Abstract
The clinical management of amyloidosis is based on the treatment of the underlying etiology, and accurate identification of the protein causing the amyloidosis is of paramount importance. Current methods used for typing of amyloidosis such as immunohistochemistry have low specificity and sensitivity. In this study, we report the development of a highly specific and sensitive novel test for the typing of amyloidosis in routine clinical biopsy specimens. Our approach combines specific sampling by laser microdissection (LMD) and analytical power of tandem mass spectrometry (MS)–based proteomic analysis. We studied 50 cases of amyloidosis that were well-characterized by gold standard clinicopathologic criteria (training set) and an independent validation set comprising 41 cases of cardiac amyloidosis. By use of LMD/MS, we identified the amyloid type with 100% specificity and sensitivity in the training set and with 98% in validation set. Use of the LMD/MS method will enhance our ability to type amyloidosis accurately in clinical biopsy specimens.
Introduction
Amyloidosis is caused by extracellular deposition of proteins in an insoluble beta-pleated sheet physical format.1 More than 25 different proteins are known to cause amyloidosis.2 Clinically, the most important amyloidogenic proteins are serum amyloid A (SAA), transthyretin (TTR), and immunoglobulin kappa (IGK) or lambda light chains (IGL; so-called primary or AL type). They account for more than 90% of systemic amyloidosis.3 The management of amyloidosis relies on treatment of the underlying etiology, often by high-risk, aggressive modalities such as high-dose chemotherapy and stem cell transplantation (for AL-type amyloidosis)4 or liver transplantation (for hereditary TTR-type amyloidosis).5,6 Given the nature of these management decisions, the accurate subtyping of amyloid deposits in clinical biopsy specimens is of paramount importance.
In routine practice, the diagnosis of amyloidosis is made by histopathologic examination and histochemical stains such as Congo red (CR).7 Further subtyping is often challenging because immunohistochemistry, the most common method used, is problematic even in the most experienced laboratories as the result of high background staining caused by serum contamination and epitope loss caused by protein cross-linking after formalin fixation.8-12 Although more sophisticated biochemical approaches have been used, they require quantities of tissue not readily available in a routine clinical setting and suffer from a lack of specificity because they contain nonamyloid tissues and serum, which are a rich source of amyloidogenic proteins.13-15
In this study, we report the development of a specific and sensitive novel test for typing of amyloidosis in routine clinical specimens. To overcome the aforementioned difficulties, our approach combines specific sampling by laser microdissection (LMD) and analytical power of tandem mass spectrometry (MS)–based proteomic analysis.
Methods
Clinical specimens
We studied 2 independent sets of amyloidosis specimens. The first set consisted of 50 diagnostic biopsy specimens from 50 patients (25 men, 25 women) and was used for the establishment of the method (training set). The specimens were fixed either in formalin (n = 42) or in B5 solution (n = 8), routinely processed, and embedded in paraffin. In each case the type of amyloid was characterized by the current gold standard approach, including an extensive clinical investigation for plasma cell disorders, serum and genetic testing for amyloidogenic TTR variants, and immunohistochemistry for TTR, SAA, IGK, IGL, and serum amyloid P component (SAP).16,17 Our study included 16 TTR (8 senile, 8 hereditary), 9 SAA, and 25 AL-type (20 IGL, 5 IGK) amyloidosis (Table 1). The second set consisted of 41 cases of cardiac amyloidosis prospectively studied by use of the LMD/MS method established (validation set). Twelve different tissues from 12 different subjects negative for amyloid by CR staining were used as control samples (supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). The study was approved by the institutional review board of the Mayo Clinic and was conducted in accordance with the Declaration of Helsinki.
Specimen preparation, microdissection, and MS-based proteomic analysis
The methods have been published previously,18 and the details are provided in the supplemental Methods. In brief, for each case, a 10-μm paraffin section was stained with CR, and then amyloid deposits were identified under fluorescent light and microdissected with LMD (supplemental Video 1). Each microdissection contained an area of 50 000 to 60 000 μm2, and 2 to 4 separate microdissections were analyzed for each specimen. Microdissected fragments were digested into tryptic peptides and analyzed by liquid chromatography electrospray tandem MS. MS raw data files were queried by the use of 3 different algorithms (Mascot, Sequest, and X!Tandem), the results were assigned peptide and protein probability scores, and they were displayed by the use of Scaffold (Proteome Software). For each case, a list of proteins based on peptides identified by MS was generated.
Data interpretation
For each case, the average number of spectra representing 4 amyloid-associated proteins (TTR, SAA, IGK, IGL) per sample (microdissection) were calculated. An algorithm was generated to determine the true disease state (amyloid type) of the specimen simply by assigning a specimen to the disease state (amyloid type) with the largest number of amyloid-associated peptide spectra hits.
Results and discussion
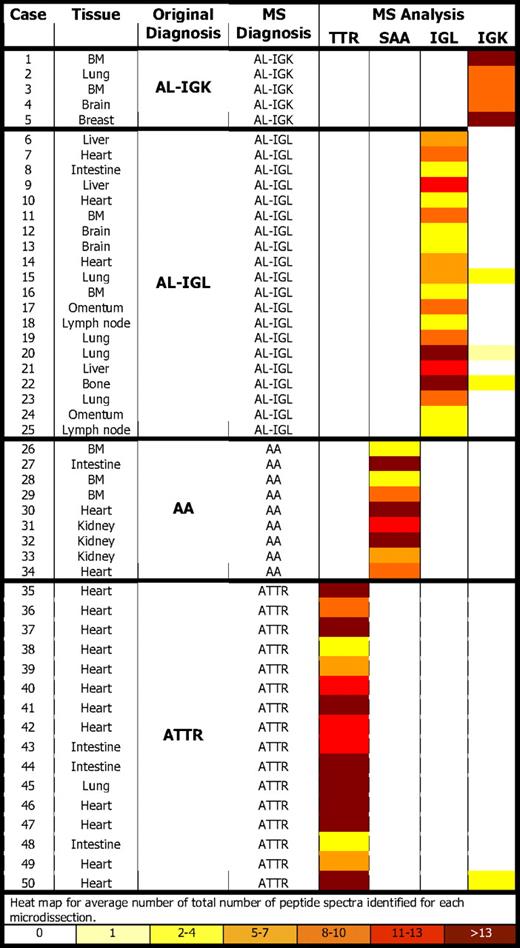
All cases contained CR-positive amyloid deposits, which were microdissected for protein extraction and tryptic digestion (Figure 1; supplemental Video 1).19 A successful proteomic profile of the amyloid deposits was obtained by MS analysis in each case. (Figure 1; supplemental Figures 1-2) In 46 of 50 cases, peptides belonging to only 1 of the 4 amyloid subtypes were detected (Figure 1; Table 1). However, interestingly, in 4 cases in addition to the dominant causative amyloid protein, a small number of spectra for IGK peptides were identified (Table 1; supplemental Figure 1). This finding was consistent between different microdissections for the same case, strongly indicating that IGK peptides were trapped within the amyloid deposits caused by other proteins. In addition, proteins known to be integrated in all types of amyloidosis, such as SAP, and apolipoprotein E, A-I, and A-IV were identified as constituents of amyloid deposits in most cases (Figure 1; supplemental Figures 1-2).
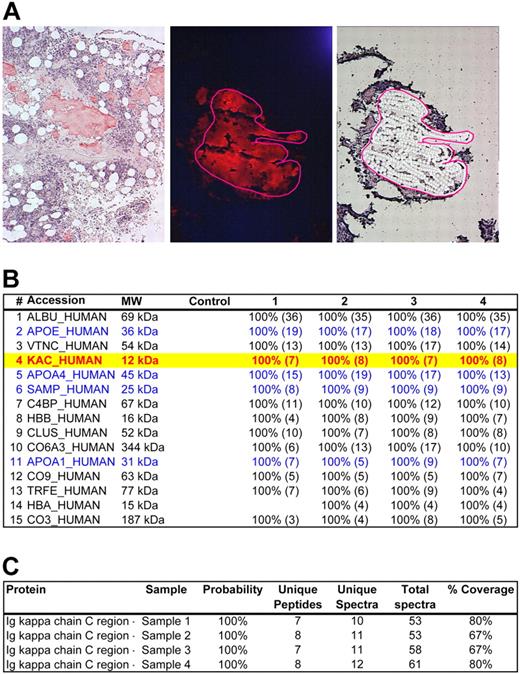
LMD/MS analysis of a case of AL-kappa amyloidosis (case 1). (A) Bone marrow specimen with interstitial nodules of amyloid deposition (Case 1). (Left) CR-stained section viewed under bright field confirming congophilia of the amyloid deposits. (Middle) CR-stained section viewed under fluorescent light source. The congophilic deposits give bright red fluorescence, confirming amyloid deposition. The area selected for microdissection is circled with a red line. (Right) Same area selected in the middle panel after microdissection of the amyloid plaque. Photograph (left) was taken with a DP70 Olympus camera (Olympus) with an Olympus BX51 microscope (Olympus); images were acquired by the use of a DP Controller 2002 (Olympus) and processed with Adobe Photoshop Version 7.0 (Adobe Systems). Other photographs (middle and right) were taken with a HV-D20 Hitachi camera (Hitachi Kokusia Electric Inc) by use of a Leica DM6000B microscope (Leica Microsystems GmbH); images were acquired with Leica Laser Microdissection LMD software (Version 6.6.0; Leica Microsystems). Fluorescence images were obtained by use of a triple band pass filter (B/G/R fluorescence filter; Leica Microsystems). Original magnifications: ×100 (left); ×200 (middle and right). (B) The list of proteins identified from the microdissected amyloid fragments are shown above in panel A. The proteins are listed according to the abundance they were represented in the sample. The panel shows the protein accession code in the UniProt database,19 the molecular weight of the protein (MW), the results of the blank control sample, and 4 different microdissections (1-4) For each protein identified in each sample, statistical probability is indicated as a percentage. The numbers in parentheses indicate the number of unique peptides identified belonging to a given protein. Of the 4 common types of systemic amyloidosis specifically studied (SAA, TTR, IGK, IGL), the samples contained peptides only belonging to IGK constant region (Accession: KAC_HUMAN, highlighted in yellow with red text). In addition, the samples contained several proteins known to be associated with amyloid deposits, such as apolipoprotein E (Accession: APOE_HUMAN), apolipoprotein A4 (Accession: APOA4_HUMAN), SAP (Accession: SAMP_HUMAN), and apolipoprotein A1 (Accession: APOA1_HUMAN). These are indicated in blue text. Other proteins seen include normal components of bone marrow stroma such as vitronectin (Accession: VTNC_HUMAN) and collagen (Accession: CO6A3_HUMAN). Given that the only amyloidogenic protein belonging to the 4 common types of amyloidosis was IGK, this case was typed AL-kappa-type by MS. This result was consistent with the previous gold standard diagnosis. (C) Detailed results of IGK constant region (Accession: IGKC_HUMAN) identified in all 4 cases samples. Probability of protein identification, the number of unique peptides, unique spectra, and total spectra and percent coverage of the protein are shown. A more detailed analysis of the proteomic data is provided in supplemental Figure 1.
LMD/MS analysis of a case of AL-kappa amyloidosis (case 1). (A) Bone marrow specimen with interstitial nodules of amyloid deposition (Case 1). (Left) CR-stained section viewed under bright field confirming congophilia of the amyloid deposits. (Middle) CR-stained section viewed under fluorescent light source. The congophilic deposits give bright red fluorescence, confirming amyloid deposition. The area selected for microdissection is circled with a red line. (Right) Same area selected in the middle panel after microdissection of the amyloid plaque. Photograph (left) was taken with a DP70 Olympus camera (Olympus) with an Olympus BX51 microscope (Olympus); images were acquired by the use of a DP Controller 2002 (Olympus) and processed with Adobe Photoshop Version 7.0 (Adobe Systems). Other photographs (middle and right) were taken with a HV-D20 Hitachi camera (Hitachi Kokusia Electric Inc) by use of a Leica DM6000B microscope (Leica Microsystems GmbH); images were acquired with Leica Laser Microdissection LMD software (Version 6.6.0; Leica Microsystems). Fluorescence images were obtained by use of a triple band pass filter (B/G/R fluorescence filter; Leica Microsystems). Original magnifications: ×100 (left); ×200 (middle and right). (B) The list of proteins identified from the microdissected amyloid fragments are shown above in panel A. The proteins are listed according to the abundance they were represented in the sample. The panel shows the protein accession code in the UniProt database,19 the molecular weight of the protein (MW), the results of the blank control sample, and 4 different microdissections (1-4) For each protein identified in each sample, statistical probability is indicated as a percentage. The numbers in parentheses indicate the number of unique peptides identified belonging to a given protein. Of the 4 common types of systemic amyloidosis specifically studied (SAA, TTR, IGK, IGL), the samples contained peptides only belonging to IGK constant region (Accession: KAC_HUMAN, highlighted in yellow with red text). In addition, the samples contained several proteins known to be associated with amyloid deposits, such as apolipoprotein E (Accession: APOE_HUMAN), apolipoprotein A4 (Accession: APOA4_HUMAN), SAP (Accession: SAMP_HUMAN), and apolipoprotein A1 (Accession: APOA1_HUMAN). These are indicated in blue text. Other proteins seen include normal components of bone marrow stroma such as vitronectin (Accession: VTNC_HUMAN) and collagen (Accession: CO6A3_HUMAN). Given that the only amyloidogenic protein belonging to the 4 common types of amyloidosis was IGK, this case was typed AL-kappa-type by MS. This result was consistent with the previous gold standard diagnosis. (C) Detailed results of IGK constant region (Accession: IGKC_HUMAN) identified in all 4 cases samples. Probability of protein identification, the number of unique peptides, unique spectra, and total spectra and percent coverage of the protein are shown. A more detailed analysis of the proteomic data is provided in supplemental Figure 1.
By assigning a specimen to an amyloid type on the basis of the largest number of amyloid-associated peptide hits, the MS test results identified the true disease state correctly with 100% specificity and sensitivity (supplemental Table 2). The results of MS-based proteomic analysis in comparison with the previous gold standard diagnosis are summarized in Table 1.
Analysis of a total of 46 microdissections from 12 CR-negative tissues identified a single IGK peptide. No peptides belonging to IGL, TTR, or SAA were found. These results suggest that amyloidogenic proteins were either present at very low levels or were absent in CR-negative solid tissues.
In every case of AL-type amyloidosis, peptides belonging to the constant region of IGL or IGK were present. Peptides of the variable region of IGK and IGL were detected in small numbers and only in a subset of the cases. This result is likely to be a consequence of the peptide identification strategy, in which publically available protein sequences are searched. In many cases, the somatic mutations affecting variable region segment of the immunoglobulin genes are unique and are not represented in public databases.20
Amyloid peptide profiles were consistent between hereditary versus senile ATTR, between different fixatives, in decalcified specimens, or between different anatomical sites (Table 1). However, several proteins specific to the microenvironment of different anatomical sites (such as myosin in heart or collagen in bone marrow) were present (Figure 1; supplemental Figures 1-2).
In the validation set, LMD/MS was successful in identifying the amyloid type in 40 (98%) of 41 of the cases, whereas immunohistochemistry was informative in only 15 (42%) of 36 of the cases (supplemental Table 3). When both methods were informative, there was 100% concordance with LMD/MS and immunohistochemistry.
In conclusion, this study demonstrates the utility of LMD and MS-based proteomic analysis for typing amyloidosis with high sensitivity and specificity in paraffin-embedded clinical biopsy specimens. The assay overcomes many of the problems presented by other methods currently used for typing of amyloidosis by combining the precision of LMD with the analytical power of MS. Although the study specifically focused on the most common types of systemic amyloidosis, the assay is not dependent on a predetermined knowledge of the patient's amyloid subtype. As a list of protein constituents of the amyloid is generated, theoretically, all known amyloid types can be identified. Since the establishment of the assay as a clinical test in our laboratories, we have witnessed 13 of the known amyloid types (data not shown) and identified a novel cause of amyloidosis.21 This finding suggests a single test that uses MS-based proteomics could replace 25 different antibody-based immunohistochemical tests that would be required to type all known amyloid types and, in the long run, would be much more cost effective. The approach reported here promises to enhance our ability to type amyloid deposits accurately in clinical biopsy specimens and to improve clinical care of amyloidosis patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank members of the Mayo Clinic Amyloid Interest Group, Dr M. A. Gertz, Dr A. Dispenzieri, and Dr S. R. Zeldenrust for their encouragement, advice, and support; Roman M. Zenka for bioinformatics support; and Douglas W. Mahoney for biostatistics support.
Authorship
Contribution: H.R.B. and A.D. designed research; J.A.V., J.D.G., B.J.M., J.D.T., H.R.B., and A.D. performed research; J.A.V., J.D.G., J.D.T., H.R.B., and A.D. analyzed data; and J.A.V. and A.D. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ahmet Dogan, MD, PhD, Professor of Laboratory Medicine and Pathology, Mayo Clinic, 200 First St SW, Rochester, MN 55905; e-mail: dogan.ahmet@mayo.edu.