Abstract
Somatic mutation of the AML1/RUNX1(RUNX1) gene is seen in acute myeloid leukemia (AML) M0 subtype and in AML transformed from myelodysplastic syndrome, but the impact of this gene mutation on survival in AML patients remains unclear. In this study, we sought to determine the clinical implications of RUNX1 mutations in 470 adult patients with de novo non-M3 AML. Sixty-three distinct RUNX1 mutations were identified in 62 persons (13.2%); 32 were in N-terminal and 31, C-terminal. The RUNX1 mutation was closely associated with male sex, older age, lower lactic dehydrogenase value, French-American-British M0/M1 subtypes, and expression of HLA-DR and CD34, but inversely correlated with CD33, CD15, CD19, and CD56 expression. Furthermore, the mutation was positively associated with MLL/PTD but negatively associated with CEBPA and NPM1 mutations. AML patients with RUNX1 mutations had a significantly lower complete remission rate and shorter disease-free and overall survival than those without the mutation. Multivariate analysis demonstrated that RUNX1 mutation was an independent poor prognostic factor for overall survival. Sequential analysis in 133 patients revealed that none acquired novel RUNX1 mutations during clinical courses. Our findings provide evidence that RUNX1 mutations are associated with distinct biologic and clinical characteristics and poor prognosis in patients with de novo AML.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous group of neoplastic disorders with great variability in the pathogenesis, clinical course, and response to therapy.1 In recent years, frequent events of acquired gene mutations, as well as deregulation of gene expression, were identified in AML.2-4 A 2-hit model proposes that cooperation between 2 classes of gene mutations contributes to the leukemogenesis.5,6 Class I mutations activate genes in the kinase signaling pathways, such as RAS, FLT3, KIT, and PTPN11, leading to cell survival and proliferation. Class II mutations, such as PML/RARA, CBFB/MYH11, RUNX1/RUNX1T1, MLL/PTD, CEBPA, and SPI1 mutations, inactivate transcriptional factors or cofactors resulting in impaired hematopoietic differentiation. Among these, FLT3/ITD and MLL/PTD are associated with inferior overall and relapse-free survival, whereas mutations in CEBPA and NPM1 (without concomitant FLT3/ITD) are linked to a favorable outcome.7-9
The AML1/RUNX1 (hereafter referred to as RUNX1) gene,10 consisting of 10 exons (exons 1-6, 7A, 7B, 7C, and 8), is one of the most frequently deregulated genes in leukemia through chromosomal translocations and point mutations.11,12 RUNX1 protein consists of 3 main structure domains: runt homology domain (RHD, residues 50-177), transcription activation domain (TAD, residues 291-371), and repression domain (residues 446-453). The RHD, a highly conserved 128–amino acid protein motif, is responsible for both DNA binding and heterodimerization with the β-subunit of CBF. The TAD is responsible for the interaction with MOZ,13 a transcription coactivator of RUNX1 currently named MYST histone acetyltransferase 3.14 RUNX1 is required for definitive hematopoiesis, and its functional dysregulation leads to leukemia.15,16 Monoallelic germline mutation of the RUNX1 gene occurs in rare cases of familial platelet disorder with predisposition to AML.17 Acquired RUNX1 mutation was mostly reported in therapy-related myelodysplastic syndrome (MDS) or MDS/AML but was also found in some de novo AMLs, especially French-American-British (FAB) subtype M0.11,18-24 This mutation has been shown to be associated with poor outcome in MDS,20,25 but its prognostic implication in de novo AML remains unclear.
In this study, we assessed the clinical implication of RUNX1 mutation in 470 unselected adults with de novo AML and its interactions with other gene alterations. Sequential analysis of RUNX1 mutation during the clinical course was also performed on 133 patients to investigate the stability and pathogenic role of this mutation in AML. To the best of our knowledge, this is the first study to address the prognostic implication of RUNX1 mutation in a large cohort of patients with de novo AML. We found that RUNX1 mutation is an independent poor-risk factor for overall survival (OS) in these patients.
Methods
Subjects
This study was approved by the institutional review board of the National Taiwan University Hospital, and written informed consent was obtained from all participants in accordance with the Declaration of Helsinki. From November 1995 to March 2007, a total of 470 adult patients who were newly diagnosed as having de novo non-M3 AML at National Taiwan University Hospital were enrolled consecutively. Patients with antecedent hematologic diseases or therapy-related AML were excluded. Diagnosis and classification of AML were made according to the FAB Cooperative Group Criteria. Patients with AML M3 subtype were excluded because their treatment and survival differ significantly from those of other AML patients. Among them, 330 patients (70.2%) received induction chemotherapy (idarubicin 12 mg/m2 per day on days 1-3 and cytarabine 100 mg/m2/d on days 1-7) and then consolidation chemotherapy with 2 to 4 courses of high-dose cytarabine (2000 mg/m2 every 12 hours on days 1-4, total 8 doses), with or without an anthracycline (idarubicin or novantrone) after achieving complete remission (CR). The remaining 140 patients received palliative therapy with supportive care or low-dose chemotherapy resulting from underlying comorbidity or based on the decision of the patients. Ninety-six patients underwent allogeneic hematopoietic stem cell transplantation (HSCT), including 11 RUNX1-mutated and 85 RUNX1-wild patients. Of the 96 allografts, 81 grafts were from related donors and 15 were from unrelated donors. HLA was matched in 87 and mismatched for at least one locus in 9. Forty-four patients were in first CR at the time of transplantation and 52, after relapse or refractory.
Cytogenetics
Bone marrow cells were harvested directly or after 1 to 3 days of unstimulated culture as described previously.26 Metaphase chromosomes were banded by the trypsin-Giemsa technique and karyotyped according to the International System for Human Cytogenetic Nomenclature.
Immunophenotype analysis
A panel of monoclonal antibodies to myeloid-associated antigens, including CD13, CD33, CD11b, CD15, CD14, and CD41a, as well as lymphoid-associated antigens, including CD2, CD5, CD7, CD19, CD10, and CD20, and lineage nonspecific antigens HLA-DR, CD34, and CD56 were used to characterize the phenotypes of the leukemia cells as previously described.27
Mutation analysis
Mutation analysis of RUNX1 exons 3 to 8 was performed by genomic DNA polymerase chain reaction (PCR) and direct sequencing as previously reported.25 Abnormal sequencing results were confirmed by at least 2 repeated analyses. Sequential analysis of RUNX1 mutation during the clinical course was performed in 294 samples from 133 patients. Mutation analysis of 11 other relevant molecular marker genes, NPM1,28 CEBPA,29 FLT3/ITD and FLT3/TKD,27 N-RAS,30 K-RAS,30 JAK2,30 KIT,25 MLL/PTD,31 PTPN11,32 and WT1,33 was performed as previously described.
TA cloning analysis
For patients with double mutations, TA cloning was performed to determine whether the 2 mutations were in the same or different alleles as previously described.25 Briefly, the cDNA was amplified to cover both mutations, the PCR products were then cloned into the Taq polymerase-amplified (TA)–cloning vector pGEM-T Easy (Promega), and 10 clones were selected for sequencing.
Statistical analysis
The discrete variables of patients with and without gene mutation were compared using the χ2 tests, but if the expected values of contingency tables were smaller than 5, Fisher exact test was used. If the continuous data were not normally distributed, Mann-Whitney tests were used to compare continuous variables and medians of distributions. To evaluate the impact of RUNX1 mutation on clinical outcome, only the 330 patients who received conventional induction chemotherapy and then consolidation chemotherapy if CR was achieved, as mentioned earlier, were included in analysis. OS was measured from the date of first diagnosis to the date of last follow-up or death from any cause, whereas disease-free status indicated that the patient achieved CR and did not relapse by the end of this study. Cox regression survival estimation was used to plot survival curves and to test the difference between groups. Multivariate Cox proportional hazard regression analysis was used to investigate independent prognostic factors for OS and disease-free survival (DFS). The proportional hazards assumption (constant hazards assumption) was examined using time-dependent covariate Cox regression before conducting multivariate Cox proportional hazard regression. The variables, including age, white blood cell count (WBC), lactic dehydrogenase (LDH), karyotype, CEBPA, WT1, RUNX1, and NPM1/FLT3-ITD, were used as covariates. Those patients who received HSCT were censored at the time of HSCT in survival analysis to ameliorate the influence of the treatment. A P value less than .05 was considered statistically significant. All statistical analyses were performed with SPSS 13 software (SPSS Inc) and Statsdirect.
Results
RUNX1 mutations in 470 adult patients with de novo non-M3 AML
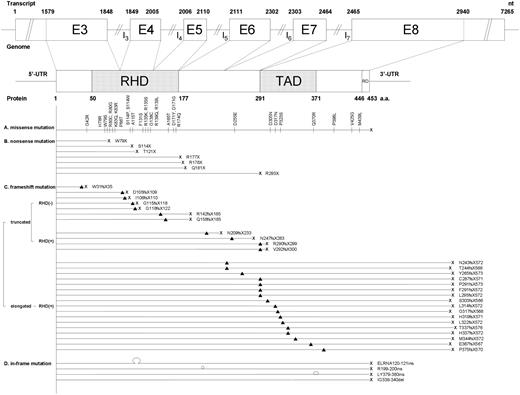
Excluding the 4 single nucleotide polymorphisms (I87I, P157P, S362S, and P436P) that were detected in 10 patients but did not alter the amino acid residues, 68 mutations were identified in 70 patients (Figure 1). Among them, 4 mutations (G42R, D317N, V425G, and M439L) in 7 patients were supposed to be silent and insignificant for 2 reasons. First, these mutations did not disappear at CR (Table 1, cases 63-66, 68-70). Second, the 4 amino acid sites are located outside RHD region and are not conserved in RUNX proteins family among different species (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Another mutation Q370R, which was found in 2 patients, was also supposed to be insignificant because it was still detectable at CR after consolidation chemotherapy and also after autologous HSCT in one child who had this mutation at diagnosis but was not recruited in this report (data not shown). Of the remaining 63 distinct mutations, 24 were missense mutations, 7 were nonsense mutations, 28 were frameshift mutations, and 4 were in-frame mutations (Figure 1). Totally, 32 mutations were located at N-terminal (exons 3-5), including 31 at RHD, and 31 were at C-terminal (exons 6-8), including 17 at TAD. Among 24 missense mutations, 20 were in the RHD region. The 4 missense mutations (D255E, D305N, P323S, and P398L) located outside the RHD region were conserved during species evolution (supplemental Figure 1). Of the 28 frameshift mutations, 7 generated truncated peptides with complete or partial deletion of the RHD, 4 generated truncated peptides with intact RHD region, and the remaining 17 generated elongated proteins with intact RHD region (Figure 1); all were suggested to abolish transactivation potential resulting from complete or partial loss of TAD,34 with the exception of P375fsX570, which retained the intact TAD. Totally, 62 patients (13.2%) had distinct RUNX1 mutations (supplemental Table 1), including 1 patient (case no. 61) who had both distinct mutation H78R on exon 3 and insignificant one Q370R on exon 8. The mutations in 6 patients (nos. 3, 4, 17, 35, 37, and 50) were double heterozygous with 2 RUNX1 mutations; the 2 mutations were in different alleles in 3 patients (nos. 4, 35, and 50) and in the same allele in 1 patient (no. 37) who had available samples for TA cloning study. All others showed only 1 mutation; 3 (nos. 27, 34, and 44) were homozygous and the remaining 53 were heterozygous.
Patterns and locations of the 68 RUNX1 mutations, including 63 distinct and 5 insignificant mutations. Four kinds of mutations were demonstrated: missense mutations, nonsense mutations, frameshift mutations, and in-frame mutations. Three kinds of frameshift mutations are noted: truncated mutations with defective RHD, truncated mutations with intact RHD, and elongated mutations with intact RHD. The numbering of nucleotide of transcript is according to the mRNA sequence from GenBank42 accession number D43968 (AML1b); on the other hand, the numbering of amino acid of protein is according to the amino acid sequence from GenBank accession number Q01196. G42R, D317N, Q370R, V425G, and M439L are insignificant mutations. ▴ and | represent the sites of mutation; X, the site of the stop codon; Ω, the site of insertion; and (hyphenated line), the site of deletion. RD indicates repression domain; E, exon; I, intron; nt, nucleotide; UTR, untranslated region; and a.a., amino acid.
Patterns and locations of the 68 RUNX1 mutations, including 63 distinct and 5 insignificant mutations. Four kinds of mutations were demonstrated: missense mutations, nonsense mutations, frameshift mutations, and in-frame mutations. Three kinds of frameshift mutations are noted: truncated mutations with defective RHD, truncated mutations with intact RHD, and elongated mutations with intact RHD. The numbering of nucleotide of transcript is according to the mRNA sequence from GenBank42 accession number D43968 (AML1b); on the other hand, the numbering of amino acid of protein is according to the amino acid sequence from GenBank accession number Q01196. G42R, D317N, Q370R, V425G, and M439L are insignificant mutations. ▴ and | represent the sites of mutation; X, the site of the stop codon; Ω, the site of insertion; and (hyphenated line), the site of deletion. RD indicates repression domain; E, exon; I, intron; nt, nucleotide; UTR, untranslated region; and a.a., amino acid.
Sequential studies in the AML patients with RUNX1 mutations*
| UPN . | Date . | Status . | Karyotype . | RUNX1 mutation . | Other mutations . | |
|---|---|---|---|---|---|---|
| Patients with distinct mutations | ||||||
| 3 | 01/26/1998 | Initial | 46,XY | MissenseFrameshift | W79XG115fsX118 | — |
| 05/16/1998 | CR | — | — | — | ||
| 05/16/2000 | Relapse | 45,XY,t(3;5)(q26;q13),-7 | Nonsense | W79X | — | |
| 4 | 04/30/1999 | Initial | 46,XY | MissenseFrameshift | A115TC287fsX571 | — |
| 10/09/2000 | CR | — | — | |||
| 8 | 07/19/1996 | Initial | 45,XY,-7 | Nonsense | Q181X | — |
| 8/26/1996 | CR | 46,XY | — | — | ||
| 9 | 06/15/1998 | Initial | 47,XY,+21 | Missense | K83Q | CEBPA |
| 07/16/1998 | CR | 46,XY | — | — | ||
| 11 | 05/20/1998 | Initial | 47,Y,inv(X)(p11q26),+del(8)(p21p23),del(8)(p22p23) | Nonsense | R178X | FLT3/ITD and NRAS |
| 10/20/1998 | CR | 46,XY | — | — | ||
| 12 | 05/21/1997 | Initial | 46,XY | Frameshift | L314fsX572 | — |
| 08/11/1997 | CR | — | — | — | ||
| 15 | 04/05/2000 | Initial | 46,XY,t(10;11)(q24;p15),add(6)(p25) | Frameshift | W31fsX35 | FLT3/ITD |
| 10/03/2000 | CR | 46,XY | — | — | ||
| 17 | 11/26/2001 | Initial | 46,XX | NonsenseMissense | R177XS114W | — |
| 04/01/2002 | CR | — | — | — | ||
| 11/15/2002 | Relapse | ND | NonsenseMissense | R117XS114W | FLT3/ITD | |
| 18 | 07/10/2002 | Initial | 47,XY,+8 | Frameshift | Q158fsX185 | — |
| 03/04/2003 | CR | 46,XY | — | — | ||
| 23 | 10/21/2002 | Initial | 46,XY | Nonsense | T121X | FLT3/ITD and FLT3/TKD |
| 02/06/2003 | CR | — | — | — | ||
| 04/30/2004 | Relapse | 46,XY | — | FLT3/ITD | ||
| 28 | 11/20/2003 | Initial | 45,XY,-7del(12)(p12p13) | Missense | D171G | PTPN11 |
| 01/28/2004 | CR | 46,XY | — | — | ||
| 29 | 06/12/2001 | Initial | 47,XX,+8 | Frameshift | L295fsX572 | — |
| 01/17/2002 | CR | 46,XX | — | — | ||
| 35 | 02/06/2004 | Initial | 46,XX | MissenseMissense | S114PA165T | FLT3/ITD |
| 06/01/2004 | CR | — | — | — | ||
| 03/07/2005 | Relapse | ND | Missense | S114P | FLT3/ITD | |
| 41 | 12/20/2004 | Initial | NM | Missense | R135K | — |
| 01/28/2005 | CR | ND | — | — | ||
| 44 | 11/15/2005 | Initial | 46,XY | Missense | R135K | — |
| 04/18/2006 | CR | — | — | — | ||
| 45 | 12/06/2005 | Initial | 46,XY | Frameshift | N209fsX233 | — |
| 01/12/2006 | CR | — | — | — | ||
| 48 | 07/21/2006 | Initial | 46,XY | Frameshift | G118fsX122 | WT1 and FLT3/TKD |
| 09/21/2006 | CR | — | — | — | ||
| 07/15/2008 | Relapse | 46,XY | — | WT1 and FLT3/TKD | ||
| 53 | 03/16/2006 | Initial | 46,XY | Frameshift | T244fsX568 | FLT3/ITD |
| 06/21/2006 | CR | — | — | — | ||
| 06/03/2008 | Relapse | ND | Frameshift | T244fsX568 | FLT3/ITD | |
| Patients with insignificant mutations | ||||||
| 63 | 09/25/1998 | Initial | 46,XY,t(7;11)(p15;p15) | V425G | — | |
| 11/20/1998 | CR | 46,XY | V425G | — | ||
| 03/18/1999 | Relapse | 46,XY,t(7;11)(p15;p15) | V425G | — | ||
| 64 | 04/25/2000 | Initial | 46,XX,del(16q) | V425G | — | |
| 01/07/2002 | CR | ND | V425G | — | ||
| 65 | 10/11/2002 | Initial | 46,XX | M439L | — | |
| 06/09/2003 | CR | — | M439L | — | ||
| 66 | 10/11/2004 | Initial | 45,X,-Y, t(7;21;8)(q32;q22;q24) | G42R | KIT(D816V) | |
| 12/22/2004 | CR | 46,XY | G42R | — | ||
| 07/28/2005 | Relapse | 45,X,-Y,t(7;21;8)(q32;q22;q24),t(8;16)(q11;p11),del(11)(q14q23) | G42R | KIT(D816V) | ||
| 68 | 02/23/2006 | Initial | 46,XY, inv(16)(p13q22) | D317N | — | |
| 04/03/2006 | CR | 46,XY | D317N | — | ||
| 09/05/2007 | Relapse | 46,XY, inv(16)(p13q22) | D317N | — | ||
| 69 | 12/25/2006 | Initial | 47,XX,+8, inv(16)(p13q22) | V425G | NRAS (Q61R) | |
| 01/25/2007 | CR | 46,XX | V425G | — | ||
| 12/20/2007 | Relapse | 46,XX | V425G | — | ||
| 70 | 08/13/2007 | Initial | 46,XX | M439L | — | |
| 09/13/2007 | CR | — | M439L | — | ||
| 01/11/2008 | Relapse | 46,XX | M439L | ND | ||
| UPN . | Date . | Status . | Karyotype . | RUNX1 mutation . | Other mutations . | |
|---|---|---|---|---|---|---|
| Patients with distinct mutations | ||||||
| 3 | 01/26/1998 | Initial | 46,XY | MissenseFrameshift | W79XG115fsX118 | — |
| 05/16/1998 | CR | — | — | — | ||
| 05/16/2000 | Relapse | 45,XY,t(3;5)(q26;q13),-7 | Nonsense | W79X | — | |
| 4 | 04/30/1999 | Initial | 46,XY | MissenseFrameshift | A115TC287fsX571 | — |
| 10/09/2000 | CR | — | — | |||
| 8 | 07/19/1996 | Initial | 45,XY,-7 | Nonsense | Q181X | — |
| 8/26/1996 | CR | 46,XY | — | — | ||
| 9 | 06/15/1998 | Initial | 47,XY,+21 | Missense | K83Q | CEBPA |
| 07/16/1998 | CR | 46,XY | — | — | ||
| 11 | 05/20/1998 | Initial | 47,Y,inv(X)(p11q26),+del(8)(p21p23),del(8)(p22p23) | Nonsense | R178X | FLT3/ITD and NRAS |
| 10/20/1998 | CR | 46,XY | — | — | ||
| 12 | 05/21/1997 | Initial | 46,XY | Frameshift | L314fsX572 | — |
| 08/11/1997 | CR | — | — | — | ||
| 15 | 04/05/2000 | Initial | 46,XY,t(10;11)(q24;p15),add(6)(p25) | Frameshift | W31fsX35 | FLT3/ITD |
| 10/03/2000 | CR | 46,XY | — | — | ||
| 17 | 11/26/2001 | Initial | 46,XX | NonsenseMissense | R177XS114W | — |
| 04/01/2002 | CR | — | — | — | ||
| 11/15/2002 | Relapse | ND | NonsenseMissense | R117XS114W | FLT3/ITD | |
| 18 | 07/10/2002 | Initial | 47,XY,+8 | Frameshift | Q158fsX185 | — |
| 03/04/2003 | CR | 46,XY | — | — | ||
| 23 | 10/21/2002 | Initial | 46,XY | Nonsense | T121X | FLT3/ITD and FLT3/TKD |
| 02/06/2003 | CR | — | — | — | ||
| 04/30/2004 | Relapse | 46,XY | — | FLT3/ITD | ||
| 28 | 11/20/2003 | Initial | 45,XY,-7del(12)(p12p13) | Missense | D171G | PTPN11 |
| 01/28/2004 | CR | 46,XY | — | — | ||
| 29 | 06/12/2001 | Initial | 47,XX,+8 | Frameshift | L295fsX572 | — |
| 01/17/2002 | CR | 46,XX | — | — | ||
| 35 | 02/06/2004 | Initial | 46,XX | MissenseMissense | S114PA165T | FLT3/ITD |
| 06/01/2004 | CR | — | — | — | ||
| 03/07/2005 | Relapse | ND | Missense | S114P | FLT3/ITD | |
| 41 | 12/20/2004 | Initial | NM | Missense | R135K | — |
| 01/28/2005 | CR | ND | — | — | ||
| 44 | 11/15/2005 | Initial | 46,XY | Missense | R135K | — |
| 04/18/2006 | CR | — | — | — | ||
| 45 | 12/06/2005 | Initial | 46,XY | Frameshift | N209fsX233 | — |
| 01/12/2006 | CR | — | — | — | ||
| 48 | 07/21/2006 | Initial | 46,XY | Frameshift | G118fsX122 | WT1 and FLT3/TKD |
| 09/21/2006 | CR | — | — | — | ||
| 07/15/2008 | Relapse | 46,XY | — | WT1 and FLT3/TKD | ||
| 53 | 03/16/2006 | Initial | 46,XY | Frameshift | T244fsX568 | FLT3/ITD |
| 06/21/2006 | CR | — | — | — | ||
| 06/03/2008 | Relapse | ND | Frameshift | T244fsX568 | FLT3/ITD | |
| Patients with insignificant mutations | ||||||
| 63 | 09/25/1998 | Initial | 46,XY,t(7;11)(p15;p15) | V425G | — | |
| 11/20/1998 | CR | 46,XY | V425G | — | ||
| 03/18/1999 | Relapse | 46,XY,t(7;11)(p15;p15) | V425G | — | ||
| 64 | 04/25/2000 | Initial | 46,XX,del(16q) | V425G | — | |
| 01/07/2002 | CR | ND | V425G | — | ||
| 65 | 10/11/2002 | Initial | 46,XX | M439L | — | |
| 06/09/2003 | CR | — | M439L | — | ||
| 66 | 10/11/2004 | Initial | 45,X,-Y, t(7;21;8)(q32;q22;q24) | G42R | KIT(D816V) | |
| 12/22/2004 | CR | 46,XY | G42R | — | ||
| 07/28/2005 | Relapse | 45,X,-Y,t(7;21;8)(q32;q22;q24),t(8;16)(q11;p11),del(11)(q14q23) | G42R | KIT(D816V) | ||
| 68 | 02/23/2006 | Initial | 46,XY, inv(16)(p13q22) | D317N | — | |
| 04/03/2006 | CR | 46,XY | D317N | — | ||
| 09/05/2007 | Relapse | 46,XY, inv(16)(p13q22) | D317N | — | ||
| 69 | 12/25/2006 | Initial | 47,XX,+8, inv(16)(p13q22) | V425G | NRAS (Q61R) | |
| 01/25/2007 | CR | 46,XX | V425G | — | ||
| 12/20/2007 | Relapse | 46,XX | V425G | — | ||
| 70 | 08/13/2007 | Initial | 46,XX | M439L | — | |
| 09/13/2007 | CR | — | M439L | — | ||
| 01/11/2008 | Relapse | 46,XX | M439L | ND | ||
UPN indicates unique patient number; ND, not done; NM, no mitosis; and —, not applicable.
The data of serial studies in another 108 patients who had no RUNX1 mutation at diagnosis were not shown in this table. None of these 108 patients acquired RUNX1 mutation at relapse after a median interval of 48 months (range, 5-160 months); 41.6% of them had karyotypic evolution at relapse.
Correlation of RUNX1 mutations with clinical features and immunophenotypes of leukemic cells
The comparison of clinical characteristics of patients with and without distinct RUNX1 mutation is shown in Table 2. Those patients with insignificant mutations were included in the RUNX1-wild group. Male patients had a higher incidence of RUNX1 mutations than female (18.4% vs 6.4%, P < .001). RUNX1-mutated patients were older (median, 62 years vs 48 years, P = .010) and had lower LDH levels than RUNX1-wild patients (P = .003). Patients with FAB M0 subtype of AML had the highest incidence (40%) of RUNX1 mutation, whereas those with M2 subtype had the lowest incidence (6.3%, P = .004). RUNX1 mutations were positively associated with the expression of HLA-DR (P < .001) and CD34 (P = .012) but inversely associated with the expression of CD33 (P < .001), CD15 (P = .003), CD19 (P = .014), and CD56 (P = .041) on the leukemic cells (supplemental Table 2). There was no difference in the expression of other antigens between the patients with and without RUNX1 mutation.
Comparison of clinical manifestation and laboratory features between AML patients with and without RUNX1 mutation
| Variable . | Total (n = 470) . | RUNX1-mutated* (n = 62, 13.2%) . | RUNX1-wild (n = 408, 86.8%) . | P . |
|---|---|---|---|---|
| Sex, no. (%) of patients | < .001 | |||
| Male | 266 | 49 (18.4) | 217 (81.6) | |
| Female | 204 | 13 (6.4) | 191 (93.6) | |
| Median age, y (range) | 52.0 (15-90) | 62.0 (15-89) | 48.0 (15-90) | = .010 |
| Laboratory data, median (range) | ||||
| WBC (/μL) | 21 850 (120-627800) | 15 225 (310-258900) | 22 430 (120-627800) | = .184 |
| Hemoglobin, g/dL | 8.0 (2.9-16.2) | 7.8 (3.8-11.9) | 8.0 (2.9-16.2) | = .855 |
| Platelets, × 1000/μL | 45.0 (3-802) | 49.5 (3-202) | 43.0 (3-802) | = .584 |
| Blasts, per μL | 9863 (0-456 725) | 7535 (102-246 602) | 10 015 (0-456 725) | = .643 |
| LDH, U/L | 890 (206-15 000) | 733 (299-8116) | 918 (206-15 000) | = .003 |
| FAB, no. (%) of patients | = .004 | |||
| M0 | 10 | 4 (40.0) | 6 (60.0) | |
| M1 | 114 | 20 (17.5) | 94 (82.5) | |
| M2 | 174 | 11 (6.3) | 163 (93.7) | |
| M4 | 126 | 19 (15.1) | 107 (84.9) | |
| M5 | 25 | 4 (16.0) | 21 (84.0) | |
| M6 | 12 | 3 (25.0) | 9 (75.0) | |
| Undetermined | 9 | 1 (11.1) | 8 (88.9) | |
| Induction response* | 330 | 37 | 293 | = .006 |
| CR | 248 (75.2) | 21 (56.8) | 227 (77.5) | |
| PR | 13 (3.9) | 5 (13.5) | 8 (2.7) | |
| Refractory | 46 (13.9) | 7 (18.9) | 39 (13.3) | |
| Induction death | 23 (7.0) | 4 (10.8) | 19 (6.5) |
| Variable . | Total (n = 470) . | RUNX1-mutated* (n = 62, 13.2%) . | RUNX1-wild (n = 408, 86.8%) . | P . |
|---|---|---|---|---|
| Sex, no. (%) of patients | < .001 | |||
| Male | 266 | 49 (18.4) | 217 (81.6) | |
| Female | 204 | 13 (6.4) | 191 (93.6) | |
| Median age, y (range) | 52.0 (15-90) | 62.0 (15-89) | 48.0 (15-90) | = .010 |
| Laboratory data, median (range) | ||||
| WBC (/μL) | 21 850 (120-627800) | 15 225 (310-258900) | 22 430 (120-627800) | = .184 |
| Hemoglobin, g/dL | 8.0 (2.9-16.2) | 7.8 (3.8-11.9) | 8.0 (2.9-16.2) | = .855 |
| Platelets, × 1000/μL | 45.0 (3-802) | 49.5 (3-202) | 43.0 (3-802) | = .584 |
| Blasts, per μL | 9863 (0-456 725) | 7535 (102-246 602) | 10 015 (0-456 725) | = .643 |
| LDH, U/L | 890 (206-15 000) | 733 (299-8116) | 918 (206-15 000) | = .003 |
| FAB, no. (%) of patients | = .004 | |||
| M0 | 10 | 4 (40.0) | 6 (60.0) | |
| M1 | 114 | 20 (17.5) | 94 (82.5) | |
| M2 | 174 | 11 (6.3) | 163 (93.7) | |
| M4 | 126 | 19 (15.1) | 107 (84.9) | |
| M5 | 25 | 4 (16.0) | 21 (84.0) | |
| M6 | 12 | 3 (25.0) | 9 (75.0) | |
| Undetermined | 9 | 1 (11.1) | 8 (88.9) | |
| Induction response* | 330 | 37 | 293 | = .006 |
| CR | 248 (75.2) | 21 (56.8) | 227 (77.5) | |
| PR | 13 (3.9) | 5 (13.5) | 8 (2.7) | |
| Refractory | 46 (13.9) | 7 (18.9) | 39 (13.3) | |
| Induction death | 23 (7.0) | 4 (10.8) | 19 (6.5) |
Only the patients with distinct RUNX1 mutations were included in this group. Those with insignificant mutations, such as G42R, D317N, Q370R, V425G, and M439L, were excluded.
Association of RUNX1 mutations with cytogenetic abnormalities
Chromosome data were available in 452 patients at diagnosis, including 56 RUNX1-mutated and 396 RUNX1-wild patients (Table 3). RUNX1 mutations occurred more frequently in patients with intermediate-risk (14.7%) or unfavorable cytogenetics (12.1%) than in those with favorable karyotype (0%, P < .001). There was no difference in the incidence of the RUNX1 mutation among patients with normal karyotype (13.9%), simple abnormalities (11.2%), and complex abnormalities (9.4%, P = .622). None of the patients with t(8;21), inv(16), or 11q23 translocation showed RUNX1 mutation. The same was also true for patients with t(15;17), who were not included in this study. There was close association of RUNX1 mutation with +8, but not other chromosomal abnormalities, including +11, +13, +21, −5/del(5q), and −7/del(7q).
Association of RUNX1 mutation with chromosomal abnormalities*
| Variable . | Total . | RUNX1-mutated . | RUNX1-wild . | P . |
|---|---|---|---|---|
| Karyotype† | < .001 | |||
| Favorable | 59 | 0 (0.0) | 59 (100.0) | |
| Intermediate | 327 | 48 (14.7) | 279 (85.3) | |
| Unfavorable | 66 | 8 (12.1) | 58 (87.9) | |
| Unknown | 18 | 6 (33.3) | 12 (66.7) | |
| Normal | 230 | 32 (13.9) | 198 (86.1) | = .622 |
| Simple | 169 | 19 (11.2) | 150 (88.8) | |
| Complex | 53 | 5 (9.4) | 48 (90.6) | |
| t(8;21) | 40 | 0 (0.0) | 40 (100.0) | = .009 |
| inv(16) | 19 | 0 (0.0) | 19 (100.0) | .149 |
| t(11q23) | 16 | 0 (0.0) | 16 (100.0) | = .239 |
| −5/5q-‡ | 1 | 0 (0.0) | 1 (100.0) | > .999 |
| −7/7q-‡ | 10 | 3 (30.0) | 7 (70.0) | = .115 |
| +8‡ | 22 | 6 (27.3) | 16 (72.7) | = .042 |
| +11‡ | 3 | 0 (0.0) | 3 (100.0) | > .999 |
| +13‡ | 1 | 0 (0.0) | 1 (100.0) | > .999 |
| +21‡ | 11 | 2 (18.2) | 9 (81.8) | .634 |
| Variable . | Total . | RUNX1-mutated . | RUNX1-wild . | P . |
|---|---|---|---|---|
| Karyotype† | < .001 | |||
| Favorable | 59 | 0 (0.0) | 59 (100.0) | |
| Intermediate | 327 | 48 (14.7) | 279 (85.3) | |
| Unfavorable | 66 | 8 (12.1) | 58 (87.9) | |
| Unknown | 18 | 6 (33.3) | 12 (66.7) | |
| Normal | 230 | 32 (13.9) | 198 (86.1) | = .622 |
| Simple | 169 | 19 (11.2) | 150 (88.8) | |
| Complex | 53 | 5 (9.4) | 48 (90.6) | |
| t(8;21) | 40 | 0 (0.0) | 40 (100.0) | = .009 |
| inv(16) | 19 | 0 (0.0) | 19 (100.0) | .149 |
| t(11q23) | 16 | 0 (0.0) | 16 (100.0) | = .239 |
| −5/5q-‡ | 1 | 0 (0.0) | 1 (100.0) | > .999 |
| −7/7q-‡ | 10 | 3 (30.0) | 7 (70.0) | = .115 |
| +8‡ | 22 | 6 (27.3) | 16 (72.7) | = .042 |
| +11‡ | 3 | 0 (0.0) | 3 (100.0) | > .999 |
| +13‡ | 1 | 0 (0.0) | 1 (100.0) | > .999 |
| +21‡ | 11 | 2 (18.2) | 9 (81.8) | .634 |
A total of 452 patients, including 56 RUNX1-mutated and 396 RUNX1-wild patients, had chromosome data at diagnosis.
Favorable consists of t(15;17), t(8;21), inv (16); unfavorable, −7, del(7q), −5, del(5q), 3q abnormality, complex abnormalities; and intermediate, normal karyotype and other abnormalities.
Includes only simple chromosomal abnormalities with 2 or fewer changes, but not those with complex abnormalities with 3 or more aberrations. There was no association of RUNX1 mutations with any of these chromosome changes, including +8, when complex abnormalities were also included in the analysis.
Association of RUNX1 mutation with other molecular abnormalities
To investigate the interaction of gene mutations in the pathogenesis of adult AML, a complete mutational screening of 11 other genes was performed in all 470 patients (Table 4). Among the 62 patients with RUNX1 mutations, 31 (50%) showed additional molecular abnormalities at diagnosis (supplemental Table 3); 16 had 1 additional change, 14 had 2, and 1 had 3. Twenty-six of them (83.9%) had at least one concurrent class I mutation. The most common associated molecular event was FLT3/ITD (14 cases), followed by MLL/PTD (9 cases), and FLT3/TKD (6 cases). Of note, 7 patients had concomitant RUNX1, FLT3/ITD, and MLL/PTD mutations (supplemental Table 3). Patients with RUNX1 mutations had a significantly higher incidence of MLL/PTD (9 of 62, 14.5%) than those with RUNX1-wild-type (19 of 408, 4.7%, P = .006). On the contrary, CEBPA and NPM1 mutations were rarely seen in patients with RUNX1 mutations (3.2%, P = .006; and 4.8%, P < .001, respectively). However, there was no difference in the incidence of FLT3/ITD and FLT3/TKD between patients with and without RUNX1 mutation. The same was also true for N-RAS, K-RAS, PTPN11, KIT, and WT1 mutations.
Association of RUNX1 mutation with other genetic mutations
| Variable . | Whole cohort, n = 470 . | RUNX1-mutated patients, n = 62 . | RUNX1-wild patients, n = 408 . | P . |
|---|---|---|---|---|
| CEBPA | 66 (14.0) | 2 (3.2) | 64 (15.7) | > .006 |
| FLT3/ITD | 110 (23.4) | 14 (22.6) | 96 (23.5) | > .999 |
| FLT3/TKD | 33 (7.0) | 6 (9.7) | 27 (6.6) | = .380 |
| N-RAS | 54 (11.5) | 5 (8.1) | 49 (12.0) | = .520 |
| K-RAS | 16 (3.4) | 2 (3.2) | 14 (3.4) | > .999 |
| PTPN11 | 21 (4.5) | 4 (6.5) | 17 (4.2) | > .504 |
| JAK2 | 4 (0.8) | 0 (0.0) | 4 (0.9) | > .999 |
| NPM1 | 106 (22.6) | 3 (4.8) | 103 (25.2) | < .001 |
| MLL/PTD | 28 (6.0) | 9 (14.5) | 19 (4.7) | = .006 |
| KIT | 14 (3.0) | 0 (0.0) | 14 (3.4) | .233 |
| WTI | 32 (6.8) | 2 (3.2) | 30 (7.4) | = .290 |
| Variable . | Whole cohort, n = 470 . | RUNX1-mutated patients, n = 62 . | RUNX1-wild patients, n = 408 . | P . |
|---|---|---|---|---|
| CEBPA | 66 (14.0) | 2 (3.2) | 64 (15.7) | > .006 |
| FLT3/ITD | 110 (23.4) | 14 (22.6) | 96 (23.5) | > .999 |
| FLT3/TKD | 33 (7.0) | 6 (9.7) | 27 (6.6) | = .380 |
| N-RAS | 54 (11.5) | 5 (8.1) | 49 (12.0) | = .520 |
| K-RAS | 16 (3.4) | 2 (3.2) | 14 (3.4) | > .999 |
| PTPN11 | 21 (4.5) | 4 (6.5) | 17 (4.2) | > .504 |
| JAK2 | 4 (0.8) | 0 (0.0) | 4 (0.9) | > .999 |
| NPM1 | 106 (22.6) | 3 (4.8) | 103 (25.2) | < .001 |
| MLL/PTD | 28 (6.0) | 9 (14.5) | 19 (4.7) | = .006 |
| KIT | 14 (3.0) | 0 (0.0) | 14 (3.4) | .233 |
| WTI | 32 (6.8) | 2 (3.2) | 30 (7.4) | = .290 |
Values are no. (%) of patients with alteration.
Impact of RUNX1 mutation on response to therapy and clinical outcome
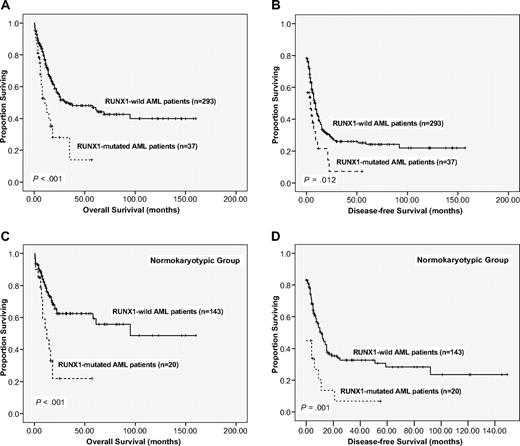
Of the 330 AML patients undergoing conventional induction chemotherapy, 248 patients (75.2%) achieved a CR. RUNX1 mutation was associated with inferior response (CR rate, 56.8% vs 77.5%, P = .009). The multivariate analysis also showed that RUNX1 mutation was an independent poor-risk factor for patients to achieve CR (hazard ratio [HR] = 0.443; 95% confidence interval [CI], 0.196-0.998, P = .050) independent from other covariates. With a median follow-up of 53 months (range, 1.0-160 months), patients with RUNX1 mutation had significantly poorer OS than those without RUNX1 mutation (median, 10.5 months vs 30.5 months, P < .001, Figure 2A). Subgroup analysis of patients with normal karyotype showed that RUNX1-mutation was also closely associated with worse OS (P = .001, Figure 2C). Similar results could be demonstrated for DFS (Figure 2B,D). There was no significant difference in survival between patients with mutations within RHD and those with mutations outside it (P = .863), and between patients with frameshift mutations and others (P = .308).
Kaplan-Meier survival curves according to RUNX1 mutation status. (A-B) A total of 330 AML patients receiving standard chemotherapy. (C-D) Those patients with normal karyotype. Data are shown for OS (A,C) and DFS (B,D). Tick marks represent patients whose data were censored at the last time they were known to be alive (A) or in CR (B) or at the time of HSCT.
Kaplan-Meier survival curves according to RUNX1 mutation status. (A-B) A total of 330 AML patients receiving standard chemotherapy. (C-D) Those patients with normal karyotype. Data are shown for OS (A,C) and DFS (B,D). Tick marks represent patients whose data were censored at the last time they were known to be alive (A) or in CR (B) or at the time of HSCT.
In multivariate analysis (Table 5), the independent poor risk factors for OS were age older than 50 years, unfavorable karyotype, RUNX1 mutation, WT1 mutation, WBC count more than 50 × 109/L, and high LDH level. CEBPA mutation and NPM1mutant/FLT3-ITD− were independent favorable prognostic factors. Multivariate analysis for DFS revealed that the independent poor risk factors for DFS included unfavorable karyotype, WT1 mutation, and age older than 50 years. There was a trend of poorer DFS in patients with RUNX1 mutation (HR = 1.426; 95% CI, 0.922-2.207, P = .111). CEBPA mutation and NPM1mutant/ FLT3-ITD− were independent favorable factors for DFS.
Multivariate analysis (Cox regression) on the DFS and OS
| Variable . | DFS, 95% CI . | OS, 95% CI . | ||||||
|---|---|---|---|---|---|---|---|---|
| RR . | Lower . | Upper . | P . | RR . | Lower . | Upper . | P . | |
| Age† | 1.520 | 1.145 | 2.017 | .004* | 2.682 | 1.867 | 3.853 | < .001* |
| Karyotype | 2.104 | 1.625 | 2.724 | < .001* | 2.325 | 1.638 | 3.300 | < .001* |
| LDH‡ | 1.203 | 0.895 | 1.616 | .221 | 1.487 | 1.018 | 2.173 | .040* |
| WBC§ | 1.336 | 0.973 | 1.834 | .073 | 1.583 | 1.077 | 2.325 | .019* |
| RUNX1 | 1.426 | 0.922 | 2.207 | .111 | 1.878 | 1.109 | 3.179 | .019* |
| CEBPA | 0.420 | 0.274 | 0.644 | < .001* | 0.423 | 0.240 | 0.746 | .003* |
| NPM1/FLT3-ITD‖ | 0.282 | 0.156 | 0.510 | < .001* | 0.276 | 0.132 | 0.578 | .001* |
| WT1 | 1.861 | 1.204 | 2.877 | .005* | 2.155 | 1.219 | 3.809 | .008* |
| Variable . | DFS, 95% CI . | OS, 95% CI . | ||||||
|---|---|---|---|---|---|---|---|---|
| RR . | Lower . | Upper . | P . | RR . | Lower . | Upper . | P . | |
| Age† | 1.520 | 1.145 | 2.017 | .004* | 2.682 | 1.867 | 3.853 | < .001* |
| Karyotype | 2.104 | 1.625 | 2.724 | < .001* | 2.325 | 1.638 | 3.300 | < .001* |
| LDH‡ | 1.203 | 0.895 | 1.616 | .221 | 1.487 | 1.018 | 2.173 | .040* |
| WBC§ | 1.336 | 0.973 | 1.834 | .073 | 1.583 | 1.077 | 2.325 | .019* |
| RUNX1 | 1.426 | 0.922 | 2.207 | .111 | 1.878 | 1.109 | 3.179 | .019* |
| CEBPA | 0.420 | 0.274 | 0.644 | < .001* | 0.423 | 0.240 | 0.746 | .003* |
| NPM1/FLT3-ITD‖ | 0.282 | 0.156 | 0.510 | < .001* | 0.276 | 0.132 | 0.578 | .001* |
| WT1 | 1.861 | 1.204 | 2.877 | .005* | 2.155 | 1.219 | 3.809 | .008* |
Statistically significant (P < .05).
Age older than 50 years relative to age 50 years or younger (reference value).
LDH greater than 920 U/L versus less than 920 U/L.
WBC greater than 50 × 109/L versus less than 50 × 109/L.
NPM1mutant/FLT3-ITD− versus other subtypes.
To confirm the prognostic significance of RUNX1 mutation in AML, we also did multivariate analysis on the subgroup of patients (n = 234) who did not undergo allogeneic HSCT. RUNX1 mutation was still an independent poor prognostic factor for OS (HR = 1.874; 95% CI, 1.075-3.269, P = .027) and a borderline poor prognostic factor for DFS (HR = 1.580; 95% CI, 0.940-2.653, P = .084) in this rather homogeneous cohort. However, among patients (n = 96) receiving allogeneic HSCT, the poor prognostic impact of RUNX1 mutation in OS and DFS was lost, and it seemed that there was a trend of better outcome in patients with RUNX1 mutation (HR = 0.465, P = .090 and HR = 0.317, P = .061, respectively). It implies that HSCT may ameliorate the poor survival impact of RUNX1 mutations. However, only 11 patients with RUNX1 mutations received HSCT in our cohort. Further study in more patients may be needed to clarify this point.
Sequential studies of RUNX1 mutations in AML patients
RUNX1 mutations were serially studied in 294 samples from 133 patients, including 18 patients with distinct RUNX1 mutations, 7 patients with insignificant RUNX1 mutations, and 108 patients without mutation at diagnosis. Among the 18 patients with distinct RUNX1 mutations, all lost their mutations at remission status (Table 1). In the 6 patients who had available samples for serial study at relapse, 2 patients (nos. 17 and 53) regained the same mutations as those detected at diagnosis. In the other 2 patients (nos. 3 and 35), one of the mutations detected at diagnosis remained, whereas the other one was lost at relapse. The remaining 2 patients (nos. 23 and 48) lost RUNX1 mutation at relapse but retained FLT3/ITD (case no. 23) or FLT3/TKD and WT1 mutation (case no. 48). In all 7 patients with insignificant mutations (nos. 63-66 and 68-70), the mutations remained the same at CR and also at relapse in the 5 patients studied, suggesting that these mutations were germline origin and might not be relevant to leukemogenesis. Among the 108 patients who had no RUNX1 mutation at diagnosis, none acquired RUNX1 mutation at relapse, whereas karyotypic evolution was noted at relapse in 41.6% of them (data not shown).
Discussion
Our results showed that RUNX1 mutation could be detected in 13.2% of adult patients with de novo non-M3 AML and was correlated with distinct clinical and biologic features. Furthermore, RUNX1 mutation was an independent risk factor for poor prognosis.
The reported incidence of RUNX1 mutation in AML varied from 2.9% to 46% depending on the patient population selected (eg, age range, history of MDS, FAB subtypes, and karyotype), the regions of RUNX1 screened, and the methods used.11,19,21,23,24,35-38 In most of the previous studies, the N-terminal exons 3 to 6 covering the RHD were selected for mutation screening based on the observation that mutations were clustered within this region in AML M0 and on the assumption that only approximately 10% of mutations were outside this region, and were seen mainly in MDS or in AML after MDS.11,20,23,24 In the present series, 31 mutations (49%) were located in the C-terminal region. This frequency was significantly higher than previously assumed. Recently, Gaidzik et al reported that 34.4% (11 of 32) of RUNX1 mutations were located in exon 8, 34.4% (11 of 32) in exon 3, and 31.2% (10 of 32) in other regions in adult AML.38 In this report, they did not distinctly mention the incidence of mutations in exons 6 and 7. More studies are necessary to clarify the true incidence of RUNX1 mutations in the C-terminal region.
Four kinds of RUNX1 mutations were demonstrated in this study: missense mutations, nonsense mutations, frameshift mutations, and in-frame mutations (Figure 1). Most RHD mutations were missense mutations (20 of 31, 64.5%), whereas most TAD mutations were of frameshift (13 of 17, 76.5%). Excluding the 5 insignificant mutations, 24 missense mutations were noted. Previous reports have verified that several residues were important for DNA binding and heterodimerization with CBF-β.39 The mutations occurring in these residues, such asR80C, R80G, K83Q, K83R, R135K, R135S, R139Q, R139L, D171Y, D171G, and R174Q detected in this study, would result in loss of hydrogen bonding interaction to DNA or disruption of the folding architecture of RUNX1; additional S114P and S114W were supposed to abolish heterodimerization with CBF-β.39 The remaining 7 RHD mutations and the 4 mutations outside the RHD region have not been reported previously but were supposed to be distinct mutations because these mutations involved residues that were highly conserved in the RUNX family (supplemental Figure 1). Of the 7 nonsense mutations, 4 mutations generated truncated RHD. The remaining 3 mutations (R178X, Q181X, and R293X), retaining the RHD region for DNA and CBF-β binding, would have a dominant negative effect on wild-type RUNX1.34 Twenty-eight mutations were frameshift mutations (Figure 1); with the exception of P375fsX570, all resulted in partial or total deletion of the TAD and loss of transactivation potential.34
In this study of a large cohort of adults with de novo non-M3 AML, we have identified distinct clinical characteristics in patients with RUNX1 mutation that either were not reported before or were different from previous studies in selected cohorts (eg, AML-M0 subtype, therapy- or MDS-related AML). We found that RUNX1-mutated patients were predominantly male, with a male-to-female ratio of 3.8, and were older than RUNX1-wild patients. The higher frequency of the leukemic cells to express CD34 and HLA-DR, but lower frequency to express CD15 in AML with RUNX1 mutations, might reflect the association of this mutation with immature subtypes of AML, M0, and M1. In previous reports, RUNX1 mutation was frequently associated with specific cytogenetic abnormalities, such as trisomies 8 (+8), +13, or +2121 ; however, we failed to find any association between RUNX1 mutation and −5/5q-, −7/7q-, +11, +13, or +21, except for +8 (Table 3). Further, RUNX1 mutation was never found in patients with t(15;17), t(8;21), or inv(16).
The role of RUNX1 mutation in the leukemogenesis of AML remains to be defined. RUNX1 mutation, a class II mutation, has been implicated as the initiating event to block differentiation of hematopoietic cells, and the subsequent class I gene mutation would synergistically provide growth advantages of these cells and lead to the development of AML.11,40 As shown in Table 4 and supplemental Table 3, 31 of the 62 RUNX1-mutated patients (50%) concurrently had other gene mutations and the majority (26 of 31, 83.9%) simultaneously showed class I mutations, most commonly FLT3/ITD, FLT3/TKD, and N-RAS, which might result in hyperactivation of the receptor tyrosine kinase-RAS signaling pathways.41 This finding was consistent with the current hypothesis of 2-hit model of leukemogenesis.5,6 However, MLL/PTD, another class II mutation, was also found in a significantly higher frequency in RUNX1-mutated patients (9 of 62, 14.5%) than in RUNX1-wild patients (19 of 408, 4.7%, P = .006). Intriguingly, 8 of the 9 patients with concurrent RUNX1 mutation and MLL/PTD also had simultaneously FLT3/ITD (7 cases) or FLT3/TKD (1 case) (supplemental Table 3). The molecular interaction among RUNX1, FLT3/ITD, and MLL/PTD deserves further investigation. Matsuno et al40 reported a higher frequency of FLT3 mutation in AML M0 patients with RUNX1 mutation (5 of 8, 63%) than in those with RUNX1-wild type (5 of 41, 12%, P = .005). Dicker et al,21 however, found that FLT3 mutations were equally distributed between RUNX1-mutated and -wild patients. In this study, although FLT3 mutation was the most frequent gene alteration cooperative with RUNX1 mutation, it occurred with similar incidence in RUNX1-mutated and -wild patients.
To the best of our knowledge, this is the first report of sequential study of RUNX1 mutation in a large number of AML patients during the clinical course. It showed that all 18 patients with distinct RUNX1 mutations studied lost the mutations at CR, indicating that these mutations were acquired in the leukemogenesis. On the other hand, all 108 patients without RUNX1 mutation at diagnosis remained RUNX1-wild at relapse. Furthermore, 2 of the 6 patients who had RUNX1 mutations at diagnosis lost the mutations at relapse (Table 1). These results suggested that, although RUNX1 mutations are important for the development of AML, they may play a small role in disease progression. Further studies are warranted to confirm the result.
So far, there has been no large cohort study to address the impact of RUNX1 mutation on treatment response and clinical outcome in primary AML. In the present study, the adult AML patients with RUNX1 mutation had inferior response to conventional chemotherapy and poorer OS and DFS than those without the mutation. The same was also true for the subgroup of patients with normal karyotype. The multivariate analysis confirmed that RUNX1 mutation was a poor prognostic factor for OS independent from other risk factors, such as age, karyotype, WBC, LDH, NPM1/FLT3-ITD, WT1, and CEBPA mutation. These results suggest that RUNX1 can be used as a new candidate molecular marker, along with NPM1/FLT3-ITD and CEBPA mutations, to stratify patients in the management of AML.
In this study, 5 types of missense mutations (G42R, D317N, Q370R, V425G, and M439L) could be detected both at diagnosis and at CR. None of these sequence alterations was recorded as polymorphisms or variants in the GenBank database,42 except for G42R, which was never reported in healthy Japanese persons.19,43 Comparison of RUNX proteins family among different species revealed that these amino acid changes might have no effect on functional alteration (supplemental Figure 1). Sequence variation in RUNX1 gene has been found not infrequently before. In normal British persons, 2 RUNX1 variations, L29S and S21syn, could be found in 5% and 3%, respectively.37 Although it was suggested that these mutations were germline mutations or variations that were functionally insignificant and might not be relevant to leukemogenesis, germline mutations of RUNX1 gene have been reported in familial platelet disorder with propensity to myeloid leukemia.44-46 However, patients with the 5 mutations mentioned did not have other hematologic diseases before, nor did their families. To clarify whether these mutations were involved in leukemogenesis, further studies, such as mutation analysis on a large cohort of normal persons or functional analyses of these mutations, need to be done.
In conclusion, this study demonstrated that RUNX1 mutations could be detected in a substantial proportion of patients with de novo AML and were closely associated with male sex, older age, immature FAB subtypes, and trisomy 8. They were mutually exclusive with CEBPA and NPM1 mutations but were closely associated with MLL/PTD. Furthermore, the RUNX1 mutation was an independent poor risk factor for OS. Sequential study during the clinical course for 133 patients showed that none of these patients acquired novel RUNX1 mutation at relapse, indicating that the mutation may play a small role in disease progression.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by the National Science Council (grant NSC 97-2314-B-002-015-MY3), Taiwan, Republic of China, and Department of Medical Research, National Taiwan University Hospital.
Authorship
Contribution: J.-L.T. and H.-A.H. were responsible for literature collection, data management and interpretation, and manuscript writing; L.-I.L. was responsible for mutation interpretation and manuscript writing; C.-Y.L was responsible for statistical analysis and interpretation of the statistical findings; C.-Y.C., W.-C.C., M.Y., S.-Y.H., B.-S.K., S.-C.H., S.-J.W., W.T., and Y.-C.C. contributed patient samples and clinical data; M.-H.T., C.-F.H., F.-Y.L., and M.-C.L. performed the laboratory research and chromosomal studies; and H.-F.T. planned, designed, wrote the manuscript, and coordinated the study over the entire period.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lang-In Lin, Department of Clinical Laboratory Sciences and Medical Biotechnology, College of Medicine, National Taiwan University, Taipei, Taiwan; e-mail: lilin@ntu.edu.tw; or Hwei-Fang Tien, Department of Internal Medicine, National Taiwan University Hospital, Taipei, Taiwan; e-mail: hftien@ntu.edu.tw.
References
Author notes
J.-L.T. and H.-A.H. contributed equally to this study and should be considered joint first authors.
L.-I.L. and H.-F.T. contributed equally to this study as the corresponding authors.