Abstract
Human plasma cells (PCs) and their precursors play an essential role in humoral immune response but are rare and difficult to harvest. We report the generation of human syndecan-1+ and immunoglobulin secreting PCs starting from memory B cells in a 3-step and 10-day (D) culture, including a 6-fold cell amplification. We report the detailed phenotypic and Affymetrix gene expression profiles of these in vitro PCs as well as of intermediate cells (activated B cells and plasmablasts) compared with memory B cells and bone marrow PCs, which is accessible through an open web ATLAS (http://amazonia.transcriptome.eu/). We show this B cell–to-PC differentiation to involve IRF4 and AICDA expressions in D4 activated B cells, decrease of PAX5 and BCL6 expressions, and increase in PRDM1 and XBP1 expressions in D7 plasmablasts and D10 PCs. It involves down-regulation of genes controlled by Pax5 and induction of genes controlled by Blimp-1 and XBP1 (unfold protein response). The detailed phenotype of D10 PCs resembles that of peripheral blood PCs detected after immunization of healthy donors. This in vitro model will facilitate further studies in PC biology. It will likewise be helpful to study PC dyscrasias, including multiple myeloma.
Introduction
Human plasma cells (PCs) and their precursors play an essential role in humoral immune response but likewise give rise to a variety of malignant B-cell disorders, including multiple myeloma. The final steps of B-cell differentiation have been extensively studied during the past 10 years.1-3 Naive B cells entering into lymph node through high endothelial venules are selected by the antigen in the germinal center reaction, yielding selection of B cells with high-affinity immunoglobulins (Igs) and differentiation into memory B cells (MBCs, CD20+CD19+CD27+CD38−), and early plasmablasts (PBs, CD20−CD19+CD27++CD38++). PBs exit into peripheral blood and may survive for a short period only unless they are recruited into mucosa or bone marrow niches, depending on their chemokine receptor expression.4-6 These niches provide these PBs the factors to survive and further differentiate into long-living mature PCs.7 CCR10-expressing IgA+ PBs are mainly recruited to the mucosa niche by the CCL28 chemokine.8 In the bone marrow, the PC niche involves SDF-1–producing cells recruiting CXCR4+ PBs and is shared by hematopoietic stem cells and pre-pro-B cells.9 The rarity of this niche explains the low amount of bone marrow PCs (BMPCs; 0.5% of bone marrow cells) and is a matter of regulation of normal Ig production.10
The differentiation of B cells into PCs involves profound molecular changes yielding a cell able to produce large amounts of Igs for a long-term period. Two sets of transcription factors (TFs) that repress each other are involved in this process.11,12 The guardian of B-cell phenotype is the PAX5 TF, which induces B-cell genes and represses genes, such as PRDM1 and XBP1, whose gene products (Blimp-1 and XBP1) are critical for PC generation and survival. The BCL6 TF in association with MTA3 maintains B-cell phenotype and proliferation, down-regulating PRDM1 expression. In germinal center, activation of B cells through BCR, CD40, and/or Toll-like receptor results in up-regulation of IRF4, down-regulation of BCL6 protein, and loss of PRDM1 gene repression. This results in down-regulation of PAX5 gene and then up-regulation of XBP1. In the centrocyte region, stimulation by interleukin-10 (IL-10), IL-21, or IL-6 results in STAT3 activation, yielding to PRDM1 overexpression.13,14 This results in the full engagement of B-cell differentiation into PBs, in particular with the switch from surface to cytoplasmic Igs, and induction of the unfold protein response driven by XBP1. The detailed hierarchy of this molecular regulation is not fully understood and is still a challenging issue. Recent data suggest that a PAX5 down-regulation and consecutive XBP1 up-regulation are the initial driving events in PC generation independently of Blimp-1 expression.15 Other data indicate a major role of IRF4, whose expression is triggered by nuclear factor-κB signaling.16 In humans, research in PC differentiation mechanisms is hampered by the rarity and lack of availability of PCs, that is, because of the necessity of bone marrow aspiration.
In current in vitro models of B-cell differentiation,17-21 mainly CD20−CD38++CD138+/− PBs have been obtained. In a recent work, Huggins et al22 have reported the possibility to obtain syndecan-1+ PCs through a 3-step culture, but a detailed phenotypic and molecular characterization of these in vitro–generated cells are not available. In the current study, we first aim to design an easy culture process making it possible to reproducibly obtain syndecan-1+ PCs. The second aim was to extensively characterize these in vitro–generated PBs and PCs using Affymetrix gene expression profiling and multicolor cytometry and to make accessible an open web atlas of the respective gene expression data.
Methods
BMPCs from healthy donors were included in the study approved by the institutional review board of the Medical Faculty of the Ruprecht-Karls-University Heidelberg, Germany. Written informed consent was obtained in accordance with the Declaration of Helsinki.
Reagents
Human recombinant IL-2, IL-12, and interferon-α (IFN-α) were purchased from R&D Systems; IL-4, IL-6, and IL-15 from AbCys SA; and IL-10 and hepatocyte growth factor (HGF) from PeproTech. Hyaluronic acid was purchased from Sigma-Aldrich. The list of monoclonal antibodies (mAbs) used for phenotype study are detailed in supplemental data (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Cell samples
Peripheral blood cells from healthy volunteers were purchased from the French Blood Center. After removal of CD2+ cells using anti-CD2 magnetic beads (Invitrogen), CD19+CD27+ MBCs were sorted by FACSAria with a 95% purity. BMPCs from healthy volunteers were purified (cell purity ≥ 80% assayed by cytometry) using anti-CD138 magnetic microbeads sorting (Miltenyi Biotec), after approval by the ethics committee and written informed consent as described.23 Cells produced in the culture system were purified by multicolor fluorescence-activated cell sorting (FACS) using fluorescein isothiocyanate (FITC)–conjugated anti-CD20 mAb and phycoerythrin (PE)–conjugated anti-CD38 mAb for day 4 activated B cells (CD20+CD38− cells), and day 4 PBs and day 7 PBs (CD20−CD38+). Day 10 PCs (CD20−CD138+) were FACS-sorted using FITC-conjugated anti-CD20 mAb and PE-conjugated anti-CD138 mAb. The purity of FACS-sorted cell populations was at least 95% as assayed by cytometry.
Cell cultures
B-cell activation.
All cultures were performed in Iscove modified Dulbecco medium (Invitrogen) and 10% fetal calf serum, supplemented with 50 μg/mL human transferrin and 5 μg/mL human insulin (Sigma-Aldrich). Purified B cells were plated at 1.5 × 105/mL and cultured with various combinations of cyokines as indicated: IL-2 (20 U/mL), IL-4 (50 ng/mL), IL-10 (50 ng/mL), and IL-12 (2 ng/mL); or IL-2 (20 U/mL), IL-10 (50 ng/mL), and IL-15 (10 ng/mL); or IL-2 (20 U/mL), and IL-4 (50 ng/mL). Cells were cultured in 5 mL/well in 6-well flat-bottomed culture plates. In respective cultures groups, 10 μg/mL phosphorothioate CpG oligodeoxynucleotide 2006 (ODN)24 (Sigma-Aldrich) and/or histidine-tagged soluble recombinant human CD40L (50 ng/mL) and anti-poly-histidine mAb (5 μg/mL; R&D Systems) were added at culture start. In respective experiments, soluble CD40L was replaced by 3.75 × 104/mL mitomycin-treated CD40L transfectant (a generous gift from S. Saeland, Schering-Plough).
PB generation.
At day 4 of culture, the cells were harvested, washed, and seeded at 2.5 × 105/mL with various combinations of cytokines: IL-2 (20 U/mL), IL-6 (50 ng/mL), IL-10 (50 ng/mL), and IL-12 (2 ng/mL); or IL-2 (20 U/mL), IL-6 (50 ng/mL), IL-10 (50 ng/mL), and IL-15 (10 ng/mL).
PC generation.
At day 7 of culture, cells were washed and cultured with IL-6 (50 ng/mL), IL-15 (10 ng/mL), and IFN-α (500 U/mL) for 3 days. In some cultures, HGF (20 ng/mL) and/or hyaluronic acid (100 μg/mL) were also added.
Flow cytometric analysis, cytology, and Ig production
Cells were stained with FITC–anti-CD20, PE–anti-CD138 (Beckman Coulter), or PE–anti-CD38 (BD Biosciences) mAbs. Isotype-matched mouse mAbs were used as control. Cytospin smears of purified CD20+CD38− cells harvested at day 4 of culture, CD20−CD38bright cells at day 7, and CD20−CD138+ cells at day 10 were stained with May-Grünwald-Giemsa. The percentage of cells in the S phase of the cell cycle was determined using propidium iodide, and data were analyzed with the ModFit LT software (Verity Software House).25 Ig production was measured in culture supernatants harvested at the end of each culture step: day 4, day 7, and day 10. IgM, IgA, and IgG levels were evaluated by nephelometry with an automated Behring Nephelometer analyser II (Siemens). The sensitivity of the assay was 2 μg/mL for IgM, 3 μg/mL for IgA, and 4 μg/mL for IgG. Ig production (picograms/cell per day) was estimated dividing Ig amount in the culture supernatant by the number of living cells and the duration of the culture period.
Immunophenotypic analysis
Cells were stained using 4- to 7-color direct immunofluorescence stain. Surface staining was performed before cell fixation and permeabilization. The Cytofix/Cytoperm kit (BD Biosciences) was used for intracellular staining of IgM, IgA, IgG, and Ki-67 antigen, according to the manufacturer's recommendations. Flow cytometric analysis was performed with a FACSAria cytometer using FACSDiva 6.1 (BD Biosciences). For data analysis, CellQuest (BD Biosciences) and Infinicit 1.3 (Cytognos SL) software were used. The fluorescence intensity of the cell populations was compared using the stain index (SI) provided by the formula: [mean fluorescence intensity (MFI) obtained from the given mAb − MFI obtained with a control mAb]/[2 times the SD of the MFI obtained with the same control mAb].26
Real-time RT-PCR analysis
Total RNA was extracted using the RNeasy Kit (QIAGEN) and reverse-transcribed with the Reverse Transcription Kit (QIAGEN). The assays-on-demand primers and probes and the TaqMan Universal Master Mix were used according to the manufacturer's instructions (Applied Biosystems). Real-time reverse-transcribed polymerase chain reaction (RT-PCR) was performed using the ABI Prism 7000 Sequence Detection System and normalized to β2-microglobulin for each sample and compared with the values obtained for a known positive control using the following formula 100/2ΔΔCt where ΔΔCt = ΔCt unknown − ΔCt positive control as described.27
Microarray hybridization and bioinformatic analysis
RNA was extracted and hybridized to human genome U133 Plus 2.0 GeneChip microarrays, according to the manufacturer's instructions (Affymetrix). Gene expression data are deposited in the ArrayExpress public database (http://www.ebi.ac.uk/microarray-as/ae/, accession number E-MEXP-2360). Gene expression data were analyzed with our bioinformatics platforms (RAGE, http://rage.montp.inserm.fr/)28 and Amazonia (http://amazonia.transcriptome.eu/).29 The clustering was performed and visualized with the Cluster and TreeView softwares.30 Genes differentially expressed between cell populations were determined with the SAM statistical microarray analysis software.31 The biologic pathways encoded by these genes were analyzed with Ingenuity software.
Statistical analysis
Statistical comparisons were made with the nonparametric Mann-Whitney test, unpaired, or paired Student t test using SPSS software. P values less than or equal to .05 were considered significant.
Results
Obtaining PCs through a 3-step culture process in vitro
Step 1. Optimization of B-cell amplification and differentiation.
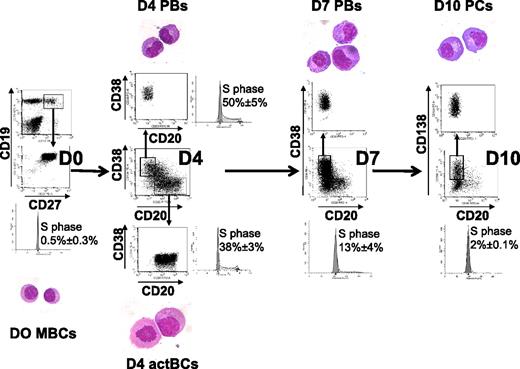
Starting from purified CD19+CD27+ MBCs, we first investigated which combination of activation signals allowed obtaining a maximum number of viable activated B cells. The best result, that is, a 6.1-fold amplification, was achieved using activations by soluble recombinant CD40L (sCD40L) and ODN and the IL-2 plus IL-10 plus IL-15 cytokine combination (Table 1). Comparable data were obtained using either a CD40L transfectant or sCD40L to trigger CD40 activation (results not shown). Activation by either sCD40L or ODN only plus in each case the same additional cytokine combination yielded a 46% or 68% lower amplification (3.3- and 2-fold stimulation, respectively, P ≤ .008; supplemental Table 1), indicating an additive effect if sCD40L and ODN are simultaneously used. Other cytokine combinations were reported to trigger B-cell activation together with CD40 activation. Using sCD40L activation and IL-2 plus IL-4 alone resulted in no cell amplification (supplemental Table 1). Adding IL-2 plus IL-4 plus IL-10 plus IL-12, as we reported initially,20 resulted in 3.4-fold amplification as with IL-2 plus IL-10 plus IL-15. In all culture conditions, except with sCD40L plus IL-2 plus IL-4, cells were at least 87% viable. Using the optimized activation combination (sCD40L + ODN + IL-2 + IL-10 + IL-15), the expanded cells at day 4 comprised 42.3% of CD20+CD38− cells, 16.4% of CD20+CD38+ cells, and 19.5% of CD20−CD38++ cells (Table 1). CD20+CD38− cells have an activated B-cell cytology and were cycling (38% ± 3% in the S phase) unlike MBCs (0.5% ± 0.3% in the S phase; Figure 1). CD20−CD38++ cells showed a typical PB morphology, with an eccentrically nucleus, relatively abundant basophilic cytoplasm with archoplasm (Figure 1). They were also highly cell cycling (50% ± 5% in the S phase). CD20+CD38− cells were termed day 4 activated B cells (D4 actBCs) and CD20−CD38++ cells PBs.
Generation of plasma cells from memory B cells
| . | B-cell amplification and differentiation (days 0-4; step 1) . | Plasmablastic differentiation (days 4-7; step 2) . | Plasma cell differentiation (days 7-10; step 3) . |
|---|---|---|---|
| Activation | sCD40L + ODN | — | — |
| Cytokines | IL-2 + IL-10 + IL-15 | IL-2 + IL-6 + IL-10 + IL-15 | IL6 + IL-15 + IFN-α |
| Mean cell amplification | 6.1 ± 1.8; n = 19 | 3.7 ± 1.3; n = 18 | 0.51 ± 0.09; n = 17 |
| Cell viability, percentage | 94 | 81 | 38 |
| CD20+CD38− activated B cells, % | 42.3 ± 12.2; n = 13 | 11.7 ± 5.1; n = 13 | 2.2 ± 1.2; n = 13 |
| CD20+CD38+ intermediate cells, % | 16.4 ± 12.5; n = 13 | 20.1 ± 8.8; n = 13 | 13.6 ± 8.5; n = 13 |
| CD20−CD38++ plasmablasts/plasma cells, % | 19.5 ± 5.5; n = 13 | 56.6 ± 7.7; n = 13 | 79.0 ± 8.8; n = 13 |
| CD20−CD38+CD138+ plasma cells, % | 2.1 ± 0.9; n = 13 | 15.9 ± 6.2; n = 13 | 54.8 ± 8.7; n = 13 |
| Yield of plasmablast/plasma cell generation for one starting memory B cells | NA | Plasmablasts /plasma cells: 12.3 ± 6.1; n = 13 | Plasma cells: 6.3 ± 3.3; n = 13 |
| . | B-cell amplification and differentiation (days 0-4; step 1) . | Plasmablastic differentiation (days 4-7; step 2) . | Plasma cell differentiation (days 7-10; step 3) . |
|---|---|---|---|
| Activation | sCD40L + ODN | — | — |
| Cytokines | IL-2 + IL-10 + IL-15 | IL-2 + IL-6 + IL-10 + IL-15 | IL6 + IL-15 + IFN-α |
| Mean cell amplification | 6.1 ± 1.8; n = 19 | 3.7 ± 1.3; n = 18 | 0.51 ± 0.09; n = 17 |
| Cell viability, percentage | 94 | 81 | 38 |
| CD20+CD38− activated B cells, % | 42.3 ± 12.2; n = 13 | 11.7 ± 5.1; n = 13 | 2.2 ± 1.2; n = 13 |
| CD20+CD38+ intermediate cells, % | 16.4 ± 12.5; n = 13 | 20.1 ± 8.8; n = 13 | 13.6 ± 8.5; n = 13 |
| CD20−CD38++ plasmablasts/plasma cells, % | 19.5 ± 5.5; n = 13 | 56.6 ± 7.7; n = 13 | 79.0 ± 8.8; n = 13 |
| CD20−CD38+CD138+ plasma cells, % | 2.1 ± 0.9; n = 13 | 15.9 ± 6.2; n = 13 | 54.8 ± 8.7; n = 13 |
| Yield of plasmablast/plasma cell generation for one starting memory B cells | NA | Plasmablasts /plasma cells: 12.3 ± 6.1; n = 13 | Plasma cells: 6.3 ± 3.3; n = 13 |
Purified memory B cells were cultured for 10 days using a 3-step culture system. In step 1, B cells were activated for 4 days with sCD40L and ODN and IL-2 + IL-10 + IL-15. In step 2, plasmablast differentiation was further promoted removing sCD40L and ODN and adding IL-6 together with IL-2 + IL-10 + IL-15. In step 3, plasma cell differentiation was induced for 3 days, removing IL-2 and IL-10 and adding IFN-α together with IL-6 + IL-15. At the end of every step, cell counts and viability were determined and cell phenotype was assayed with fluorochrome-conjugated anti-CD20, CD38, or CD138 mAbs, or isotype-matched control mAbs. Flow cytometry was performed with a FACScan device. Results are shown as the mean ± SD of n experiments.
— indicates no sCD40L or ODN; and NA, not applicable.
Three-step in vitro model of PC generation. Peripheral blood human MBCs were purified and cultured with sCD40L, ODN, and IL-2 + IL-10 + IL-15, then with IL-2 + IL-6 + IL-10 + IL-15 for 3 days, and then with IFN-α + IL-6 + IL-15 for 3 days. Cells were labeled with anti-CD20, CD38, and anti-CD138 mAbs, CD20+CD38− D4 actBCs, CD20−CD38++ D4 or D7 PBs, and CD20−CD38++CD138+ D10 PCs were FACS sorted and stained with May-Grünwald-Giemsa (original magnification, ×1000). The percentage of cells in the S hase of the cell cycle was determined using propidium iodide, and data were analyzed with the ModFit LT software. Histograms are those of 1 experiment representative of 3.
Three-step in vitro model of PC generation. Peripheral blood human MBCs were purified and cultured with sCD40L, ODN, and IL-2 + IL-10 + IL-15, then with IL-2 + IL-6 + IL-10 + IL-15 for 3 days, and then with IFN-α + IL-6 + IL-15 for 3 days. Cells were labeled with anti-CD20, CD38, and anti-CD138 mAbs, CD20+CD38− D4 actBCs, CD20−CD38++ D4 or D7 PBs, and CD20−CD38++CD138+ D10 PCs were FACS sorted and stained with May-Grünwald-Giemsa (original magnification, ×1000). The percentage of cells in the S hase of the cell cycle was determined using propidium iodide, and data were analyzed with the ModFit LT software. Histograms are those of 1 experiment representative of 3.
Step 2. Cell amplification and plasmablastic differentiation.
Step 2 aims to promote further PC differentiation. sCD40L was removed because it partially blocks PC differentiation.20 We also found that the presence of ODN blocked PC differentiation (results not shown). IL-6 was added together with IL-2 plus IL-10 plus IL-15 because it promotes PC differentiation and survival,18 in particular through STAT3 activation and Blimp-1 induction.14 After 3 days of culture, a 3.7-fold cell expansion with at least 80% viable cells could be found if cells were cultured in step 1 with sCD40L and ODN (Table 1). The expansion in step 2 was 57% and 32% lower (P ≤ .05), respectively, if cells were cultured with ODN or sCD40L only in step 1 (supplemental Table 2). At day 3 of step 2 culture (day 7 of the whole culture), the percentage of CD20+CD38− cells decreased from 42.3% at day 4 to 11.7% (P < .0001, n = 13) with an increase in the percentage of CD20−CD38++ cells (56.6%, P < .0001, n = 13). In addition, 15.9% of CD20−CD38++CD138+ cells were detected. Day 7 CD20−CD38++ were sorted and show the same plasmablastic morphology as day 4 CD20−CD38++ cells and were termed day 7 PBs (D7 PBs; Figure 1). They had a reduced number of cycling cells compared with D4 PBs (13% ± 4% vs 50% ± 5% in the S phase, paired t test, n = 3, Figure 1) and an increased CD38 density (anti-CD38 SI, 125 vs 20, P = .0005, n = 5). Thus, starting from 1 MBC, 12.3 plus or minus 6.1 CD20−CD38++ D7 PBs could be generated using the optimal step 1 and 2 culture conditions (Table 1). This step 2 culture could not be extended longer than 3 days as a rapid PB death occurred on day 4 or day 5, despite addition of fresh cytokines. Prolonging the first step 1 culture for 4 additional days with fresh sCD40L plus ODN plus IL-2 plus IL-10 plus IL-15 yielded to a further B-cell amplification, but to a rapid cell death in step 2 and a lower number of overall generated PCs (results not shown).
Step 3. PC differentiation.
To avoid the rapid cell death occurring after 3 days in step 2, cells were washed and cultured with IL-6 plus IL-15 plus IFN-α for 3 days; 60% of the cells died at this stage. Adding hepatocyte growth factor and/or hyaluronic acid, as suggested,22 did not improve cell survival (results not shown). Differentiation within this last step 3 was independent on the initial step 1 conditions (supplemental Table 3). Surviving cells were composed mostly of CD20−CD38++ cells (79%), including 54.8% CD20−CD38++CD138+ (Table 1). FACS-sorted CD138+ cells had PC cytology and were termed day 10 PCs (D10 PCs). These cells were rarely cell cycling with 2% of cells in the S phase, compared with D7 or D4 PBs (13% and 50%, respectively). Thus, this 3-step culture process made it possible to generate 12.3 CD20−CD38++ D7 PBs (at step 2) and 6.3 CD20−CD38++CD138+ D10 PCs (at step 3) starting from 1 MBC (Table 1). The density of CD38 expression was increased in D10 PCs compared with D7 PCs (SI 276 vs 125, P = .006, n = 5).
Expression of surface, cytoplasmic IgM, IgG, IgA, and Ig production
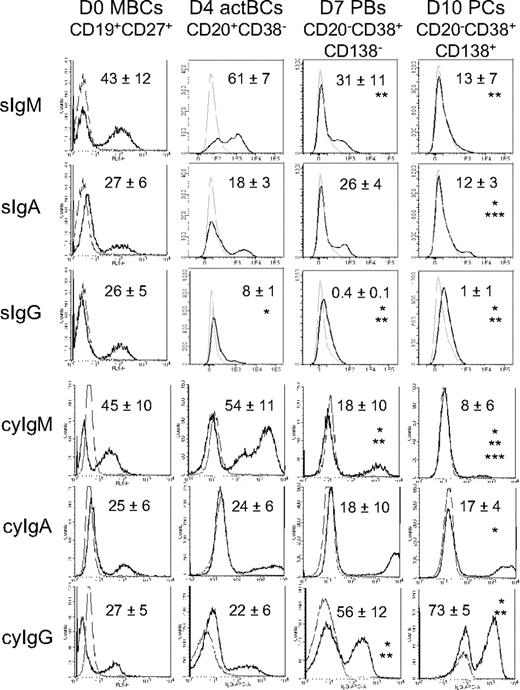
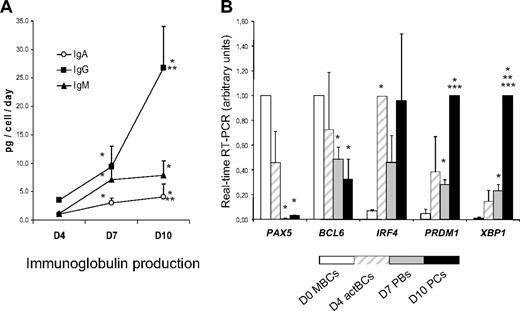
Surface (s) Igs were detected by labeling cells with anti–Ig heavy chain antibodies (IgM, IgA, and IgG) without permeabilization and cytoplasmic (cy) and surface Igs after cell permeabilization. MBCs used to start culture were composed of 43% plus or minus 12% sIgM+, 27% plus or minus 6% sIgA+, and 26% plus or minus 5% sIgG+ cells (n = 5, Figure 2). Permeabilization of MBCs yielded similar percentages of cyIgM+, cyIgA+, and cyIgG+ cells with similar MFIs. CD20+CD38− D4 actBCs were composed of 61% plus or minus 7% sIgM+ and 18% plus or minus 3% sIgA+ cells (not significantly different from MBCs) and a 3-fold–reduced percentage of sIgG+ cells (8% vs 26%, P ≤ .05, n = 5; Figure 2). D4 actBC cells were preparing to secrete Igs, as permeabilization resulted in detection of 22% cyIgG+ actBCs and a 20-fold and 2.5-fold, respectively, significantly increased MFI (P ≤ .05, n = 5) for cyIgM and cyIgA labeling (Figure 2). The differentiation of D4 actBCs into D7 PBs and consecutively D10 PCs was associated with a loss of cyIgM+ cells (from 54% in D4 actBCs to 18% in D7 PBs and 8% in D10 PCs, P ≤ .05, n = 5), an increase in cyIgG+ cells (from 22% in D4 actBCs to 56% in D7 PBs and 73% in D10 PCs, P ≤ .05, n = 5), with no significant difference in the percentage for cyIgA+ cells. In agreement with detection of cytoplasmic Igs and expression of PC markers by flow cytometry, the rate of IgG production/cell per day increased 8-fold at day 10 compared with day 4 (P = .003, n = 5; Figure 3A). The rates of IgA and IgM production also significantly increased (P ≤ .005, n = 5; Figure 3A).
Expression of surface and cytoplasmic Ig heavy chain isotypes by B cells and PCs generated in the 3-step culture system. MBCs were cultured as described in Figure 1. Starting MBCs, D4 actBCs, D7 PBs, and D10 PCs were labeled with fluorochrome-conjugated anti-CD20, CD38, and CD138 mAbs and with fluorochrome-conjugated anti–human IgM, IgA, IgG mAbs, or isotype-controlled mAbs before or after cell permeabilization. The bold histograms represent labeling with anti-IgM, IgA, or IgG mAb and the light ones with the control mAb. Histograms are those of 1 experiment representative of 5. The numbers in the panels are the mean ± SD of the percentage of labeled cells (ie, ≥ MFI + SD of the control mAb). *The mean percentage of labeled cells is different from that in D0 MBCs. **The mean percentage of labeled cells is different from that in D4 actBCs. ***The mean percentage of labeled cells is different from that in D7 PBs.
Expression of surface and cytoplasmic Ig heavy chain isotypes by B cells and PCs generated in the 3-step culture system. MBCs were cultured as described in Figure 1. Starting MBCs, D4 actBCs, D7 PBs, and D10 PCs were labeled with fluorochrome-conjugated anti-CD20, CD38, and CD138 mAbs and with fluorochrome-conjugated anti–human IgM, IgA, IgG mAbs, or isotype-controlled mAbs before or after cell permeabilization. The bold histograms represent labeling with anti-IgM, IgA, or IgG mAb and the light ones with the control mAb. Histograms are those of 1 experiment representative of 5. The numbers in the panels are the mean ± SD of the percentage of labeled cells (ie, ≥ MFI + SD of the control mAb). *The mean percentage of labeled cells is different from that in D0 MBCs. **The mean percentage of labeled cells is different from that in D4 actBCs. ***The mean percentage of labeled cells is different from that in D7 PBs.
Ig production and gene expression of TFs involved in B cells to PC differentiation. (A) MBCs were cultured as described in Figure 1, and culture supernatants were harvested at day 4, day 7, and day 10 to assay for IgM, IgA, and IgG concentrations using nephelometry. The rate of Ig production per cell and per day was calculated by dividing the amount of Igs in the culture supernatant by the number of viable cells at the time of culture supernatant harvesting and by the number of days of culture. Data are the mean ± SD of the rates of Ig production determined in 5 separate experiments. *The rate of Ig productions is different from those at day 4. **The rate of Ig productions is different from those at day 7. (B) D0 MBCs, D4 actBCs, D7 PBs, and D10 PCs were FACS sorted, and the expression of PAX5, BCL6, IRF4, PRDM1, and XBP1 genes was evaluated by real-time RT-PCR. The gene expression in the different cell populations was compared assigning the arbitrary value 1 to the maximal expression. Data are the mean value ± SD of gene expression determined in 5 separate experiments. *The mean expression is different from that in D0 MBCs. **The mean expression is different from that in D4 actBCs. ***The mean expression is different from that in D7 PBs.
Ig production and gene expression of TFs involved in B cells to PC differentiation. (A) MBCs were cultured as described in Figure 1, and culture supernatants were harvested at day 4, day 7, and day 10 to assay for IgM, IgA, and IgG concentrations using nephelometry. The rate of Ig production per cell and per day was calculated by dividing the amount of Igs in the culture supernatant by the number of viable cells at the time of culture supernatant harvesting and by the number of days of culture. Data are the mean ± SD of the rates of Ig production determined in 5 separate experiments. *The rate of Ig productions is different from those at day 4. **The rate of Ig productions is different from those at day 7. (B) D0 MBCs, D4 actBCs, D7 PBs, and D10 PCs were FACS sorted, and the expression of PAX5, BCL6, IRF4, PRDM1, and XBP1 genes was evaluated by real-time RT-PCR. The gene expression in the different cell populations was compared assigning the arbitrary value 1 to the maximal expression. Data are the mean value ± SD of gene expression determined in 5 separate experiments. *The mean expression is different from that in D0 MBCs. **The mean expression is different from that in D4 actBCs. ***The mean expression is different from that in D7 PBs.
Phenotype of B cells, D7 PBs, and D10 PCs
D4 actBCs expressed CD19, C27, CD45, and human leukocyte antigen (HLA) class II (Figure 4A). D7 PBs and D10 PCs were CD19+, CD45+, and HLA class II+ but with a 2.5-, 3.0-, and 5-fold, respectively, lower expression for D7 PBs and 2.5-, 3.4-, and 12-fold, respectively, for D10 PCs (P ≤ .05, n = 5) compared with D4 actBCs. CD27 expression was increased 2.5- and 3-fold in D7 PBs and D10 PCs, respectively, compared with D4 actBCs (P ≤ .05, n = 5). In agreement with S-phase data in Figure 1, only D4 actBCs and D7 PBs were Ki-67+. CD43 was expressed in 66% plus or minus 8% of D7 PBs and 57% plus or minus 9% of D10 PCs. Regarding homing molecules, PC differentiation was characterized by a disappearance of CXCR5, a progressive reduction in CXCR4 (1.8- and 3-fold, respectively, decrease in D7 PBs and D10 PCs compared with D4 actBCs, P ≤ .05, paired t test, n = 3), induction of CCR10, and increased CD62L/L-selectin (Figure 4B).
Phenotype and expression of homing molecules of B cells and PCs generated in the 3-step culture system. MBCs were cultured as described in Figure 1. Cells were stained for CD20, CD38, and CD138. The cell phenotype was analyzed by gating on CD20+CD38− lymphocytes, CD20−CD38++CD138− D7 PBs, and CD20−CD38++CD138+ D10 PCs. (A) Black histograms represent FACS labeling with anti-CD19, CD27, CD45, HLA class II, Ki-67 (after cell permeabilization), and CD43. Gray histograms represent the corresponding negative control mAbs. Data from 1 experiment representative of 3 are shown. Numbers in panels indicate mean values ± SD of the percentage of positive cells of 3 separate experiments, and numbers in brackets indicate the mean staining indexes ± SD. (B) Black histograms represent FACS labeling with anti-CXCR5, CXCR4, CCR10, and CD62L mAbs. Gray histograms represent the corresponding negative control mAbs. Data from 1 experiment representative of 3 are shown. Numbers in panels indicate mean values ± SD of the percentage of positive cells, and numbers in brackets the mean staining indexes ± SD of 3 separate experiments. *Value is different from that in D0 MBCs using a paired t test. **Value is different from that in D4 actBCs. ***Value is different from that in D7 PBs.
Phenotype and expression of homing molecules of B cells and PCs generated in the 3-step culture system. MBCs were cultured as described in Figure 1. Cells were stained for CD20, CD38, and CD138. The cell phenotype was analyzed by gating on CD20+CD38− lymphocytes, CD20−CD38++CD138− D7 PBs, and CD20−CD38++CD138+ D10 PCs. (A) Black histograms represent FACS labeling with anti-CD19, CD27, CD45, HLA class II, Ki-67 (after cell permeabilization), and CD43. Gray histograms represent the corresponding negative control mAbs. Data from 1 experiment representative of 3 are shown. Numbers in panels indicate mean values ± SD of the percentage of positive cells of 3 separate experiments, and numbers in brackets indicate the mean staining indexes ± SD. (B) Black histograms represent FACS labeling with anti-CXCR5, CXCR4, CCR10, and CD62L mAbs. Gray histograms represent the corresponding negative control mAbs. Data from 1 experiment representative of 3 are shown. Numbers in panels indicate mean values ± SD of the percentage of positive cells, and numbers in brackets the mean staining indexes ± SD of 3 separate experiments. *Value is different from that in D0 MBCs using a paired t test. **Value is different from that in D4 actBCs. ***Value is different from that in D7 PBs.
B-cell and PC TFs
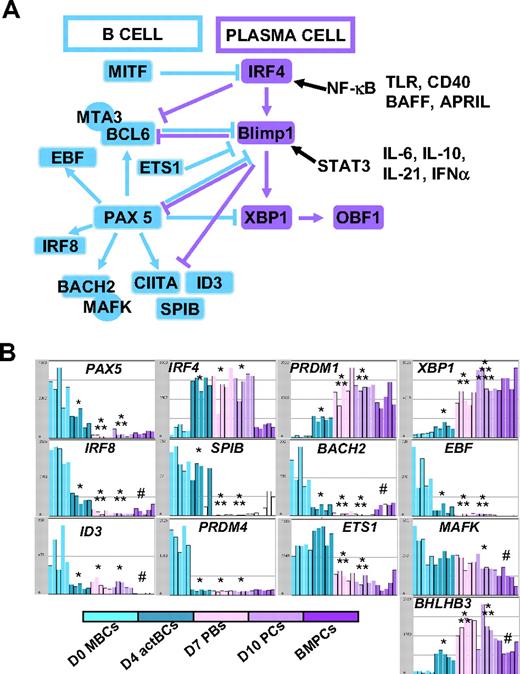
In Figure 3B, the gene expression of 5 major TFs that control B-cell to PC differentiation is shown. A clear-cut difference was the lack of expression of PAX5, the guardian of B-cell phenotype11 in D7 PBs and D10 PCs, unlike D0 MBCs and D4 actBCs. BCL6 and PRDM1, whose gene products mutually repress gene expression of the other,12 showed a correlated inverse pattern. BCL6 expression progressively decreased and PRDM1 progressively increased from D0 MBCs to D10 PCs. PRDM1, XBP1, and high IFR4 expressions were already found in D4 actBCs, suggesting that these cells, with a B-cell phenotype (Figures 1, 3B), were already on the way toward plasmablastic differentiation.
Gene expression atlas of B-cell to PC differentiation
Genome-wide gene expression profiling of the 4 cell populations identified (D0 MBCs, D4 actBCs, D7 PBs, and D10 PCs) above and of purified BMPCs were performed using Affymetrix U133 Plus 2.0 microarrays. First, an unsupervised clustering with 2000 probe sets after filtering with an SD of 520.8 or higher defined 5 clusters grouping the samples of 5 populations with a strong correlation: D0 MBCs (r = 0.74), D4 actBCs (r = 0.76), D7 PBs (r = 0.41), D10 PCs (r = 0.59), and BMPCs (r = 0.75; Figure 5A). D7 PBs and D10 PCs clusters were correlated together (r = 0.27, P = .05), unlike other clusters (Figure 5A). Approximately one-third of these genes delineated a PC cluster (D7 PBs, D10 PCs, and BMPCs) versus a B-cell cluster (MBCs and D4 actBCs). To extract optimally this PC versus B-cell gene signature, a supervised analysis was run comparing D0 MBCs plus D4 actBCs and D7 PBs plus D10 PCs plus BMPCs (Wilcoxon statistic, 1000 permutations, 2-fold ratio) yielding to 676 probe sets on the basis of a 0% false discovery rate. They corresponded to 459 unique genes (202 PC and 257 BC genes) using Ingenuity analysis, separating B cells from PCs (Figure 5B; supplemental Table 4). The resulting networks encoded by these PC and BC genes were analyzed and scored with Ingenuity. The PC and BC networks and detailed data including gene lists associated with networks are shown in supplemental Table 5. The highest scoring PC network mainly is composed of genes induced by XBP1 TF (supplemental Figure 1).
Gene expression profiles of B cells and PCs generated in the 3-step culture system, of MBCs and BMPCs. The gene expression profile of purified B cells or PC populations (5 separate samples for each population) was determined with Affymetrix U133 Plus 2.0 microarrays. (A) An unsupervised hierarchical clustering was run with the 2000 probe sets with the highest SD (log transform, center genes and arrays, uncentered correlation, and average linkage). The dendrogram shows that all samples of a given population (D0 MBCs, D4 actBCs, D7 PBs, D10 PCs, and BMPCs) strongly cluster together (r ≥ 0.5) and that D7 PBs and D10 PCs are correlated together unlike other populations. (B) The probe sets differentially expressed between D0 MBCs + D4 actBCs and D7 PBS + D10 PCS + BMPCs were determined with a SAM-supervised analysis (Wilcoxon statistic, 2-fold ratio, 0% false discovery rate), identifying 459 unique genes with Ingenuity software. When a gene was assayed by several probe sets, the probe set with the highest variance was used. An unsupervised hierarchical clustering was run on this 459 unique gene list. The normalized expression value for each gene is indicated by a color: red represents high expression; green, low expression.
Gene expression profiles of B cells and PCs generated in the 3-step culture system, of MBCs and BMPCs. The gene expression profile of purified B cells or PC populations (5 separate samples for each population) was determined with Affymetrix U133 Plus 2.0 microarrays. (A) An unsupervised hierarchical clustering was run with the 2000 probe sets with the highest SD (log transform, center genes and arrays, uncentered correlation, and average linkage). The dendrogram shows that all samples of a given population (D0 MBCs, D4 actBCs, D7 PBs, D10 PCs, and BMPCs) strongly cluster together (r ≥ 0.5) and that D7 PBs and D10 PCs are correlated together unlike other populations. (B) The probe sets differentially expressed between D0 MBCs + D4 actBCs and D7 PBS + D10 PCS + BMPCs were determined with a SAM-supervised analysis (Wilcoxon statistic, 2-fold ratio, 0% false discovery rate), identifying 459 unique genes with Ingenuity software. When a gene was assayed by several probe sets, the probe set with the highest variance was used. An unsupervised hierarchical clustering was run on this 459 unique gene list. The normalized expression value for each gene is indicated by a color: red represents high expression; green, low expression.
The changes in gene expression at different stages of B-cell to PC differentiation can be quickly visualized using our Amazonia “B to PC” Atlas (http://amazonia.transcriptome.eu/). Regarding genes coding for membrane markers of B cells and PCs, the PC Atlas is in agreement with FACS data of Figure 4A and B (supplemental Figure 2A-B). The 3 PC populations did not express CD20 and CD22 genes and expressed weakly HLA class II genes in agreement with decreased CIITA compared with D0 MBCs and D4 actBCs. The 3 PC populations still expressed CD19, although at a lower level, and CD45 gene expression progressively decreased from MBCs to BMPCs. CD24 expression was lost on D7 PBs and D10 PCs but expressed in BMPCs again. PC differentiation is evidenced by increased Ig heavy chain (IgH) gene expression, increased expression of CD27, expression of CD38 and its ligand CD31/PECAM1, and of CD138. CD9 gene was highly expressed only on BMPCs, and CD40 was highly expressed in BMPCs. Fas/CD95 expression was increased in MBCs, D4 actBCs, D7 PBs, and D10 PCs compared with BMPCs. CD23 expression was rapidly lost on D4 actBCs. Of interest, only D4 actBCs highly expressed AICDA gene, suggesting that these cells could be in a process of Ig hypermutations and/or switch. Both D4 actBCs and D7 PCs expressed MKi67 gene, in agreement with cell-cycle cytometry data. Among TFs, which have been shown to control the B-cell and PC phenotype, the expression of 13 of these could be investigated with Affymetrix U133 Plus 2.0 microarrays. The current knowledge of the mechanisms of action of these TFs are shown in Figure 6A. Affymetrix data for the main TF genes (Pax 5, IRF4, PRDM1, and XBP1) are confirmed by real-time RT-PCR data (Figures 3B,6B). In agreement with PAX5 down-regulation in D7 PBs, D10 PCs, and BMPCs, the following PAX5-regulated genes11 were down-regulated in these cells: IRF8, SPIB, BACH2, EBF, ID3, and CIITA, in association with increased expression of PAX5-inhibited genes,11 PRDM1 and XBP1. Regarding genes coding for homing molecules (supplemental Figure 2B), PC differentiation was associated with loss of expression of genes coding for lymph node chemokine receptors (CCR7, CXCR5). CXCR4 expression was decreased and CCR10 expression increased in D7 PBs and D10 PCs in agreement with flow cytometric data. CCR2 gene, which is down-regulated by PAX5, was not expressed in MBCs but highly in BMPCs. CD62L was highly expressed in D7 PBs and D10 PCs unlike BMPCs. PC differentiation is associated with increased expression of ITGA4 and ITGB1, coding for the VLA4 heterodimer, increased expression of ICAM2, coding for a VLA4 ligand. The gene coding for sphingosine phosphate receptor (EDG1/S1PR1), which is involved in cell exit from tissues,32 was decreased in D10 PCs and BMPCs. ITGAL, a gene coding for CD11a, is increased in D4 actBCs, D7 PBs, and D10 PCs. Finally, the gene coding for ERN1, which induces XBP1 mRNA splicing, and the genes coding for XBP1-driven unfold protein response were up-regulated throughout B to PC differentiation (supplemental Figure 3).
Visualization of gene expression of TFs using Amazonia “B cell to PC Atlas.” The gene expression of the 54 613 Affymetrix probe sets in MBCs, D4 actBCs, D7 PBs, D10 PCs, and BMPCs can be visualized using the Amazonia web site (http://amazonia.transcriptome.eu/). The known interactions of these TFs are displayed in panel A. Data are the expression of genes coding for TFs controlling B-cell and PC fate (B). *The mean expression is different from that in D0 MBCs. **The mean expression is different from that in D4 actBCs. ***The mean expression is different from that in D7 PBs. #The mean expression is different between D10 PCs and BMPCs.
Visualization of gene expression of TFs using Amazonia “B cell to PC Atlas.” The gene expression of the 54 613 Affymetrix probe sets in MBCs, D4 actBCs, D7 PBs, D10 PCs, and BMPCs can be visualized using the Amazonia web site (http://amazonia.transcriptome.eu/). The known interactions of these TFs are displayed in panel A. Data are the expression of genes coding for TFs controlling B-cell and PC fate (B). *The mean expression is different from that in D0 MBCs. **The mean expression is different from that in D4 actBCs. ***The mean expression is different from that in D7 PBs. #The mean expression is different between D10 PCs and BMPCs.
Discussion
Human PCs and their precursors play an essential role in humoral immune response but likewise give rise to a variety of malignant B-cell neoplasias. They are difficult to obtain, as they are rare cells located in specific niches in the bone marrow and mucosa,10 hindering the understanding of their physiology and pathophysiology. The aim of the current study was to provide a full phenotypic and molecular characterization of in vitro–generated PCs and of the intermediate cells. We have first compared the activation signals and cytokine combinations reported in various methodologies for in vitro PB and PC generation17,20,22 to get a maximum of PCs while limiting the number of activation signals and cytokines. Here, a mean number of 6.3 viable PCs could be generated in a 3-step culture system starting from one MBC. PCs show a PC morphology, secrete Igs, express PC markers (CD38, CD31, and CD138), and lack B-cell markers (CD20, CD21, CD22, and CD23). This PC phenotype was associated with expression of PC TF genes (PRDM1, XBP1) and decreased expression of B-cell ones (PAX5, BCL6).
The current strategy mimics the activation and differentiation process occurring in the germinal center reaction using activation of CD40 (mimicking T-cell help) and Toll-like receptor activation (mimicking Ag activation) with a combination of cytokines produced by T helper cells, dendritic cells, and macrophages.2 These activation signals trigger nuclear factor-κB signaling that induces IRF4 expression, resulting in down-regulation of BCL6, being critical to maintain the centroblast phenotype.2 This is what was observed in D4 actBCs, activated by sCD40L and CpG ODN, that express highly IRF4 and a lower level of BCL6 compared with MBCs. The IRF4 expression is associated with high AICDA expression in D4 actBCs in agreement with data showing IRF4 to control AICDA gene expression.33 AID controls the process of Ig variable gene mutation. AID also controls heavy chain isotype switching, which may explain the progressive loss of IgM+ cells and appearance of IgG+ cells in this in vitro model. Alternatively, the shift from B cells expressing IgM to PCs expressing mainly IgG could be the result of a selective proliferation of IgG+ starting B cells. IRF4, when expressed at high level, also induces PRDM1 and XBP1 expression.33 The high IRF4 expression in D4 actBCs may explain why these cells are already in the way toward PC differentiation, expressing weakly PRDM1 and XBP1 and cytoplasmic Igs while still highly proliferating and expressing B-cell markers. The second step of culture consists of removing CD40 activation and CpG ODN that block the full process of PC generation and adding IL-6 to further promote STAT3 activation. STAT3 induces PRDM1 expression34 together with IRF433 and also further down-regulates BCL6 expression. In particular, a knockout of STAT3 abrogates PC differentiation.35 The final step consists of removing cytokines inducing proliferation (IL-2 and IL-10) and adding IFN-α, IL-6, and IL-15, yielding to PCs that express syndecan-1 and secrete higher amounts of Igs, as measured in the culture supernatants. Both IFN-α and IL-6 highly stimulate STAT3 pathway, resulting in the observed increased PRDM1 expression in D10 PCs (Figure 5). This likely explains syndecan-1 expression and increased Ig secretion in D10 PCs as the PRDM1 gene product (Blimp-1) induces syndecan-1 gene expression and splicing of Ig RNA yielding to Ig secretion in B cells.36 Huggins et al22 have reported that the addition of hyaluronic acid, to stimulate CD44, and hepatocyte growth factor further improved differentiation of PBs into PCs, but we found no benefit of adding hyaluronic acid or HGF. Adding IL-21 and/or a proliferation-inducing ligand did not result also in improvement of PC generation and survival (results not shown), and these in vitro–generated PCs progressively died in culture. The identification of signals promoting long-term PC survival is a major unresolved issue. PC long-term survival and differentiation may require cell-to-cell contacts, mimicking what is occurring in the putative PC niches. Tokoyoda et al9 reported that murine PCs home in contact to SDF-1–producing cells in the bone marrow, sharing the same niche with hematopoietic stem cells and pre-pro-B cells. In mucosa, a recent report has shown that tissue PCs are located in a proliferation-inducing ligand-rich niches, composed of myeloid cells.37 Thus, the current model will make it possible to further identify the niche that can promote long-term survival of human PCs.
An important question is what the differences are between these in vitro D10 PCs compared with the most studied human PCs in vivo, that is, tonsil PCs, BMPCs, and peripheral blood PCs. Tonsil PCs, present in either germinal centers or follicular and parafollicular zones, express CD20, CD19, HLA class II, CD45, CD22, CD9, and highly CD38 and do not express CD62L and CD138.38,39 Peripheral blood PCs detected in healthy persons after tetanus toxoid immunization are CD20−CD19+CD45+CD62L+HLA class II+CD9−CD38high and half of them express CD138.38 BMPCs express CD138, highly CD38, and CD31, lack CD20, and express weakly CD19, and half of them express CD45 and HLA class II.38 The phenotype of in vitro–generated D10 PCs is different from that of tonsil PCs that are more immature through the expression of CD20 and CD22 and lack of expression of CD138. It could correspond to the phenotype of the fraction of CD45+ HLA class II+ BMPCs because they all express these 2 molecules, but at reduced levels compared with B cells. A difference is that D10 PCs express CD62L, weakly CXCR4 and CCR2, and did not express CD9. Actually, the phenotype of these D10 PCs fits well with that of peripheral blood PCs induced by tetanus toxoid immunization of healthy persons.38 These circulating PCs do express CD62L, intermediate levels of CD138, and weakly CXCR4 and CCR2, and do not express CD9, unlike BMPCs. They are considered to be newly generated PCs, leaving the lymphoid organs to home to bone marrow or tissue, as is the case for in vitro–generated D10 PCs. As both these circulating PCs and in vitro–generated D10 PCs express weakly CXCR4, other molecules could be involved in their homing to bone marrow or mucosa. VLA4 could be this homing molecule because hematopoietic stem cells can home to BM through VLA4 in a CXCR4-independent manner.40 Of interest, both peripheral blood PCs and D10 PCs highly express VLA4, making possible their homing to BM or tissues.
Besides studying B-cell differentiation, this in vitro model is likewise of major interest to further understand the biology of multiple myeloma by introducing genes deregulated in multiple myeloma cells and looking for their ability to induce long-term survival and proliferation of (malignant) PCs. It would also be of interest to look for whether the multiple myeloma bone marrow environment could trigger the survival of these PCs in vitro.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the staff of IRB Affymetrix platform (http://irb.montp.inserm.fr/en/index.php?page = Plateau&IdEquipe = 6; John De Vos, Veronique Pantesco, Jennifer Torrent, and Tanguy Le Carrour) for their assistance with the microarray assay.
This work was supported by Ligue Nationale Contre le Cancer (équipe labellisée 2009), Paris, France, Institut National du Cancer, and Myeloma Stem Cell Network European strep (E06005FF).
Authorship
Contribution: M.J. designed research, performed the experiments, and wrote the paper; G.F. provided technical assistance; J.D.V. designed the Amazonia web site; A.C. and M.L. provided assistance for cytometry experiments; C.B. performed the cytology analysis; C.C. performed the Ig production determination; C.D. provided assistance for cytometry experiments; D.H. provided GEP data for BMPCs; D.H. and A.C. participated in the writing of the paper; and B.K. is the senior investigator who designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bernard Klein, Inserm U847, Institute for Research in Biotherapy, Centre Hospitalier Universitaire Montpellier, Hospital St Eloi, Av Augustin Fliche, 34295 Montpellier, France; e-mail: bernard.klein@inserm.fr.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal