Abstract
Despite widespread use of the anti-CD20 monoclonal antibody (mAb), rituximab, in treating B-cell lymphomas, its efficacy remains variable and often modest. A better understanding of rituximab-mediated killing mechanisms is essential to develop more effective therapeutic agents. In this study, we modulated the binding property of rituximab by introducing several point mutations in its complementarity-determining regions. The data showed that changing the binding avidity of rituximab in the range from 10−8 to 10−10 M could regulate its antibody-dependent cellular cytotoxicity but not affect its complement-dependent cytotoxicity and apoptosis-inducing activity in B-lymphoma cells. Contradictory to previous findings, we found that the complement-dependent cytotoxicity potency of CD20 mAb was independent of the off-rate. Despite still being a type I CD20 mAb, a rituximab triple mutant (H57DE/H102YK/L93NR), which had a similar binding avidity to a double mutant (H57DE/H102YK), was unexpectedly found to have extremely potent apoptosis-inducing activity. Moreover, this triple mutant, which was demonstrated to efficiently initiate both caspase-dependent and -independent apoptosis, exhibited potent in vivo therapeutic efficacy, even in the rituximab-resistant lymphoma model, suggesting that it might be a promising therapeutic agent for B-cell lymphomas.
Introduction
The CD20 molecule is a 30- to 35-kDa integral membrane protein expressed by B lymphocytes in early stages of differentiation and by most B-cell lymphomas.1,2 CD20 is an ideal target for monoclonal antibodies (mAbs), as it is expressed at high levels on most B-cell malignancies but does not become internalized or shed from the plasma membrane after mAb treatment.3,4 The mouse/human chimeric anti-CD20 antibody, rituximab, is the first therapeutic mAb approved for the treatment of relapsed/refractory low-grade or follicular B-cell non-Hodgkin lymphomas.5,6 Previous studies have suggested that several mechanisms might be involved in providing therapeutic efficacy, including complement-dependent cytotoxicity (CDC), antibody-dependent cellular cytotoxicity (ADCC), and the induction of apoptosis.4,7 The relative contributions of these different mechanisms of action are still a matter of debate.4,7 Anti-CD20 mAbs are usually defined as either type I or II, based on their ability to redistribute CD20 into lipid rafts.8,9 Type I mAbs (rituximab and most anti-CD20 mAbs) are able to efficiently shift CD20 complexes into rafts, but the type II mAbs (B1 and 11B8) are not. The in vitro assays further indicate that type I mAbs usually exhibit potent CDC activity and relatively low level of apoptosis unless extensively cross-linked by antibody,8 whereas type II mAbs are relatively inactive in complement activation but tend to promote more apoptosis.9,10 Both types of mAb are equally potent in ADCC with FcR-bearing myeloid effectors.
Although rituximab has been widely used in the treatment of lymphoma, only 48% of patients respond to the treatment, and complete responses are less than 10%.6,11 A better understanding of anti-CD20 mAb-mediated killing mechanisms should make it possible to develop new and more effectively therapeutic agents. Many researchers have made substantial efforts to address this issue.12-15 Previous studies by Teeling et al16 indicated that the anti-CD20 mAb 2F2 with an unusually slow off-rate showed significantly more potent ability to activate complement compared with rituximab. They concluded that slow off-rates could influence the activity of CD20 mAbs to induce complement activation and complement-mediated lysis. However, it might not be suitable to illustrate the relationship between binding off-rates and CDC potency by directly comparing rituximab and 2F2, which have been demonstrated to recognize different epitopes on the CD20 molecule.17 Moreover, the epitope mapping data have suggested that most anti-CD20 mAbs recognize the epitopes on the larger extracellular loop (only 44 amino acids) of the CD20 molecule.18-20 Despite apparently similar specificity, these anti-CD20 mAbs show different effector functions and different efficacies.17,20-22 Therefore, through comparison of these currently available anti-CD20 mAbs recognizing different epitopes, it is difficult to elucidate what factors affect the potency of the anti-CD20 mAb-mediated killing mechanisms (eg, CDC, ADCC, and the induction of apoptosis).
Tools to rationally alter and manipulate antibody binding characteristics offer great promise for understanding and delineating the mechanisms of antibody action when binding to the target molecule. In this study, we have redesigned rituximab with different binding avidities to CD20 antigen by a computational design method we have recently developed (B.L. and L.Z., manuscript in preparation). Our results indicate that manipulation of the binding avidity of rituximab can only handle its ADCC activity but is unable to influence the CDC and apoptosis-inducing activity. Unexpectedly, an unusual apoptosis-inducing activity was observed for a triple variant. We further evaluated the in vitro and in vivo antitumor activity of the triple variant in both B-lymphoma cells and rituximab-resistant (RR) B-lymphoma cells.
Methods
Cell lines, antibodies, and animals
Two human Burkitt lymphoma cell lines, Raji and Daudi, were obtained from the ATCC. The anti-HER2 humanized antibody trastuzumab (anti-HER2) was purchased from Roche Ltd. F(ab′)2 fragments of IgG were produced by bromelain digestions and were further purified by passing material through an antihuman Fc column. Rituximab and its mutants were labeled with fluorescein isothiocyanate (FITC) to produce FITC-conjugated antibodies, respectively. Rituximab-resistant cell lines (Raji-R and Daudi-R) were generated from Raji and Daudi cells as described previously.23,24 Eight-week-old female BALB/c SCID mice were housed in pathogen-free conditions and were treated in accordance with guidelines of the Committee on Animals of the Second Military Medical University. The study using human peripheral blood mononuclear cells (PBMCs) from the donors was approved by the Institutional Review Board of the Second Military Medical University.
Computational redesign of the binding avidity of rituximab
The crystal structure of the rituximab Fab-CD20 peptide complex (PDB code 2osl) was determined in our laboratory previously.25 Hydrogen-atom positions were assigned using the Biopolymer module of Insight II (Accelrys). The computational mutation was carried out on rituximab. Docking was performed using Monte Carlo Simulated Annealing26,27 for random generation of a maximum of 60 structures through the Affinity module of Insight II (CVFF force field28 ). The lowest energy complexes presenting lower root mean square deviation were selected for the binding-free energy calculations. Briefly, the protein-protein complexes generated were minimized using the CHARMM force field (CHARMM version 34b1 program29 ) and the Generalized Born with a simple Switching implicit solvent model.30 Finally, the binding-free energy was calculated using the Molecular Mechanics Poisson-Boltzmann surface area (MM/PBSA) method.31 The simulation procedure was described in detail in another manuscript (B.L. and L.Z., manuscript in preparation).
Construction, expression, and purification of rituximab mutants
The heavy chain variable region gene and the light chain gene of rituximab32 were synthesized by the Sangon Biological Engineering Technology Company. The point mutations were introduced into rituximab using overlap extension polymerase chain reaction. Rituximab and the rituximab mutants were expressed using the identical procedures described previously.33 Finally, mAbs were purified by affinity chromatography on protein A-Sepharose (GE Healthcare) from the serum-free culture supernatants.
Binding activity assays
All the avidity constants of rituximab mutants were accomplished by radioimmunoassay.8 Briefly, purified rituximab mutants fragments, F(ab′)2, were radiolabeled with 125I by the iodobead method. The 125I-labeled F(ab′)2 fragments of rituximab mutants were incubated with Daudi cells for 2 hours at 37°C. The cell-bound and free 125I-labeled mAbs were then separated by centrifugation through phthalate oils, and the cell pellets together with bound antibody counted for radioactivity. The dissociation constants were determined by nonlinear least-squares regression analyses.34
Off-rate measurements
To determine the off-rate of CD20 mAbs from Raji cells, cells were pelleted and resuspended in medium containing 10 μg/mL FITC-labeled CD20 mAb IgG. Cells were incubated for 1 hour at room temperature, pelleted, and resuspended in medium containing 1 mg/mL of the unlabeled IgG. After different time intervals, the samples were taken, washed, and analyzed by flow cytometry (FCM) using a FACScan flow cytometer (BD Biosciences) to determine the percentage of the remaining cells that were still stained.
Cytotoxicity assays
CDC and ADCC assays were performed as described previously.33 Briefly, the cells were incubated with antibodies for 1 hour in phenol red-free Dulbecco modified Eagle medium culture medium in a 5% CO2 incubator at 37°C, followed by the addition of either normal human serum (NHS, 10% vol/vol) as a source of complement (for CDC assay) or human PBMCs as effector cells (for ADCC assay). After an additional incubation for 4 hours at 37°C, the cell lysis was determined by measuring the amount of lactate dehydrogenase (LDH) released into the culture supernatant. Maximum LDH release was determined by lysis in 0.2% Triton X-100.
Binding of complement subcomponent C1q to CD20 mAbs
To assess the extent of cellular C1q deposition, various concentrations of mAbs were added to cells (105/well in a 96-well plate) and allowed to bind for 10 to 15 minutes at room temperature. Human serum was then added to a final concentration of 1% (vol/vol) followed by incubation at 37°C for 10 minutes. After washing, FITC-labeled sheep anti–human Clq mAb (Serotec) was added and samples were incubated for 30 minutes at 4°C and then analyzed by FCM.
Assessment of raft-associated antigen by Triton X-100 insolubility
As a rapid assessment of the presence of CD20 in raft microdomains, we used a flow cytometry method based on their Triton X-100 insolubility at low temperatures as described previously.16 In brief, cells were incubated with FITC-conjugated mAbs (10 μg/mL) for 15 minutes at 37°C. After washing, one-half of the sample was maintained on ice to allow calculation of 100% surface antigen levels; the other half was treated with 0.5% Triton X-100 for 15 minutes on ice to determine the proportion of antigens remaining in the insoluble raft fraction. Cells were maintained at 4°C throughout the assay, washed once in phosphate-buffered saline (PBS)/bovine serum albumin/azide, and assessed by FCM.
Apoptosis assay
The cells were incubated with different concentrations of CD20 mAbs at 37°C for 16 hours. After washing, cells were treated with annexin V–FITC (BD Biosciences), washed again, and analyzed by FCM. F(ab′)2 fragment of goat anti–human IgM (anti-IgM; Jackson ImmunoResearch Laboratories) was used as a positive control for the induction of apoptosis.
Cytosolic calcium flux
Cells were incubated in RPMI containing 4 μg/mL fluo-3AM (Invitrogen) for 30 minutes at room temperature followed by washing and resuspension in RPMI containing 10% fetal calf serum. Cells were excited at 480 nm and emission was measured at 530 nm. Samples were incubated at 37°C for 1 to 2 minutes and assessed by FCM to establish a baseline fluorescence for unstimulated cells. Cells were then stimulated with the desired treatment. The data were acquired using a FACScan flow cytometer and analyzed using CellQuest software (BD Biosciences).
Immunotherapy
Groups of 10 8-week-old female SCID mice were injected via the tail vein with 3.5 × 106 Daudi or Daudi-R cells on day 0, followed 7 days later by the intravenous injection of CD20 mAb IgG (100 μg/mouse). For F(ab′)2 fragment treatment groups, groups of 10 SCID mice were injected with 3.5 × 106 Daudi-R cells intravenously on day 0 and then treated with 100 μg F(ab′)2 fragments intravenously on day 7. Additional 100-μg injections of F(ab′)2 were given intraperitoneally on day 7, intravenously on day 8, and intraperitoneally on day 9, to a total of 400 μg. The mice were observed daily and killed at the onset of hind-leg paralysis.
Statistical analysis
Statistical analysis was performed by Student unpaired t test to identify significant differences unless otherwise indicated. Differences were considered significant at a P value of less than .05.
Results
Design and characterization of rituximab mutants
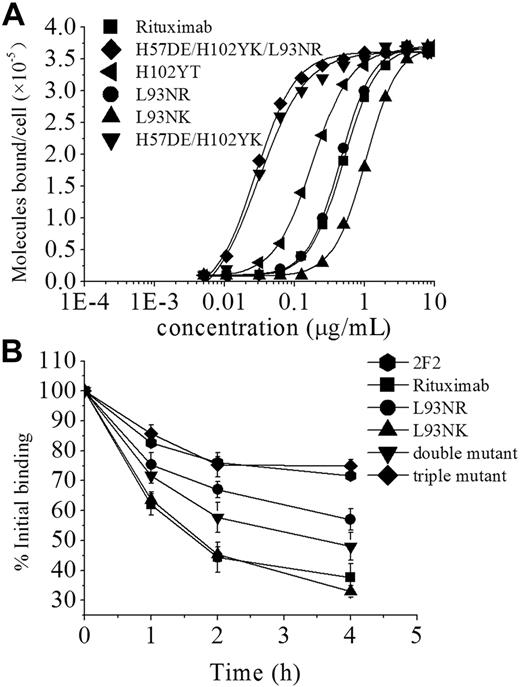
Based on the crystal structure of rituximab Fab-CD20 epitope peptide complex we determined previously,25 a computational method was used to rationally manipulate the binding avidity of rituximab to CD20 antigen. Depending on the calculated binding energy, rituximab mutants with different binding avidities were designed and constructed by introducing several point mutations in the CDRs. The avidity of rituximab mutants was evaluated by analyzing direct cell surface saturation binding to Daudi cells (Figure 1A). As shown in Table 1, the dissociation constant of wild-type rituximab (4.96 ± 0.21nM) is quantitatively consistent with the previous report by Reff et al.32 The mutations (H102YK and L93NR) are additive with the best single mutation H57DE, generating a triple mutant with an avidity of 0.27 plus or minus 0.02nM, which is an approximately 18.4-fold improvement over wild-type (Table 1).
Characterization of rituximab mutants. (A) Binding of 125I-labeled F(ab′)2 fragments of rituximab mutants to Daudi cells. 125I-labeled F(ab′)2 fragments of mAbs were incubated with Daudi cells for 2 hours at 37°C. The cell-bound and free 125I-labeled mAbs were then separated by centrifugation through phthalate oils and the cell pellets together with bound antibody counted for radioactivity. Data from saturation-binding experiments were analyzed by nonlinear least-squares regression for curve-fitting and dissociation constant estimation. (B) Dissociation of FITC–anti-CD20 mAbs from Raji cells. Cells were incubated with FITC-labeled anti-CD20 mAbs (10 μg/mL) at 37°C for 1 hour, washed twice, and resuspended. Samples of cells were taken at 0, 1, 2, and 4 hours and then washed and analyzed by FCM. Data are mean ± SD of at least 3 experiments.
Characterization of rituximab mutants. (A) Binding of 125I-labeled F(ab′)2 fragments of rituximab mutants to Daudi cells. 125I-labeled F(ab′)2 fragments of mAbs were incubated with Daudi cells for 2 hours at 37°C. The cell-bound and free 125I-labeled mAbs were then separated by centrifugation through phthalate oils and the cell pellets together with bound antibody counted for radioactivity. Data from saturation-binding experiments were analyzed by nonlinear least-squares regression for curve-fitting and dissociation constant estimation. (B) Dissociation of FITC–anti-CD20 mAbs from Raji cells. Cells were incubated with FITC-labeled anti-CD20 mAbs (10 μg/mL) at 37°C for 1 hour, washed twice, and resuspended. Samples of cells were taken at 0, 1, 2, and 4 hours and then washed and analyzed by FCM. Data are mean ± SD of at least 3 experiments.
Experimental binding avidities of rituximab mutants to CD20 molecule
| Rituximab mutant . | KD, nM . |
|---|---|
| WT | 4.96 ± 0.21 |
| H57DE | 1.13 ± 0.07 |
| H102YT | 1.81 ± 0.09 |
| H57DE/H102YK/L93NR | 0.27 ± 0.02 |
| L93NR | 4.48 ± 0.18 |
| L93NK | 10.36 ± 0.45 |
| H57DE/H102YK | 0.31 ± 0.03 |
| Rituximab mutant . | KD, nM . |
|---|---|
| WT | 4.96 ± 0.21 |
| H57DE | 1.13 ± 0.07 |
| H102YT | 1.81 ± 0.09 |
| H57DE/H102YK/L93NR | 0.27 ± 0.02 |
| L93NR | 4.48 ± 0.18 |
| L93NK | 10.36 ± 0.45 |
| H57DE/H102YK | 0.31 ± 0.03 |
Experimental error is the SD from 3 independent experiments.
WT indicates wild-type.
The binding “off-rate” experiments using FITC-labeled IgG were performed to compare the dissociation of rituximab, rituximab mutants, and 2F2 from Raji cells. As shown in Figure 1B, a single mutation (L93NR) in the light chain CDR3 markedly reduced the off-rate of rituximab but could not significantly alter its functional affinity. The data showed that, although the double mutant (H57DE/H102YK) had a similar avidity to the triple mutant, it exhibited a relatively faster off-rate. It can be clearly seen in Figure 1B that the triple mutant (H102YK/L93NR/H57DE) has a significantly reduced off-rate, which is comparable with that of 2F2. Approximately 44% of the rituximab, and more than 80% of triple mutant or 2F2, remained bound to the cells after 2 hours. In addition, the similar results were achieved with F(ab′)2, which excluded an interaction with FcγR on target cells influencing mAb dissociation (data not shown).
The affinity (avidity) of a protein complex is a function of the rates of association (kon) and dissociation (koff). Previous studies have shown that the rate of association (kon) between a pair of proteins can be specifically enhanced without affecting the rate of dissociation (koff), which have demonstrated that the kon and the koff are 2 independent factors.35,36 Thus, it is possible that the antibody variants with different off-rates can be generated without changing their binding avidity.
Antibody off-rate and CDC
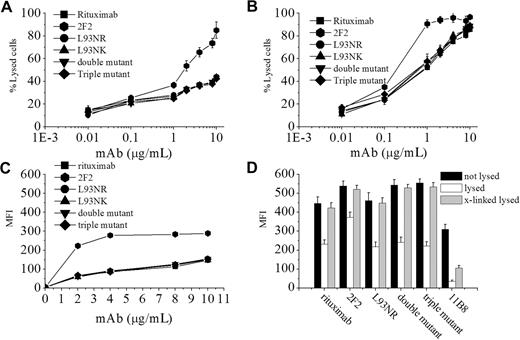
In initial functional experiments, the cytotoxic activities of these rituximab mutants were assessed against 2 CD20+ human lymphoma cell lines, Daudi and Raji. The results were obtained with the single mutant (L93NK), the single mutant (L93NR), the double mutant (H57DE/H102YK), and the triple mutant (H102YK/L93NR/H57DE), which represent IgG molecules with different binding avidities. As illustrated in Figure 2A-B, rituximab variants with various avidities in the range 10−8 to 10−10 M exhibited approximately the same level of CDC activity as wild-type rituximab. Surprisingly, we found that these variants (L93NR, double mutant, and triple mutant) with a slower off-rate could not display a significant enhancement in CDC activity in both Raji and Daudi cells. These results clearly show that the CDC potency of rituximab cannot be affected by either binding avidity or off-rate. We subsequently investigated the abilities of these variants to fix C1q, the first component of the complement cascade. The results shown in Figure 2C indicated that all of the variants with different binding avidities bound equal amount of C1q, and the off-rate seemed to also have no effect on the amount of C1q binding. In addition, further studies indicated that all rituximab mutants exhibited similar capability as rituximab to translocate CD20 into lipid rafts (Figure 2D), suggesting that these rituximab variants still belonged to type I CD20 mAb.
CDC induced by CD20 mAbs on B-cell lines. Raji (A) and Daudi (B) cells were incubated with increasing concentrations CD20 mAbs in the presence of human complement at 37°C for 4 hours. CDC activity was determined by a standard LDH assay as described in “Cytotoxicity assays.” Data are mean ± SD (n = 5). (C) To detect C1q binding to CD20 mAbs coated on the cell surface, Raji cells were incubated with CD20 mAbs at 2 μg/mL rituximab variants at 37°C for 15 minutes. After washing, the cells were incubated with NHS for 10 minutes at 37°C. Deposition of complement components was assessed by FCM. Points represent mean (n = 3); bars represent SD. (D) Translocation of CD20 into Triton X-100-insoluble membrane fraction. Daudi cells were incubated with FITC-labeled anit-CD20 mAbs (10 μg/mL) for 15 minutes at 37°C. After washing, the cells were resuspended and treated with goat anti–human κ F(ab′)2 fragment (x-link; Southern Biotechnology Associates Inc) or not for 30 minutes on ice. After chilling on ice, half of each sample was treated with 0.5% Triton X-100 for 15 minutes on ice. All of samples were washed and analyzed by FCM to assess bound FITC-CD20 mAbs. The graphs are representative of at least 3 experiments, each of which showed similar results.
CDC induced by CD20 mAbs on B-cell lines. Raji (A) and Daudi (B) cells were incubated with increasing concentrations CD20 mAbs in the presence of human complement at 37°C for 4 hours. CDC activity was determined by a standard LDH assay as described in “Cytotoxicity assays.” Data are mean ± SD (n = 5). (C) To detect C1q binding to CD20 mAbs coated on the cell surface, Raji cells were incubated with CD20 mAbs at 2 μg/mL rituximab variants at 37°C for 15 minutes. After washing, the cells were incubated with NHS for 10 minutes at 37°C. Deposition of complement components was assessed by FCM. Points represent mean (n = 3); bars represent SD. (D) Translocation of CD20 into Triton X-100-insoluble membrane fraction. Daudi cells were incubated with FITC-labeled anit-CD20 mAbs (10 μg/mL) for 15 minutes at 37°C. After washing, the cells were resuspended and treated with goat anti–human κ F(ab′)2 fragment (x-link; Southern Biotechnology Associates Inc) or not for 30 minutes on ice. After chilling on ice, half of each sample was treated with 0.5% Triton X-100 for 15 minutes on ice. All of samples were washed and analyzed by FCM to assess bound FITC-CD20 mAbs. The graphs are representative of at least 3 experiments, each of which showed similar results.
Binding avidity regulates the capacity to promote ADCC
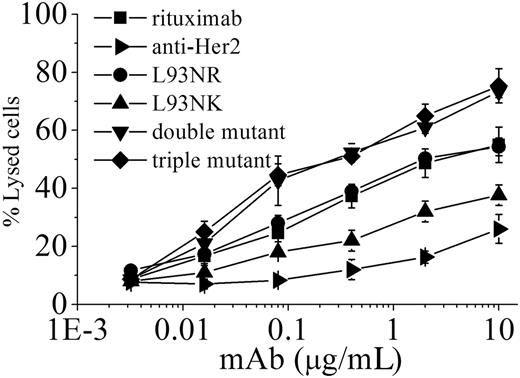
To determine whether antibody-binding avidity affected the capacity of PBMCs to mediate ADCC, a standard LDH assay was performed. Purified human PBMCs from healthy donors were used as effector cells and Daudi cells were used as target. Assays were conducted at effector/target (E/T) ratios of 50:1, 25:1, 5:1, and 1:1 using antibody concentrations ranging from 0.003 to 10 μg/mL (Figure 3; supplemental Figure 1A-C, available on the Blood website; see the Supplemental Materials link at the top of the online article). In these assays, which used inactivated human PBMCs, a higher E/T ratio increased the amount of cytotoxicity, but similar avidity-dependent patterns were observed at all E/T ratios (Figure 3; supplemental Figure 1A-C). To facilitate comparative analysis of the data, we evaluated the concentrations of antibody needed for 50% cytotoxicity at E/T ratios of 25:1. Our data showed a clear avidity-dependent susceptibility to ADCC, as more than 10 μg/mL L93NK IgG was required to achieve the targeted lysis level, compared with 2 μg/mL rituximab and only 0.08 μg/mL triple variant (Figure 3). Similar results were also obtained with Raji cells (data not shown).
ADCC activity of avidity variants. ADCC activity against Daudi cells using human PBMCs as effector cells at E/T ratio of 25:1. The ADCC activity of rituximab variants at various concentrations was measured using a standard LDH assay as described in “Cytotoxicity assays.” Data are mean ± SD (n = 3).
ADCC activity of avidity variants. ADCC activity against Daudi cells using human PBMCs as effector cells at E/T ratio of 25:1. The ADCC activity of rituximab variants at various concentrations was measured using a standard LDH assay as described in “Cytotoxicity assays.” Data are mean ± SD (n = 3).
The triple variant efficiently induces both caspase-dependent and -independent apoptosis
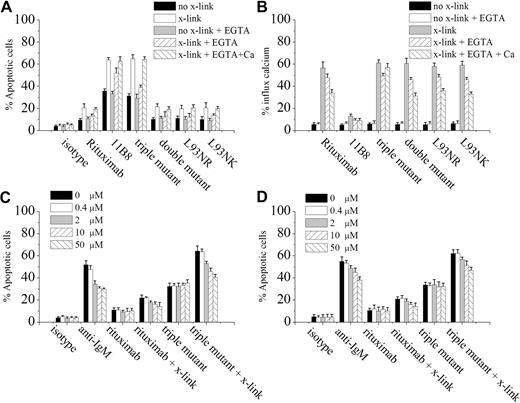
Induction of apoptosis was evaluated by FITC–annexin V assays in Raji cells. As indicated in Figure 4A, without cross-linking, rituximab and all of the variants (except for triple variant) triggered a similar low level of apoptosis (< 15%) in Raji cells at the concentration of 10 μg/mL. Surprisingly, despite showing similar binding avidity, the triple variant induced a substantially higher level of apoptosis than that induced by double variant, even comparable with that evoked by 11B8 (a type II CD20 mAb). When adding the F(ab′)2 anti-κ cross-linker, all of the anti-CD20 mAbs exhibited a markedly enhanced apoptosis-inducing activity in Raji cells, but the increased extent of apoptosis induced by either 11B8 or the triple variant was much higher than that of other rituximab mutants (Figure 4A). Based on our aforementioned results indicating that the triple variant is type I CD20 mAb, the data shown here suggest that type I mAb has the potential to induce a similar high level of apoptosis as type II mAb.
Apoptosis induced by anti-CD20 mAbs. (A) Raji cells were either preincubated with ethyleneglycoltetraacetic acid (1.5 mM, Sigma-Aldrich) or not for 15 minutes at room temperature before addition of different rituximab variants (10 μg/mL). The cells were immediately split into 3 aliquots. Goat anti–human κ F(ab′)2 fragment (20 μg/mL) was added to one aliquot, goat anti–human κ F(ab′)2 fragment (20 μg/mL) plus excessive CaCl2 to the second, and medium to the third. After 16 hours of incubation, apoptotic cells were scored by single staining of annexin V–Fluos on a flow cytometer. (B) Intracellular calcium mobilization initiated by anti-CD20 mAb with or without hyper-cross-linking. Fluo-3–loaded Raji cells were either preincubated 1mM ethyleneglycoltetraacetic acid or not for 15 minutes at room temperature before addition of distinct rituximab variants (10 μg/mL). The cells were immediately split into 3 aliquots. Goat anti–human κ F(ab′)2 fragment (20 μg/mL) was added to 1 aliquot and medium to the second. The third aliquot was incubated with goat anti–human κ F(ab′)2 fragment for 5 minutes, followed by the addition of excessive calcium. Samples were analyzed using a FACScan flow cytometer with excitation at 480 nm. Data are mean ± SD (n = 3). (C-D) Effect of ZVAD (C) and VDVAD (D) on the apoptosis induced in 16 hours by inhibitor alone, 10 μg/mL rituximab, and triple variant in the presence or absence of cross-linker (goat anti–human κ F(ab′)2 fragment, 20 μg/mL) in Daudi cells. Inhibitors (ZVAD and VDVAD) were added over a range of different concentrations for 2 hours before the addition of mAbs. Data are mean ± SD of at least 3 experiments.
Apoptosis induced by anti-CD20 mAbs. (A) Raji cells were either preincubated with ethyleneglycoltetraacetic acid (1.5 mM, Sigma-Aldrich) or not for 15 minutes at room temperature before addition of different rituximab variants (10 μg/mL). The cells were immediately split into 3 aliquots. Goat anti–human κ F(ab′)2 fragment (20 μg/mL) was added to one aliquot, goat anti–human κ F(ab′)2 fragment (20 μg/mL) plus excessive CaCl2 to the second, and medium to the third. After 16 hours of incubation, apoptotic cells were scored by single staining of annexin V–Fluos on a flow cytometer. (B) Intracellular calcium mobilization initiated by anti-CD20 mAb with or without hyper-cross-linking. Fluo-3–loaded Raji cells were either preincubated 1mM ethyleneglycoltetraacetic acid or not for 15 minutes at room temperature before addition of distinct rituximab variants (10 μg/mL). The cells were immediately split into 3 aliquots. Goat anti–human κ F(ab′)2 fragment (20 μg/mL) was added to 1 aliquot and medium to the second. The third aliquot was incubated with goat anti–human κ F(ab′)2 fragment for 5 minutes, followed by the addition of excessive calcium. Samples were analyzed using a FACScan flow cytometer with excitation at 480 nm. Data are mean ± SD (n = 3). (C-D) Effect of ZVAD (C) and VDVAD (D) on the apoptosis induced in 16 hours by inhibitor alone, 10 μg/mL rituximab, and triple variant in the presence or absence of cross-linker (goat anti–human κ F(ab′)2 fragment, 20 μg/mL) in Daudi cells. Inhibitors (ZVAD and VDVAD) were added over a range of different concentrations for 2 hours before the addition of mAbs. Data are mean ± SD of at least 3 experiments.
Further studies indicated that rituximab and all of the mutants, including the triple variant, induced apoptosis in a calcium-independent manner, but the increased apoptosis induction caused by hyper-cross-linking was dependent on calcium as chelating calcium by ethyleneglycoltetraacetic acid significantly inhibited this process (Figure 4A). These data suggest that there may be 2 different apoptotic pathways based on the dependence of extracellular calcium. Next, we investigated the role of calcium influx in the apoptotic process induced by the triple variant. In agreement with recent data,37 we found that extracellular calcium chelation had no effect on the magnitude of the calcium flux induced by cross-linked CD20 mAbs, indicating that cross-linking of CD20 mAbs could evoke a calcium flux predominantly through intracellular calcium release (Figure 4B). When extracellular calcium was reintroduced by the addition of CaCl2, a second calcium flux was observed with all of the rituximab variants and the triple variant was shown to evoke the strongest calcium flux (Figure 4B). Furthermore, the apoptosis inhibited by chelating calcium was recovered by reintroducing excessive calcium and the most potent apoptosis-inducing activity was also observed with the cross-linked triple variant (Figure 4A). These data suggest that the increase of apoptotic activity of the triple variant after adding cross-linker may be associated with the extracellular calcium influx.
We next asked whether the apoptosis triggered by the triple variant was in a caspase-dependent manner. The cell-permeable caspase inhibitor ZVAD-FMK was used, and our experimental results revealed that ZVAD-FMK over a range of concentrations from 0.5 to 50 μM was unable to prevent the triple variant-induced apoptosis in the absence of cross-linker but was significantly effective in reducing apoptosis when adding cross-linker (Figure 4C). As ZVAD-FMK was a poor inhibitor of caspase-2, we also assessed the efficacy of a specific caspase-2 inhibitor (VDVAD) in Raji cells, and similar results were observed (Figure 4D). The 2 inhibitors in the same range of concentrations were also assessed in Daudi cells, and the results were similar to those obtained for Raji cells (data not shown). These results clearly demonstrated that the triple variant could efficiently activate both caspase-dependent and -independent apoptotic pathways.
The triple variant effectively induces ADCC and apoptosis in RR lymphoma cells
Because of the unusual characteristics of the triple variant in terms of potent ADCC and apoptotic activity in vitro, we further investigated its ability to kill RR lymphoma cells. Two RR cell lines (Raji-R and Daudi-R) were established and used in this study. In agreement with previous reports,23,24 the 2 RR cell lines had the characteristics of diminished surface CD20 expression and failure to respond to rituximab-mediated CDC (Figure 5A-B). As measured by mean fluorescence intensity, wild-type cells show significant CD20 surface expression, whereas the RR clones exhibit approximately 55% reduction in surface CD20 (Figure 5A). Similar to rituximab, the CDC activity of the triple variant was not detectable in the 2 RR cell lines (Figure 5B). However, the triple variant exhibited markedly enhanced ADCC activity relative to rituximab in the 2 RR cell lines in vitro (Raji-R, 32.67% ± 2.52% vs 16.1% ± 2.21%; Daudi-R, 46.67% ± 3.05% vs 23.34% ± 2.59%; Figure 5C). Moreover, the triple variant could trigger significant levels of apoptosis in Raji-R (29.33% ± 1.53%) and Daudi-R (34.27% ± 3.12%) cells, whereas rituximab was ineffective in inducing apoptosis (Figure 5D). After cross-linking, a pronounced increase in apoptosis in the 2 RR cell lines was observed with the triple variant (Figure 5D). Rituximab was only able to induce detectable apoptosis in Raji-R and Daudi-R cells even when the cross-linker was added (Figure 5D). This suggested that the triple variant could effectively mediate apoptosis in the lymphoma cells, which had developed higher threshold and did not efficiently response to rituximab. Next we evaluated the ability of the triple variant to displace rituximab from Raji cells. Our data clearly showed that, in the presence of the triple variant, approximately 80% of rituximab could be replaced in 1 hour from the target cell (supplemental Figure 2A). Our data also demonstrated that, even in the presence of rituximab (10 μg/mL), the triple variant still exhibited potent ADCC and apoptosis-inducing activity (supplemental Figure 2B-D).
The triple variant effective kills RR cells in vitro. (A) Surface expression of CD20 on RR cells. Raji or Raji-R cells (2 × 106) were incubated with different concentrations of FITC-labeled rituximab or triple variant for 1 hour on ice. Then, the cells were washed and then analyzed by FCM. The intensity of surface CD20 expression was measured by the mean fluorescence intensity. (B-C) Rituximab-resistant cells derived from Raji and Daudi cells were exposed to different rituximab variants (10 μg/mL), followed by the addition of NHS (B) or PBMCs at an E/T ratio of 40:1 (C), respectively. A standard LDH assay was performed. (D) Cross-linked triple mutant induced more apoptosis relative to rituximab in RR cells in vitro. Cells (2 × 106) were treated with isotype control antibody, rituximab, or triple variant (10 μg/mL) with or without cross-linker (20 μg/mL) for 16 hours. The apoptotic cells were determined by single staining of annexin V–Fluos on FCM. Columns represent mean (n = 3); bars represent SD.
The triple variant effective kills RR cells in vitro. (A) Surface expression of CD20 on RR cells. Raji or Raji-R cells (2 × 106) were incubated with different concentrations of FITC-labeled rituximab or triple variant for 1 hour on ice. Then, the cells were washed and then analyzed by FCM. The intensity of surface CD20 expression was measured by the mean fluorescence intensity. (B-C) Rituximab-resistant cells derived from Raji and Daudi cells were exposed to different rituximab variants (10 μg/mL), followed by the addition of NHS (B) or PBMCs at an E/T ratio of 40:1 (C), respectively. A standard LDH assay was performed. (D) Cross-linked triple mutant induced more apoptosis relative to rituximab in RR cells in vitro. Cells (2 × 106) were treated with isotype control antibody, rituximab, or triple variant (10 μg/mL) with or without cross-linker (20 μg/mL) for 16 hours. The apoptotic cells were determined by single staining of annexin V–Fluos on FCM. Columns represent mean (n = 3); bars represent SD.
Therapeutic efficacy of the triple variant in vivo
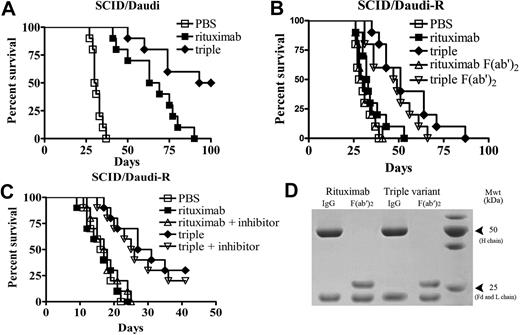
The therapeutic efficacy of the triple variant and rituximab was evaluated in Daudi and Daudi-R lymphoma-bearing SCID mice (SCID/Daudi and SCID/Daudi-R). The survival curves were plotted according to the Kaplan-Meier method and compared using the log-rank test.38 As shown in Figure 6A, when mAbs were administered to mice at a dose of 100 μg/mouse, both rituximab and the triple variant were shown to significantly improve the survival of SCID mice bearing disseminated Daudi tumor cells (P < .001 for each compared with the PBS control). However, a pronounced difference in survival was observed between rituximab and the triple variant treatment groups (P < .01), and the triple variant showed more potent antitumor activity.
The survival of tumor-bearing SCID mice treated with anti-CD20 mAbs. Groups of 10 SCID mice were injected intravenously with 3.5 × 106 Daudi (A) or Daudi-R cells (B). Five days after tumor cell inoculation, the mice were treated with rituximab or triple variant at a dose of 100 μg. For F(ab′)2 fragment treatment groups, groups of 10 SCID mice were injected with 3.5 × 106 Daudi-R cells intravenously on day 0 and then treated with 100 μg F(ab′)2 fragments intravenously on day 7. Additional 100-μg injections of F(ab′)2 were given intraperitoneally on day 7, intravenously on day 8, and intraperitoneally on day 9, to a total of 400 μg. (C) Groups of 10 SCID mice were injected intravenously with 107 Daudi-R cells. Five days after tumor cell inoculation, the mice were treated with rituximab or triple variant at the dose of 100 μg. The SCID/Daudi-R mice were treated with pan caspase inhibitor (Q-VD(OMe)-Oph) at a dose of 200 μg/day intraperitoneally 5 days a week for 3 weeks. (D) Samples of the IgG and F(ab′)2 fragments of the rituximab and the triple variant used in panel B were assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis under reducing conditions. Note the absence of contaminating heavy (H) chains in both F(ab′)2 preparations.
The survival of tumor-bearing SCID mice treated with anti-CD20 mAbs. Groups of 10 SCID mice were injected intravenously with 3.5 × 106 Daudi (A) or Daudi-R cells (B). Five days after tumor cell inoculation, the mice were treated with rituximab or triple variant at a dose of 100 μg. For F(ab′)2 fragment treatment groups, groups of 10 SCID mice were injected with 3.5 × 106 Daudi-R cells intravenously on day 0 and then treated with 100 μg F(ab′)2 fragments intravenously on day 7. Additional 100-μg injections of F(ab′)2 were given intraperitoneally on day 7, intravenously on day 8, and intraperitoneally on day 9, to a total of 400 μg. (C) Groups of 10 SCID mice were injected intravenously with 107 Daudi-R cells. Five days after tumor cell inoculation, the mice were treated with rituximab or triple variant at the dose of 100 μg. The SCID/Daudi-R mice were treated with pan caspase inhibitor (Q-VD(OMe)-Oph) at a dose of 200 μg/day intraperitoneally 5 days a week for 3 weeks. (D) Samples of the IgG and F(ab′)2 fragments of the rituximab and the triple variant used in panel B were assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis under reducing conditions. Note the absence of contaminating heavy (H) chains in both F(ab′)2 preparations.
No statistical difference in survival was observed between the PBS- and rituximab-treated SCID/Daudi-R mice (Figure 6B). Rituximab-treated SCID/Daudi-R mice had a median survival time of 32 days after tumor inoculation. Median survival in the triple variant treatment group was extended to 51 days, with statistically significant survival extension in this model by log-rank analysis (P < .005 compared with the rituximab treatment group). The survival of SCID/Daudi-R mice treated with the triple variant in the presence of rituximab was evaluated. These results indicated that the triple variant still exhibited potent antitumor activity, significantly prolonging the survival of SCID/Daudi-R mice (supplemental Figure 3). Next, we investigated the role of caspase in the in vivo antitumor activity of the triple variant. The results showed that, in the presence of pan-caspase inhibitor, the triple variant was still effective in prolonging the survival of SCID/Daudi-R mice and pan-caspase inhibitor could not significantly affect the therapeutic efficacy of the triple variant (Figure 6C). These data suggested that caspase-independent apoptosis and ADCC might play a major role in the in vivo antitumor activity of the triple variant in SCID/Daudi-R mice. To remove the potential for any conventional effector functions, we produced F(ab′)2 fragment of the triple variant. The resulting highly purified F(ab′)2 fragment was assessed on sodium dodecyl sulfate-polyacrylamide gel electrophoresis to confirm a lack of contaminating IgG (Figure 6D), and then injected into SCID/Daudi-R mice on days 7, 8, and 9 after tumor inoculation. The results showed that F(ab′)2 fragment from triple variant still possessed strong antitumor activity, effectively prolonging the survival of SCID/Daudi-R mice (P < .001 compared with PBS-treated mice; Figure 6B). These data suggested that the potent caspase-independent apoptosis induced by triple variant might be at least partially responsible for its therapeutic efficacy in the RR lymphoma mouse model.
Discussion
In the present study, rituximab mutants with different off-rates were generated and their CDC activities were compared. We clearly show that all of the rituximab variants, even those with much slower off-rate than rituximab, do not have the capability to activate more complement and induce more tumor cell lysis, suggesting that CDC potency of CD20 mAbs is independent of the off-rate. On initial inspection, our data appear to contradict those reported earlier by Teeling et al,16,17 which have indicated that CDC potency of CD20 mAbs is directly related to binding off-rates. But it is noteworthy that they drew the relationship between binding off-rate and CDC potency by comparing those CD20 mAbs recognizing different epitopes.16,17 In this study, our conclusion about the relationship between binding off-rate and CDC potency seems to be more reasonable, which is based on analysis of those rituximab variants that have different off-rates but recognize the same epitope. Moreover, we also first demonstrate that the change of binding avidities in a certain range cannot affect the CDC potency of rituximab.
Up to now, the mechanism of cell death induced by CD20 ligation still remains controversial. The most striking finding in our present study is that the triple variant, which has been defined as type I mAb based on its ability to efficiently translocate CD20 into rafts, exhibits unusual activity in the induction of apoptosis even without cross-linking. It is difficult to understand why the avidity-enhanced binding of triple variant generated by introducing only 3 point mutations in the CDRs can induce significantly more apoptosis than rituximab because avidity enhancement has proven to be unable to influence the apoptotic activity of rituximab. The CD20 molecule, which is predicted to have 2 extracellular loops,1,2 may have the potential to undergo conformational changes when binding to the antibody. Compared with the complex structure of rituximab with the same epitope peptide, the residues SerP177 and ProP178 in the peptide and C-terminal region of the peptide in the C2H7-CD20 peptide complex displayed substantially different conformations.39 The in vitro assays showed that, even recognizing the same epitope,17 C2H7 still exhibited a measurable difference in functions compared with rituximab.39 Furthermore, recent research showed that, after making an unspecified change to the sequence of the framework during the humanization process, the GA101, which was developed from a type I CD20 mAb (Bly-1), converted to a type II mAb.15 Considering that GA101 and the triple variant recognize the same epitope as their respective parental antibodies and the conclusion we have drawn that manipulation of avidity or the off-rate of CD20 mAb does not affect the apoptotic activity, a reasonable explanation is that the conformation change of CD20 induced by those CD20 mAbs might play a role in affecting their functions.
Immunotherapy with rituximab has significantly improved the treatment outcome of lymphoma patients.6,11 However, a subpopulation of patients, via an elusive mechanism, does not respond to rituximab and/or acquires resistance on long-term rituximab therapy.40,41 Previous report by Jazirehi et al23 described the establishment of RR non-Hodgkin lymphoma cell lines, which shows that repeated rituximab exposure results in a reduced CD20 surface expression, overexpression of resistant factors, and increased apoptosis threshold. In the present study, we established RR lymphoma cell lines following the method described previously23,24 and then investigated the ability of the triple variant to kill these RR lymphoma cell lines. The data showed that the triple variant still exhibited potent ADCC and apoptotic activity in both Daudi-R and Raji-R cells. Immunotherapeutic study further demonstrated that the triple variant was effective in prolonging the survival of SCID/Daudi-R mice, whereas rituximab was not. In addition, F(ab′)2 fragment of the triple variant, which was prepared to remove the potential for any conventional effector functions, also exhibited a substantial proportion of therapeutic activity in vivo. Although a larger dose of F(ab′)2 than IgG was given in these experiments, this was necessary to allow for the far shorter half-life of F(ab′)2 compared with IgG in vivo. It has been previously demonstrated that the half-life of F(ab′)2 fragments is at least 3- to 4-fold lower than the whole IgG molecule.21 Taken together, it could be concluded that the in vivo antitumor effect of the triple variant might be at least partially attributable to its potent caspase-independent apoptosis.
Previous studies have demonstrated that the type II mAb (such as tositumomab), with its greater tendency to promote caspase-independent apoptosis but not CDC, is more effective than rituximab in depleting malignant B cells in vivo.42,43 One obvious explanation for the in vivo potency of tositumomab is that it relates directly to the ability to induce nonclassic apoptosis.43 Increasing evidence is becoming available to suggest that, in situations where classic apoptotic pathways may be crippled, eg, in Bcl-2 overexpressing or highly chemorefractory tumors, the ability to kill cells by alternative death pathways may be of critical clinical importance.44,45 As shown in our report, binding of rituximab to CD20 is not sufficient to kill RR lymphoma cells, indicating that there are mechanisms of resistance. Ongoing investigations show that multiple mechanisms of action and of resistance may be operative, suggesting that a multipronged attack on resistance will be required.40,41,46,47 Our data further demonstrate that the rituximab triple variant has not only the capability to mediate potent ADCC but strongly trigger both caspase-dependent and -independent apoptosis, showing a potent therapeutic efficacy in SCID mice bearing RR lymphoma. It can be speculated that multiple mechanisms of action may contribute to overcoming rituximab resistance, although we are unable to discern the relative importance of these mechanisms.
In conclusion, the rituximab variants presented here offer exciting reagents for studying CD20 mAb-mediated killing mechanisms. In addition, the triple variant (H57DE/H102YK/L93NR) exhibits unusual apoptotic and ADCC activity and has potent antitumor activity, even in RR lymphoma mouse models, which suggests that it may serve as a potential therapeutic agent for the treatment of human B-cell lymphoproliferative disorders.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from National Natural Science Foundation of China, Ministry of Science & Technology of China (973 and 863 program projects), National Key project for New Drug Creation and Manufacture, and Shanghai Commission of Science & Technology.
Authorship
Contribution: Y.G. and B.L. designed research, analyzed data, and wrote the paper; and L.Z., H.G., C.W., X.Z., L.W., L.C., Q.T., W.Q., and H.W. performed experiments and analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yajun Guo, International Joint Cancer Institute and 301 General Hospital Cancer Center, Second Military Medical University, 800 Xiang Yin Rd, Shanghai 200433, People's Republic of China; e-mail: yjguo@smmu.edu.cn.
References
Author notes
B.L. and L.Z. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal