washed, pelleted, and stored at −80°C until performing GC/MS.
Ki-67
Cells from CLL 452 and CLL 625 were labeled with anti–CD38-PE, –CD3-PerCP, –CD19-APC, and –CD5-PE cyanin 7 (all from BD Biosciences), permeabilized and fixed (Cytofix/Cytoperm solution; BD Biosciences), and then stained intracellularly with murine monoclonal antibody (mAb) to Ki-67–FITC (BD Biosciences). Fractions were sorted and stored as for CD38.
Determination of Ki-67, ZAP-70, and chemokine receptor levels
Ki-67 expression was determined by flow cytometry after labeling cells with the panel of aforementioned mAbs. For ZAP-70, cells were incubated with murine monoclonal IgG1 anti–human CD5-PE, CD3-PerCP, and CD19-APC (BD Biosciences), permeabilized and fixed (Cytofix/Cytoperm solution; BD Biosciences), and then stained intracellularly with mAb to ZAP-70-FITC (eBiosciences). For chemokine receptors, the following murine mAbs were used: CD38-APC, CD19-PerCP, and CD5-FITC (BD Biosciences); C-C chemokine receptor type 1 (CCR1), CCR2, CCR7, CXCR1, CXCR4 (clone 12G5), and CXCR5 (R&D Systems); CCR4, CCR5, CXCR2, and CXCR3 (BD Biosciences). All anti–chemokine receptor mAbs were conjugated to phycoerythrin (PE). All data were acquired with a BD LSRII flow cytometer (Becton Dickinson Immunocytometry Systems) and analyzed by FlowJo V7.2.4 software (TreeStar).
Culture conditions to analyze 2H dilution in proliferating cells
PBMCs from CLL 355, taken when the highest 2H enrichment was found (Table 2), were labeled with carboxyfluorescein succinimidyl ester (CFSE; Molecular Probes/Invitrogen) at a final concentration of 10 μM/107 cells. An aliquot of cells was analyzed as a measure of CFSE intensity on day 0, and the remaining cells were cultured for 5 days (4 × 106/mL) over a layer of irradiated CD32-transfected murine fibroblasts (ATCC) at a fibroblast/PBMC ratio of 1:10. Interleukin-15 (IL-15), IL-2 (both 5 ng/mL; R&D Systems), and CpG 2006-G5 (0.5 μg/mL; InvivoGen) were added. When harvested, the absolute number of cells was determined with Countbright beads for flow cytometry (Invitrogen) according to instructions. B cells were identified using CD19-APC cyanin 7 (BD Biosciences) and sorted on a BD FACSAria based on CFSE labeling.
Fraction (%) of CD38+ and CD38− 2H-labeled cells (f) in the clones of CLL patients involved in this study
| CLL patient no. . | 1st time point . | 2nd time point . | 3rd time point . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CD38+, f . | CD38−, f . | CD38+, f/CD38−, f . | CD38+, f . | CD38−, f . | CD38+, f/CD38−, f . | CD38+, f . | CD38−, f . | CD38+, f/CD38−, f . | |
| 189 | 30.0 | 17.0 | 1.8 | 31.0 | 26.0 | 1.2 | 24.0 | 29.0 | 0.8 |
| 280 | 7.5 | 8.5 | 0.9 | 15.6 | 12.0 | 1.3 | 16.9 | 12.6 | 1.3 |
| 321 | 19.5 | 14.1 | 1.4 | 26.5 | 19.2 | 1.4 | 20.4 | 22.5 | 0.9 |
| 332 | 2.4 | 1.5 | 1.6 | 3.2 | 2.1 | 1.5 | 16.0 | 14.0 | 1.1 |
| 355 | 2.1 | 1.9 | 1.1 | 8.9 | 7.3 | 1.2 | 26.6 | 21.3 | 1.2 |
| 452 | 16.0 | 2.7 | 5.9 | 25.0 | 8.4 | 3.0 | 9.1 | 7.3 | 1.2 |
| 546 | 28.0 | 4.7 | 5.9 | 34.4 | 10.7 | 3.2 | 10.7 | 9.5 | 1.1 |
| 569 | 24.0 | 8.7 | 2.8 | 40.7 | 16.3 | 2.5 | 21.7 | 21.0 | 1.0 |
| 606 | 46.2 | 22.1 | 2.8 | 73.3 | 8.9 | 8.2 | 4.8 | 5.1 | 0.9 |
| 625 | 1.8 | 1.5 | 1.2 | 9.7 | 6.4 | 1.5 | 13.4 | 10.0 | 1.3 |
| 822 | 26.5 | 5.3 | 5.0 | 44.0 | 11.0 | 4.0 | 20.8 | 13.3 | 1.6 |
| 875 | 3.6 | 1.8 | 2.0 | 6.6 | 5.3 | 1.2 | 4.7 | 5.0 | 0.9 |
| 931 | 8.7 | 3.3 | 2.6 | 18.4 | 12.1 | 1.5 | 8.1 | 7.5 | 1.1 |
| Average ± SE | 16.5 ± 3.9* | 7.3 ± 1.8* | 2.7 ± 0.5 | 25.9 ± 5.4† | 11.2 ± 1.7† | 2.4 ± 0.5 | 15.2 ± 2‡ | 13.7 ± 2.1‡ | 1.1 ± 0.1 |
| CLL patient no. . | 1st time point . | 2nd time point . | 3rd time point . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CD38+, f . | CD38−, f . | CD38+, f/CD38−, f . | CD38+, f . | CD38−, f . | CD38+, f/CD38−, f . | CD38+, f . | CD38−, f . | CD38+, f/CD38−, f . | |
| 189 | 30.0 | 17.0 | 1.8 | 31.0 | 26.0 | 1.2 | 24.0 | 29.0 | 0.8 |
| 280 | 7.5 | 8.5 | 0.9 | 15.6 | 12.0 | 1.3 | 16.9 | 12.6 | 1.3 |
| 321 | 19.5 | 14.1 | 1.4 | 26.5 | 19.2 | 1.4 | 20.4 | 22.5 | 0.9 |
| 332 | 2.4 | 1.5 | 1.6 | 3.2 | 2.1 | 1.5 | 16.0 | 14.0 | 1.1 |
| 355 | 2.1 | 1.9 | 1.1 | 8.9 | 7.3 | 1.2 | 26.6 | 21.3 | 1.2 |
| 452 | 16.0 | 2.7 | 5.9 | 25.0 | 8.4 | 3.0 | 9.1 | 7.3 | 1.2 |
| 546 | 28.0 | 4.7 | 5.9 | 34.4 | 10.7 | 3.2 | 10.7 | 9.5 | 1.1 |
| 569 | 24.0 | 8.7 | 2.8 | 40.7 | 16.3 | 2.5 | 21.7 | 21.0 | 1.0 |
| 606 | 46.2 | 22.1 | 2.8 | 73.3 | 8.9 | 8.2 | 4.8 | 5.1 | 0.9 |
| 625 | 1.8 | 1.5 | 1.2 | 9.7 | 6.4 | 1.5 | 13.4 | 10.0 | 1.3 |
| 822 | 26.5 | 5.3 | 5.0 | 44.0 | 11.0 | 4.0 | 20.8 | 13.3 | 1.6 |
| 875 | 3.6 | 1.8 | 2.0 | 6.6 | 5.3 | 1.2 | 4.7 | 5.0 | 0.9 |
| 931 | 8.7 | 3.3 | 2.6 | 18.4 | 12.1 | 1.5 | 8.1 | 7.5 | 1.1 |
| Average ± SE | 16.5 ± 3.9* | 7.3 ± 1.8* | 2.7 ± 0.5 | 25.9 ± 5.4† | 11.2 ± 1.7† | 2.4 ± 0.5 | 15.2 ± 2‡ | 13.7 ± 2.1‡ | 1.1 ± 0.1 |
P < .01, paired t test.
P = .013, paired t test.
P = .25, paired t test.
Measurement of body 2H2O enrichment
Measurement of 2H enrichment in deoxyadenosine of DNA
Calculation of fraction of new cells
The fraction (%) of labeled new cells, f, was calculated from EM1 and the precursor enrichment (p), which was the average body water percentage of 2H2O at plateau. The asymptotic or maximal EM1* was calculated using the following equation: EM1* = −7.7268p2 + 3.5291p + 0.0023.22 f was then calculated as f = EM1/EM1*.
Analysis of IGHV mutations in CLL cells
IGHV mutation status was determined as described.8
Quantification of mean telomere lengths
A flow cytometry–fluorescence in situ hybridization (flow-FISH) protocol was used as published.29
Cytogenetic analysis
Cytogenetic analyses were performed by FISH using the following probes: chromosome 11, LSI ATM at 11q22.3; chromosome 12, CEP-12 centromere; chromosome 13, LSI D13S319 at 13q14.3 and LSI 13q34 at 13q34; and chromosome 17, LSI p53 at 17p13.1 (all from Vysis Inc).
Statistical analyses
A mixed model repeated measures analysis was used to determine whether the ratio of the percentage of labeled CD38+ to percentage of labeled CD38− cells (expressed as log of the ratio) differed across the 3 time points and whether the patterns of change over time of this ratio were significantly different across the 3 protocol groups (Figure 1). All pairwise comparisons were carried out using a Bonferroni adjustment. For data in Table 2, a paired t test was used within each time point. Classification of patients into group A and group B (Figure 3) was based on slopes of the lines generated by the percentage of labeled cells in CD38+ and CD38− fractions from time points 2 to 3, and declaring slopes as concordant positive, if both CD38+ and CD38− lines exhibited positive slopes, or non–concordant positive, otherwise. The Fisher exact test was used to compare clinical data between group A and group B patients. White blood cell (WBC) counts were compared using a mixed model repeated measures analysis. The Mann-Whitney test was used for data in Figure 5.
Results
Intraclonal heterogeneity: CD19+CD5+CD38+ cells proliferate more rapidly than CD19+CD5+CD38− cells
In this study we asked whether all cells in a CLL clone divide at the same rate and, if not, how to identify those subpopulations that divide more rapidly. Given the known biologic and clinical value of CD38,6,15 we measured 2H incorporation into CD38+ and CD38− fractions of leukemic clones from 13 patients who participated in 2H2O-labeling studies. 2H enrichment was measured in genomic DNA from CD19+CD5+CD38+ and CD19+CD5+CD38− cells at 3 time points (Figure 1A-B), and these values were used to calculate f, percentage of newly produced CLL cells.
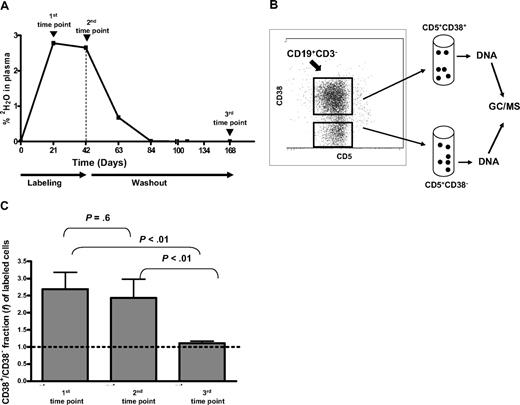
Procedure for determining 2H enrichments in CD38+ and CD38− CLL cells and time point selection in relation to 2H availability in body water. (A) Deuterium enrichment measured in plasma of body water compartment of a representative case (CLL 875). CLL cells were flow-sorted at the 3 time points indicated by arrows. Vertical dashed line indicates time when 2H2O intake was ended. (B) After gating CD19+CD3− cells, CD5+ cells were flow-sorted based on their expression of CD38. 2H in genomic DNA was then measured by GC/MS. (C) The ratio of CD38+/CD38− fractions (f) of labeled cells (Table 2) was averaged. Bars represent SEs of the 3 time points studied. Significant differences in 2H incorporation between the CD38− and CD38+ B-CLL subpopulations were achieved early and maintained during labeling period, whereas they disappeared during washout.
Procedure for determining 2H enrichments in CD38+ and CD38− CLL cells and time point selection in relation to 2H availability in body water. (A) Deuterium enrichment measured in plasma of body water compartment of a representative case (CLL 875). CLL cells were flow-sorted at the 3 time points indicated by arrows. Vertical dashed line indicates time when 2H2O intake was ended. (B) After gating CD19+CD3− cells, CD5+ cells were flow-sorted based on their expression of CD38. 2H in genomic DNA was then measured by GC/MS. (C) The ratio of CD38+/CD38− fractions (f) of labeled cells (Table 2) was averaged. Bars represent SEs of the 3 time points studied. Significant differences in 2H incorporation between the CD38− and CD38+ B-CLL subpopulations were achieved early and maintained during labeling period, whereas they disappeared during washout.
We found (Table 2) a 2-fold higher percentage of newly produced cells in the CD38+ compared with the CD38− compartment at both time points during labeling period (16.5% ± 3.9% vs 7.3% ± 1.8% at first time point, P < .01; 25.9% ± 5.4% vs 11.2% ± 1.7% at second time point, P = .013). When analyzed on an individual patient basis, a larger f was found in the CD38+ population of all but 1 patient (CLL 280 at only the first time point), with maximum numbers ranging from 6.6% to 73% (CLLs 875 and 606, respectively).
When presented as a ratio of all patients (Figure 1C), newly labeled cells (f) in the CD38+ fractions are approximately 2.5 times greater than in the CD38− fraction at both time points. Of note, observed differences did not correlate with the number of CD38+ cells in individual CLL clones prior to fractionation (range: 1%–98%; Table 1). For example, when analyzing data from cells taken at the first time point, CD38+ to CD38− labeled fraction ratios were more than 2 in 6 patients (CLLs 452, 546, 569, 606, 822, 931; Table 2) and the percentage of CD38+ cells in these clones ranged from 7% to 98% (Table 1).
Within each CLL clone, the Ki-67+ fraction is enriched in CD38+ cells and has more 2H incorporated in genomic DNA
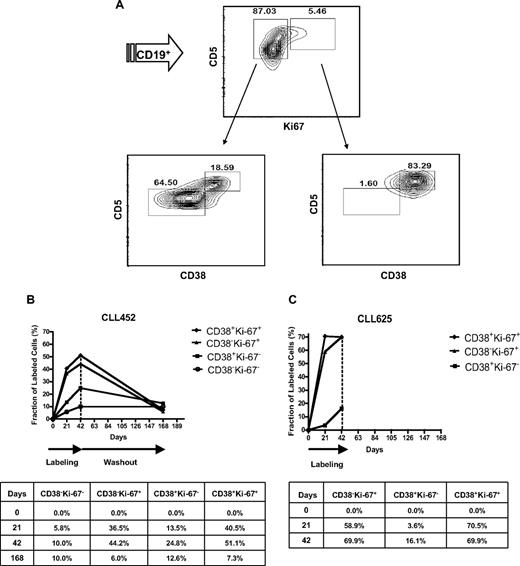
Ki-67 is a cell cycle–related molecule,30 and Ki-67–expressing cells are enriched in CD38+ fractions of individual CLL clones.21 As illustrated by CLL 822, the Ki-67+ fraction (5.5%; Figure 2A top right) was markedly enriched in CD38+ cells (83%; Figure 2A bottom right). Similar enrichments of CD38+ cells in Ki-67+ fractions were found in all patients tested (supplemental Figure 1).
CD38 expression in Ki-67+ and Ki-67− CLL cells and 2H enrichment in CLL clonal fractions sorted based on CD38 and Ki-67. (A) PBMCs from CLL 822 were incubated with fluorochrome-labeled mAbs reactive with CD19, CD5, and CD38, and, after cell permeabilization, with Ki-67. Cells were first gated for CD19 and then analyzed as reported in the figure. (Top plot) CLL clone contains ∼ 5% Ki-67+ cells. (Bottom plots) Percentage of CD38+ cells in the CD19+CD5+Ki-67+ fraction (83%; bottom right) is much higher compared with that observed in the CD19+CD5+Ki-67− fraction (18%; bottom left). CLL cells from CLL 452 (B) and CLL 625 (C) were flow-sorted based on CD38 and Ki-67 expression, and 2H-labeled DNA was measured in the sorted fractions. Curves represent the fraction (f) of labeled cells reported in the tables. Ki-67+ cells, both CD38− and CD38+, incorporated more 2H than their Ki-67− counterparts.
CD38 expression in Ki-67+ and Ki-67− CLL cells and 2H enrichment in CLL clonal fractions sorted based on CD38 and Ki-67. (A) PBMCs from CLL 822 were incubated with fluorochrome-labeled mAbs reactive with CD19, CD5, and CD38, and, after cell permeabilization, with Ki-67. Cells were first gated for CD19 and then analyzed as reported in the figure. (Top plot) CLL clone contains ∼ 5% Ki-67+ cells. (Bottom plots) Percentage of CD38+ cells in the CD19+CD5+Ki-67+ fraction (83%; bottom right) is much higher compared with that observed in the CD19+CD5+Ki-67− fraction (18%; bottom left). CLL cells from CLL 452 (B) and CLL 625 (C) were flow-sorted based on CD38 and Ki-67 expression, and 2H-labeled DNA was measured in the sorted fractions. Curves represent the fraction (f) of labeled cells reported in the tables. Ki-67+ cells, both CD38− and CD38+, incorporated more 2H than their Ki-67− counterparts.
To directly prove that Ki-67 expression represents proliferating cells in CLL, CD19+CD5+ cells from CLL 452 obtained at the 3 time points were sorted into 4 fractions based on expression of Ki-67 and CD38 (Figure 2B). Consistent with phenotypic data,31 f was higher among cells expressing Ki-67 at both time points during the labeling period, with a hierarchy of incorporation: CD38+Ki-67+ > CD38−Ki-67+ > CD38+Ki-67− > CD38−Ki-67−. Of note, the CD38+Ki-67+ fraction contained approximately 50% newly divided cells and the Ki-67+ cells represented a highly proliferating subset, defined by 2H incorporation in DNA, regardless of the expression of CD38. Similar findings were obtained from CLL 625 (Figure 2C). In this case, 70% of cells in the CD38+Ki-67+ fraction were new and already at plateau by 21 days of labeling, suggesting a minimum estimated cellular turnover rate of 3.3% per day (70%/21 days), approximately 2- to 30-fold higher than the rates previously observed for whole CLL clones.3,24
Differences in proliferative rates between CD19+CD5+CD38+ and CD19+CD5+CD38− cells are no longer evident after 2H2O washout
The rate at which the fraction of 2H-labeled cells of a population with a high turnover (eg, polymorphonuclear leukocytes) disappears from blood after 2H2O intake cessation (washout period) is relatively rapid and follows the decline of available 2H2O4. In this study, at the end of the washout period we observed a diminution in the fraction of 2H-containing CD38+ cells, accompanied by stability or increase in the CD38−, 2H-labeled fraction in many cases (Table 2). As a result, f was no longer different (CD38+ vs CD38−: 15.2% ± 2 vs 13.7% ± 2.1; P = .25) and the ratio of f of the 2 fractions was essentially 1. Mean ratios recorded during labeling periods (time points 1 and 2) were significantly different from those recorded at time point 3 (Figure 1C). A decrease in the ratios of CD38+ to CD38− newly produced cells between second and third time point was observed in 11 of 13 patients (Table 2).
Telomere length measurements of CD38+ and CD38− fractions
The 2H incorporation data presented in Table 2 indicate that within each CLL clone, CD38+ cells divide faster than CD38−. We wanted to determine whether the differences in newly produced cells were reflected by differences in telomere lengths.
Notably, an average of the mean telomere lengths of the 13 CLL clones and their CD38-defined fractions did not correspond with proliferation data obtained by 2H incorporation (Table 3). CD38+ subsets had shorter telomere lengths compared with CD38− ones in only 5 of 13 patients. Moreover, some patients in whom higher 2H fractional enrichments in CD38+ cells were most evident (CLLs 546 and 569) had longer telomere lengths, compared with CD38− cells. Thus, no differences in the average of mean telomere lengths between the 2 fractions of the 13 patients were found (Table 3).
Mean telomere lengths (kilobases) measured in CD19+CD5+, CD19+CD5+CD38+, and CD19+CD5+CD38− fractions
| CLL patient no. . | CD19+ . | CD38+ . | CD38− . |
|---|---|---|---|
| 189 | 6.61 | 5.18 | 4.75 |
| 280 | 5.16 | 8.76 | 4.17 |
| 321 | 4.7 | 4.23 | 3.76 |
| 332 | 0.56 | 0.99 | 0.9 |
| 355 | 3.72 | 3.31 | 4.51 |
| 452 | 1.19 | 1.11 | 2.13 |
| 546 | 7.54 | 8.27 | 8.04 |
| 569 | 2.35 | 2.79 | 1.39 |
| 606 | 3.64 | 3.22 | 3.4 |
| 625 | 1.94 | 2.11 | 1.19 |
| 822 | 2.76 | 2.72 | 4.48 |
| 875 | 3.77 | 4.02 | 3.74 |
| 931 | 4.16 | 3.89 | 4.34 |
| Average ± SE | 3.7 ± 0.56 | 3.9 ± 0.66 | 3.6 ± 0.53 |
| CLL patient no. . | CD19+ . | CD38+ . | CD38− . |
|---|---|---|---|
| 189 | 6.61 | 5.18 | 4.75 |
| 280 | 5.16 | 8.76 | 4.17 |
| 321 | 4.7 | 4.23 | 3.76 |
| 332 | 0.56 | 0.99 | 0.9 |
| 355 | 3.72 | 3.31 | 4.51 |
| 452 | 1.19 | 1.11 | 2.13 |
| 546 | 7.54 | 8.27 | 8.04 |
| 569 | 2.35 | 2.79 | 1.39 |
| 606 | 3.64 | 3.22 | 3.4 |
| 625 | 1.94 | 2.11 | 1.19 |
| 822 | 2.76 | 2.72 | 4.48 |
| 875 | 3.77 | 4.02 | 3.74 |
| 931 | 4.16 | 3.89 | 4.34 |
| Average ± SE | 3.7 ± 0.56 | 3.9 ± 0.66 | 3.6 ± 0.53 |
Interclonal kinetic heterogeneity of CD38+ and CD38− fractions
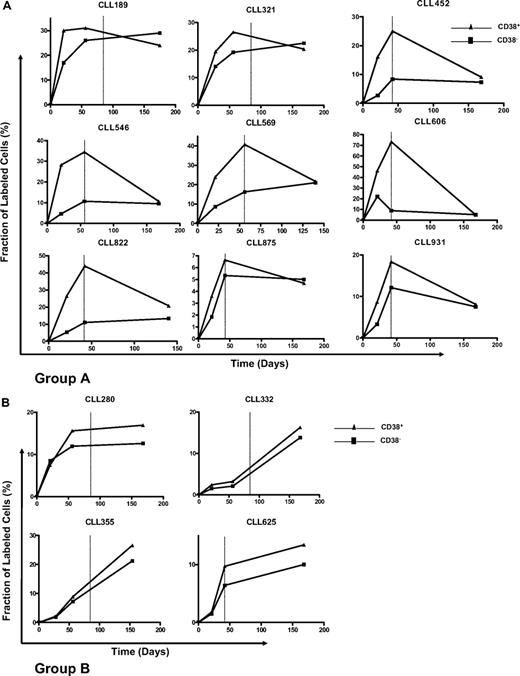
There were clear differences in CD38+ and CD38− kinetics among patients (Figure 3). For most of our cases (9/13), defined as group A (Figure 3A), CD38+ cells rapidly achieved a maximal fraction of 2H-containing cells at the end of the labeling period. These fractions exceeded, in some instances dramatically (eg, CLLs 452, 546, and 822), those reached by CD38− cells. 2H incorporation in CD38+ cells then fell during washout. Patient CLL 606 was peculiar because of the loss of 2H-labeled CD38− cells before the end of the labeling period, although the kinetics for CD38+ cells resemble those observed in the other members of group A.
Kinetics of CD38+ and CD38− CLL cells in each patient studied. Curves represent the fraction (f) of labeled cells at the 3 time points studied. Note that different levels of 2H enrichment were observed among patients, and the graphs have different scales. Vertical dotted lines indicate the end of 2H2O assumption. Based on the slopes of the CD38+ and CD38− kinetic curves, 2 groups of patients were defined, A and B. (A) In group A, CD38+ cells rapidly achieve maximal 2H enrichment levels at the end of the labeling period; 2H incorporation in these cells subsequently falls during washout. (B) Patients in group B are characterized by similar kinetic patterns of both CD38+ and CD38− fractions with delayed appearance and slow achievement of maximal levels of labeled cells.
Kinetics of CD38+ and CD38− CLL cells in each patient studied. Curves represent the fraction (f) of labeled cells at the 3 time points studied. Note that different levels of 2H enrichment were observed among patients, and the graphs have different scales. Vertical dotted lines indicate the end of 2H2O assumption. Based on the slopes of the CD38+ and CD38− kinetic curves, 2 groups of patients were defined, A and B. (A) In group A, CD38+ cells rapidly achieve maximal 2H enrichment levels at the end of the labeling period; 2H incorporation in these cells subsequently falls during washout. (B) Patients in group B are characterized by similar kinetic patterns of both CD38+ and CD38− fractions with delayed appearance and slow achievement of maximal levels of labeled cells.
However, the kinetics of patients 280, 332, 355, and 625 (Figure 3B group B) differed significantly from the 9 patients in Figure 3A. The fraction of 2H-labeled CD38+ cells of these patients reached maximum levels only at the end of the washout period. Moreover, CD38+ and CD38− fractions in these patients had very similar kinetics, compared with the usually clearly divergent and more heterogeneous curves of the other patients.
These observed differences were quantified. The slopes recorded between time points 2 and 3 were concordant positive in group B and nonconcordant positive in group A (see also “Statistical analyses”).
CD38+ cells may be diluted more rapidly with unlabeled, newly divided cells during the 2H2O washout period
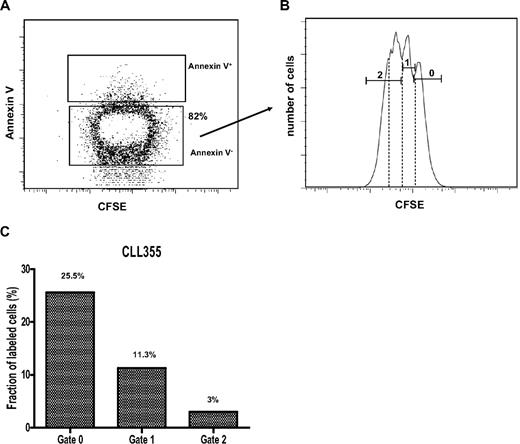
In the presence of decreasing amounts of 2H, unlabeled CD38+ cells, because of their faster proliferative rate, could more rapidly dilute labeled CD38+ cells than unlabeled CD38− cells could dilute labeled CD38− cells. To test this possibility, cells from CLL 355, taken when they were maximally enriched in 2H in vivo (Table 2), were loaded with CFSE and stimulated in vitro with CpG 2006-G5 plus IL-2 and IL-15; these conditions are known to induce proliferation of human B cells.32 Four million cells were cultured initially, and a total of 1.8 × 106 cells were recovered at day 5. Based on annexin V labeling (Figure 4A), 82% of these (∼ 1.5 × 106) were scored as viable. These live cells were segregated according to CFSE expression peaks, thereby defining 3 generations (Figure 4B): generation 0 or undivided (CFSE mean fluorescence intensity [MFI]: 1600), generation 1 (CFSE MFI: 780), and generation 2 (CFSE MFI: 350). These 3 annexin V−CFSE+ fractions were sorted and analyzed for 2H-labeled DNA. Enrichment of 2H in cellular DNA decreased as each new generation arose in vitro in the absence of 2H2O (Figure 4C). The gated generation 2 probably included a small generation 3 (Figure 4B dotted lines), thus explaining an MFI and 2H content lower than the expected reduction from those of generation 1.
2H content decreases generation after generation among cells that proliferate in absence of 2H2O. (A) PBMCs from CLL 355 at the third time point during 2H2O protocol (Table 2) were loaded with CFSE and cultured with CpG 2006-G5 (5 μg/mL), IL-2 (5 ng/mL), and IL-15 (5 ng/mL) to induce proliferation. Cells were harvested after 5 days and stained with annexin V–APC to exclude dying/dead cells. (B) Annexin V− cells were analyzed for CFSE content and 3 generations of cells were flow-sorted based on the indicated gates (0, 1, 2). Gate 0 represents undivided cells. Note that whereas gates 0 and 1 contained discreet CFSE peaks, gate 2 probably contained a main peak and a smaller one (dotted lines), possibly representing another generation of progeny. (C) A decreased fraction (f) of labeled cells was found generation after generation.
2H content decreases generation after generation among cells that proliferate in absence of 2H2O. (A) PBMCs from CLL 355 at the third time point during 2H2O protocol (Table 2) were loaded with CFSE and cultured with CpG 2006-G5 (5 μg/mL), IL-2 (5 ng/mL), and IL-15 (5 ng/mL) to induce proliferation. Cells were harvested after 5 days and stained with annexin V–APC to exclude dying/dead cells. (B) Annexin V− cells were analyzed for CFSE content and 3 generations of cells were flow-sorted based on the indicated gates (0, 1, 2). Gate 0 represents undivided cells. Note that whereas gates 0 and 1 contained discreet CFSE peaks, gate 2 probably contained a main peak and a smaller one (dotted lines), possibly representing another generation of progeny. (C) A decreased fraction (f) of labeled cells was found generation after generation.
Differences in rates at which CD38+ and CD38− cells leave solid tissues after replication
Differences in 2H labeling of CD38+ and CD38− cells could relate to the time cells take to exit solid tissues where division and 2H incorporation into DNA most likely occurred. Because chemokines and their receptors are involved in cell retention, migration, and homing,33-36 we analyzed several chemokine receptors in all 13 patients to determine whether a link existed between these receptors and CLL cell kinetics (Figure 5).
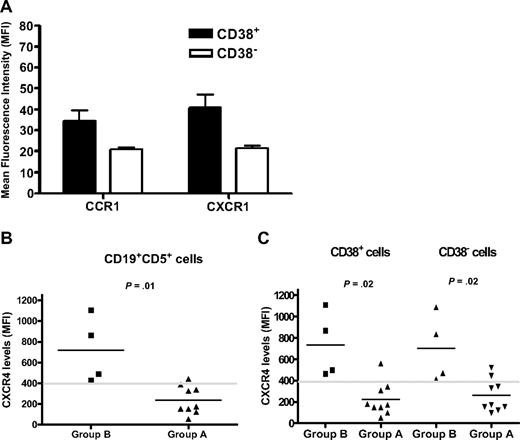
Chemokine receptor levels on CLL cells of patients in the study. Surface membrane expression of a panel of chemokine receptors was evaluated in the 13 patients in the study. All the samples were analyzed in the same experiment. (A) Of the chemokine receptor levels studied (see text for the list), only 2 (CCR1 and CXCR1) were expressed at significantly higher densities (MFI) in CD38+ compared with CD38− cells (P < .05). Error bars represent SEs of measurements. (B) Group B patients (n = 4) had significantly higher CXCR4 levels (MFI) compared with group A (n = 9; P = .01). Comparisons of the other chemokine receptors revealed no significant differences between the 2 groups. (C) Both CD38+ (left) and CD38− (right) fractions from group B patients exhibited significantly higher CXCR4 levels compared with the same fractions from group A (P = .02).
Chemokine receptor levels on CLL cells of patients in the study. Surface membrane expression of a panel of chemokine receptors was evaluated in the 13 patients in the study. All the samples were analyzed in the same experiment. (A) Of the chemokine receptor levels studied (see text for the list), only 2 (CCR1 and CXCR1) were expressed at significantly higher densities (MFI) in CD38+ compared with CD38− cells (P < .05). Error bars represent SEs of measurements. (B) Group B patients (n = 4) had significantly higher CXCR4 levels (MFI) compared with group A (n = 9; P = .01). Comparisons of the other chemokine receptors revealed no significant differences between the 2 groups. (C) Both CD38+ (left) and CD38− (right) fractions from group B patients exhibited significantly higher CXCR4 levels compared with the same fractions from group A (P = .02).
CLL cells were evaluated for CCR1, CCR2, CCR4, CCR5, CCR7, CXCR1, CXCR3, CXCR4, and CXCR5, both as percentage of positive cells as well as MFIs. The latter (MFI) was necessary because chemokine receptors such as CXCR4, CXCR5, and CCR7 are expressed on virtually every cell in a CLL clone,33 albeit over a range of densities. Only CCR1 and CXCR1 were expressed at greater densities in the CD38+ fractions in all 13 CLL clones studied (P < .05; Figure 5A).
We then compared data for patients in group A with those in group B, because the latter had a slower appearance of 2H-marked cells in the blood, suggesting longer retention in solid tissues. No differences were found in the percentage of chemokine receptor–expressing cells in group A versus group B (not shown).
However, when analyzed for cell surface density, a significant difference for CXCR4 was seen between the 2 groups (MFI group B: 720 ± 160 vs group A: 238 ± 47; P = .01; Figure 5B). In addition, all 4 patients in group B had a CXCR4 MFI higher than 400, compared with only 1 of 9 in group A. Furthermore, consistent with the observation that in group B CD38+ and CD38− fractions had similar kinetics (Figure 5B), both CD38+ and CD38− subsets exhibited higher CXCR4 densities compared with the same fractions in group A (Figure 5C). Thus, higher CXCR4 densities were a property of CLL clones exhibiting delayed appearance in the periphery.
Kinetic patterns of CD38+ and CD38− subsets relate to clinical outcome
Clinical data for the patients in this report are provided in Table 1. Of the 13 patients studied, 8 experienced progression in Rai stage, required treatment, or had a fatal course. This was the case for all patients in group B (CLLs 280, 332, 355, 625) but only for 4 (44.4%) of 9 of those in group A. Group B patients had a shorter, albeit not statistically significant, median time to treatment compared with group A (57.6 vs 119.1 months; P = .06). In addition, all group B patients had involvement of a lymphoid organ detectable by physical examination, whereas this was the case again for only 4 of 9 patients of group A. Mean WBC counts averaged over the course of the study for group B patients were significantly higher (P = .044) than those of group A. Lastly, only 1 patient (CLL 280) in group B had a leukemic clone expressing a mutated IGHV. Among the IGHV-mutated cases in the entire cohort, CLL 280 is the only patient who progressed in terms of a change in Rai stage, had spleen and liver involvement, and the highest WBC and ZAP-70+ CLL cell counts.
Discussion
In this study we refined our understanding of the proliferative compartment in CLL. We determined that (1) different subsets of cells within a clone divide at different rates, (2) these cells can be identified by surface membrane phenotype, and (3) patients can differ in kinetics of the proliferating subsets. We defined these by measuring 2H incorporation into genomic DNA, a direct indicator of cell division,22,23 and by focusing on purified populations of cells expressing surface membrane CD38. We chose CD38 because the CD38+ subset within CLL clones is enriched in cells that have entered G1,21 and CD38 has biologic relevance and clinical value in the disease.6,15,17
Using this approach, we found clear evidence for interclonal and intraclonal kinetic heterogeneity. Interclonal heterogeneity, based on distinct CD38+ and CD38− kinetic patterns, identified 2 groups of patients, A and B (Figure 3). In group A cases (9/13 subjects), the fraction of 2H-labeled CD38+ cells rose quickly to plateau levels and then declined, whereas the fraction of labeled CD38− cells rose slowly to a maximum and declined at a slower rate during washout such that the CD38+ and CD38− washout curves often intersected (Figure 3A). Conversely, in group B patients, CD38+ and CD38− cells displayed similar kinetics, with continuous increases in the fraction of 2H-labeled cells throughout study periods, including the washout phase (Figure 3B). After 2H2O has washed out, label is no longer available to mark subsequently born cells. Therefore, if there is a continued increase in the percentage of labeled cells in the circulation during the washout period, this must represent a release of previously labeled/divided cells from the extravascular space.
We also identified intraclonal heterogeneity. In every patient a larger enrichment of newly produced CD38+ than CD38− cells was found during the period when 2H2O was present in body water (Table 2). This was evident throughout the labeling period for both group A and B patients and persisted at the end of labeling in group B. Importantly, the higher fraction of newly divided cells was a general characteristic of the CD38+ compartment, regardless of the percentage of CD38+ cells present in a leukemic clone (range: 1%–98%; Table 1). However in group A patients, we observed a decrease in the percentage of CD38+, 2H-labeled cells, which in 4 cases (CLLs 189, 321, 569, and 822) was paralleled by an increase in the percentage of CD38−-labeled cells (Table 2).
Loss of circulating CD38+, 2H-labeled cells after ceasing 2H2O intake can be explained by different, not mutually exclusive, mechanisms.
Higher death rates of rapidly proliferating cells
Cell death of activated, proliferating T and B cells occurs,37,38 thereby maintaining steady-state cell numbers over time. In studies of T-cell kinetics using 2H incorporation, this mechanism can explain why curves of appearance and disappearance of labeled cells have similar slopes.39 Thus decay of labeled CD38+ cells, characteristically observed in group A patients (Figure 3A), could indicate higher death rates of the more proliferative fraction.
Change in surface membrane phenotype over time with some CD38+ cells becoming CD38−
In 4 of 9 group A patients, decay of CD38+, 2H-labeled cells was paralleled by an increase in CD38−-labeled cells (Figure 3A and Table 2). For example, for CLL 822, between the end of labeling and end of washout, the f of 2H-labeled CD38+ cells decreased approximately 50% (44%-20.8%), whereas the f of labeled CD38− increased by approximately 15% (11%-13%; Table 2). These reciprocal changes may indicate that a portion of CD38+ cells became CD38−, and the remaining either died or left the circulation and were retained in a solid tissue, thereby becoming undetectable.
Such a phenotypic conversion of some CD38+ cells is also suggested by experiments with sorted Ki-67+CD38+ and Ki-67+CD38− cells (Figure 2 and Table 2). These fractions tended to equalize in terms of newly divided cells (Figure 2B-C), due either to (1) an early equilibration of 2H-labeled cells among the 2 compartments as a result of proliferating CD38+ cells becoming CD38−, or (2) CD38+Ki-67+ and CD38−Ki-67+ fractions constituting 2 distinct compartments proliferating at very similar (but different) rates. We favor the first explanation, for at least the majority of CD38+ fractions. In fact, if CD38+ and CD38− were 2 distinct compartments and the CD38+ compartment proliferated more rapidly the percentage of CD38+ cells within a clone would increase at the end of washout or over longer periods of time (unless all newly divided cells died). Although the percentage of CD38+ cells within a clone can change, it is not common (Damle et al,6 Hamblin et al,40 and supplemental Figure 1). Moreover, if CD38+ and CD38− cells were distinct stable subgroups, CD38+ cells should exhibit features of longer proliferative histories. However, this was not the case because mean telomere lengths were similar among CD38+ and CD38− cells of each clone (Table 3, Damle et al,21 Pepper et al41 ). These findings support the concept that at least a portion of the CD38+ and CD38− fractions is a continuum of the same cell population.
In this regard, 2 recent intraclonal comparisons of chromosomal aberrations in CD38+ and CD38− fractions reached different conclusions, one in support of and one against a change of CD38 phenotype (Lin et al42 and Grubor et al43 ; note that the latter reference includes 4 patients enrolled in this study—CLLs 355, 452, 625, and 931). If the phenotype of all CD38+ cells could change over time, similar chromosomal aberrations should exist in each fraction. However, if such a transition occurred in only a fraction of CD38+ cells and the size of this fraction differed among patients, then genomic changes might prevail in one fraction and not another; in such a situation, detection of these differences would depend on the size of a fraction and the sensitivity of the method measuring genomic aberrations. Our kinetic data provide indications of clonal heterogeneity, the extent of which is still not known but that might explain these apparent inconsistencies.
Dilution of a labeled population by unlabeled cells when proliferation occurs in absence of 2H2O
When labeled CLL cells proliferated ex vivo in the absence of 2H2O, 2H content among daughter cells decreased with each generation (Figure 5). This was likely due to the production of newly synthesized DNA in the absence of 2H, thereby “diluting” labeled DNA by unlabeled DNA (Figure 5C). Thus, at least a part of the decay curve for CD38+ cells is due to ongoing proliferation in the absence of 2H2O. This phenomenon would have a greater effect in the population that proliferates faster, that is, the CD38+ subset.
Differential rates of entering and leaving the blood from solid lymphoid tissues
In humans, bromodeoxyuridine-labeled T cells equilibrate between blood and lymphoid tissues within 24 hours.44 The same rapid equilibration was not found for proliferating B cells; in fact, there were 6 times higher numbers in lymphoid tissues than blood at all time points considered.44 In vivo rates of CLL cells flowing from peripheral blood into solid lymphoid tissues and vice versa are poorly understood, although a few studies have attempted to dissect the former trafficking.45-47
In regard to the latter situation, CD38− cells in every patient except CLL 931 reached plateaus later than CD38+ cells (Figure 3A-B). This could represent differences in activation states and in chemokine receptor expression between the 2 subpopulations. Indeed, higher levels of CCR1 and CXCR1 were found among CD38+ cells (Figure 5A), probably due to a more activated state of this proliferating population. CCR1 is overexpressed on normal B cells activated in vitro48 ; this issue has not been addressed directly for CXCR1.
In addition, a relationship between CLL cell kinetics and CXCR4 levels was found between group A and group B patients (Figure 5B-C). In this interclonal comparison, delayed appearance of labeled cells in group B suggested longer retention within lymphoid tissues. Consistent with this idea, group B clones had significantly higher densities of surface membrane CXCR4 compared with group A. Longer retention in solid tissues would give cells a prolonged exposure to survival factors and stimulatory ligands, and CXCR4 may have a crucial role in regulating this process.25,26 Proliferation centers are characteristically found in solid lymphoid tissues of patients, and it appears that leukemic cell proliferation and CD38 up-regulation occur within these structures.49 Our observed proliferation rates, together with CXCR1 and CCR1 expression levels, suggest that CD38+ cells have more recently left a solid tissue where they proliferated, either spontaneously or after receiving activating stimuli. Within each clone, moreover, the fraction expressing lower levels of surface CXCR4 was enriched in CD38+ cells and CD5+ cells and incorporated more 2H than the fraction with higher levels of CXCR4 (C.C. et al, manuscript in preparation). These findings also support a model in which, within the context of a lymphoid solid tissue, CLL cells down modulate CXCR4 (after CXCL12/SDF-1 ligation and activation stimuli) and ornate cell membranes with more CD38 and CD5.
Remarkably, and in line with the relationship between CXCR4 expression, bone marrow infiltration, and Rai stage,50 all group B patients experienced disease progression, regardless of IGHV mutation status (Table 1). Of interest, an in vivo study performed in the 1960s analyzing CLL cell kinetics using 3H-thymidine incorporation45 described 2 groups of patients: group 1 in which labeled cells rapidly peaked and then disappeared (similar to our group A) and group 2 with a flat slope of labeled cells (similar to our group B). In that study, group 2 patients had more organomegaly and higher WBC counts. In line with this, all group B patients had lymphoid organ involvement compared with 4 of 9 group A patients. Furthermore, group B patients had a shorter time to treatment (57.6 vs 119.1 months) and higher WBC counts than group A patients. Thus, differences in CXCR4 levels between leukemic clones relate well to differences in CLL cell kinetics and possibly clinical outcome.
A relationship between CLL kinetics, based on 2H incorporation, and disease activity was previously suggested, with higher birth rates associating with more aggressive disease.3 In addition, higher numbers of CD38-expressing cells correlate with greater risk of a poor clinical course and shortened survival.6 Because DNA replication is a prerequisite for the accumulation of new DNA lesions that could result in disease progression, our data suggest a link between percentage of CD38+ cells and clinical deterioration and outcome. Surprisingly, however, a clear correlation between the percentage of CD38+ proliferating cells in CLL clones and disease progression and survival was not found. Further studies with additional patients will be necessary to test this link.
In summary, this study demonstrates both intraclonal and interclonal heterogeneity in the proliferation of CLL cells. Some of this heterogeneity may be specious but still important, resulting from changes in surface membrane phenotype and differences in proliferative rates between the CD38+ and CD38− fractions in the absence of 2H2O. Other heterogeneity likely reflects fundamental differences in CLL subset survival and trafficking, for example, CXCR4 expression. Therefore, classifying patients on the basis of differences in CLL cell kinetics (eg, group A vs group B) can reveal molecular differences in leukemic cells and may eventually pinpoint differential balances between prosurvival, proliferative, and apoptotic mechanisms within solid tissues. Combining the 2H-labeling approach with more extensive cell surface and genetic analyses may help to further dissect these complexities. Indeed, a refined definition of the CLL proliferative compartment might allow us to focus our therapeutic strategies on the aggressive core of the clone and possibly identify leukemic stem cells in CLL, should they exist.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the FACS core facility at The Feinstein Institute for Medical Research for technical help.
This study was supported in part by grants from the CLL Global Research Foundation and the National Center for Research Resources of the National Institutes of Health (NIH; M01 General Clinical Research Center Grant, RR018535). In addition, the Karches Foundation, the Prince Foundation, the Marks Foundation, the Jerome Levy Foundation, the Leon Levy Foundation, the Tebil Foundation Inc, and the Joseph Eletto Leukemia Research Fund provided support.
Authorship
Contribution: C.C. performed research, analyzed data, and wrote the paper; R.N.D. and C.M. assisted with the experiments and reviewed work; G.H., E.J.M., and M.K.H. performed gas chromatography/mass spectrometry analyses and reviewed the paper; C.S. contributed analytical tools; M.S.K., J.E.K., S.L.A., and K.K.R. coordinated the clinical work; and N.C. designed research and wrote the paper.
Conflict-of-interest disclosure: M.K.H. has ownership interest in KineMed Inc. G.H., E.J.M., and N.C. own stock options in KineMed Inc. The remaining authors declare no competing financial interests.
Correspondence: Nicholas Chiorazzi, The Feinstein Institute for Medical Research, 350 Community Dr, Manhasset, NY 11030; e-mail: nchizzi@nshs.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal