Abstract
Specific genetic alterations in multiple myeloma (MM) may cause more aggressive diseases. Paired gene array analysis on 51 samples showed that retinoic acid (RA) receptor α (RARα) expression significantly increased at relapse compared with diagnosis. RARα encodes 2 major isoforms: RARα1 and RARα2. In this study, we examined the function of RARα2 in MM. Reverse transcription–polymerase chain reaction (RT-PCR) revealed ubiquitous RARα1 expression in MM cells, but RARα2 was expressed in 26 (32%) of 80 newly diagnosed patients and 10 (28%) of 36 MM cell lines. Patients with RARα2 expression had a significantly shorter overall survival on identical treatments. The presence of RARα2 remained significant on multivariate analysis. Knockdown of RARα2 but not RARα1 induced significant MM cell death and growth inhibition, and overexpressing RARα2 activated STAT3 and mitogen-activated protein kinase kinase (MEK)/extracellular signal–regulated kinase (ERK) signaling pathways. Interestingly, all-trans retinoic acid (ATRA) treatment induced potent cell death and growth inhibition in RARα2+ but not RARα2− MM cells; overexpressing RARα2 in RARα2-deficient MM cells restored sensitivity to ATRA. Furthermore, ATRA treatment significantly inhibited the growth of RARα2-overexpressing MM tumors in severe combined immunodeficiency (SCID) mouse model. These findings provide a rationale for RA-based therapy in aggressive RARα2+ MM.
Introduction
Multiple myeloma (MM) is a plasma cell malignancy.1,2 Although high-dose therapy and tandem bone marrow autotransplantation can produce higher response rates and longer survival than standard chemotherapy, MM remains largely incurable by current therapeutic strategies.3,4 Furthermore, clinical outcomes of patients with MM are extremely heterogeneous, with survival ranging from only a few months to more than 15 years.4-6 Increasing evidence suggests that genetic heterogeneity in MM cells largely accounts for the divergent clinical outcomes.6 We have been pursuing the genetic characteristics of MM for nearly 10 years, which has contributed to the classification of MM diseases.7-11 However, it remains unknown how to transform MM into a curable disease. Therefore, identification of new targets and genetic alterations, especially those for which there are available drugs, is important.
All-trans retinoic acid (ATRA)–based regimens have been used in the therapy of acute promyelocytic leukemia (APL) for more than 20 years and have dramatically raised the 5-year disease-free survival to more than 70%. Furthermore, the toxicity of ATRA has been limited compared with conventional cytotoxic chemotherapy.12 The high efficacy of retinoic acid (RA)–based therapy in APL and its well-documented safety profile have stimulated considerable interests in the treatment of other malignancies,13-15 including myeloma.16,17 Despite many efforts, however, clinical benefits of ATRA treatment in other tumors have been minor. RARα has 2 major isoforms, RARα1 and RARα2, which differ in their expression patterns and N-terminal AF-1 functional domains.18 Both RARα1 and RARα2 are considered specific receptors for RA-based reagents. However, the specific cellular roles of RARα1 and RARα2 are still unclear. In this study, we found that RARα2 plays a crucial role in myeloma progression, and more importantly, in mediating RA-based therapy of RARα2+ MM diseases.
Methods
Study subjects
CD138+ myeloma cell samples were obtained from patients who were enrolled on the National Institutes of Health (NIH)–sponsored clinical trials UARK 98-026 (Total Therapy 2 [TT2]) and UARK 03-033 (TT3). The Institutional Review Board of the University of Arkansas for Medical Sciences approved the research studies, and all subjects provided written informed consent in accordance with the Declaration of Helsinki.
We have an inventory of more than 30 myeloma cell lines in our laboratory. Cells from these cell lines were cultured in RPMI 1640 containing 10% heat-inactivated fetal calf serum (FCS), 2 mM l-glutamine (Gibco), penicillin (100 U/mL), and streptomycin (100 μg/mL) in a humidified incubator at 37°C in 5% CO2.
Gene expression profiling
Plasma cell purifications and gene expression profiling, using the Affymetrix U133Plus2.0 microarray, were performed as previously described.9,11 Signal intensities were preprocessed and normalized by GCOS1.1 software (Affymetrix).9,11 Myeloma cell samples from 80 newly diagnosed patients with myeloma were used, including 54 patients without RARα2 and 26 patients with RARα2.
RT-PCR
Regular reverse transcriptase–polymerase chain reaction (RT-PCR) was used to detect RARα1 and RARα2 expression in MM cells. MM cell samples from a total of 80 patients newly diagnosed with MM and 36 cell lines were examined. Detailed protocol was described previously.19 Primer sets used for these analyses were as follows: RARα1 forward, 5′-CCA TCT AGG AGT GGC ATC TTT T, reverse, 5′-ACT AAC TGG AAG CTG GTG CTG; RARα2 forward, 5′-GAG GAG GAA GTG CTC GTT GAC, reverse, 5′-GCC CGG TTC TGG TTA TAA AAA T; and GAPDH forward, 5′-GAC ACC CAC TCC TCC ACC T, reverse, 5′-ATG AGG TCC ACC ACC CTG T. GAPDH transcript levels were used to normalize the amount of cDNA in each sample.
Specific gene silencing or overexpressing by lentivirus expression vector system
Specific gene knockdown was previously described.20,21 Similar methods are used to overexpress RARα2 by this system. Two synthetic double-stranded oligonucleotide sequences specific for RARα2 silencing (5′-GAT CCC CGC TTT GGA ATG GCT CAA ACC ATT CAA GAG ATG GTT TGA GCC ATT CCA AAG CTT TTT A-3′ and 5′-AGC TTA AAA AGC TTT GGA ATG GCT CAA ACC ATC TCT TGA ATG GTT TGA GCC ATT CCA AAG CGG G-3′) were obtained from OligoEngine. A scrambled oligonucleotide was used as a control. Primers used for RARα2 cDNA clones (forward, 5′-ATG TAC GAG AGT GTA GAA GT, and reverse, 5′-CGG TAG AAA GGC AGA GAA AA) were purchased from Invitrogen. RARα2 short hairpin RNA (shRNA) double-stranded oligonucleotides and RARα2 cDNA were cloned into lentiviral vectors (kindly provided by Dr Didier Trono, School of Life Sciences and Frontier Genetics, National Center for Competence in Research, Lausanne, Switzerland). Recombinant lentivirus were produced by transient transfection of 293T cells following a standard protocol.21 Efficiency of viral transfection was determined by counting the number of green fluorescent protein (GFP)–expressing cells using flow cytometry, and 95% transduction efficiency of MM cells was achieved.
Treatment of MM cells with ATRA
RARα2+ (ARK, KMS11, and OPM2) and RARα2− cells (ARP1, OCI-MY5, CAG, and XG1) were maintained in RPMI 1640 with 10% fetal calf serum (FCS). Cells were continuously treated for 4 days with ATRA at 10−6 M (Sigma-Aldrich). Cultured cells were counted with a hemocytometer to evaluate cellular proliferation, and dead cells were determined by trypan blue staining, from which a dead cell fraction was calculated.
To determine the role of RARα2 in ATRA-induced MM cell death and growth inhibition, RARα2− ARP1, OCI-MY5, and XG-1 cells were transfected with RARα2 cDNA to overexpress RARα2 in these cell lines by lentivirus expression vector system. Cells were continuously treated for 4 days with ATRA at 10−6 M in the cultures, and cell proliferation and cell death were evaluated as described in the previous paragraph.
Western blots
Western blots were performed to examine the protein levels in MM cells as previously described.20 All primary antibodies were purchased from Cell Signaling Technology. β-actin was used to normalize the amount of proteins in each sample.
ATRA treatment of myeloma xenograft tumors in SCID mice
Male C.B-17/IcrHsd severe combined immunodeficiency (SCID) mice, 5 to 6 weeks old, were purchased from Harlan Sprague Dawley. Experiments were under an approved protocol of the Institutional Animal Care and Use Committee of the University of Utah. To examine the therapeutic effects of ATRA on the growth of RARα2-overexpressing myeloma tumors, RARα2-transfected OCI-MY5 cells were injected subcutaneously into mice (n = 5) at a dose of 2 × 106 cells/mouse; myeloma tumors were allowed to develop for 10 days, and ATRA (10 mg/kg, 3 times per week) was given subcutaneously around myeloma tumors in 100 μL of phosphate-buffered saline (PBS) for 3 weeks. ATRA-treated tumors developed from empty vector (EV)–transfected myeloma cells and tumors developed from parent OCI-MY5 cells receiving PBS alone will be used as controls. Tumor size was measured twice a week in 2 dimensions using a caliper, and tumor volume (mm3) was calculated as 4π/3 × (tumor diameter/2).3 Mice were humanely killed by CO2 asphyxiation when moribund or when subcutaneous tumors reached 15 mm in diameter. Survival was evaluated from the day of tumor inoculation until death.
Statistical analysis
Overall survival (OS) distributions of patients with myeloma with or without RARα2 expression were estimated by the Kaplan-Meier method. Survival of mice was evaluated from the day of tumor inoculation until death, and the Kaplan-Meier test was used to compare survival between the groups. One-way analysis of variance (> 3 groups) and the Student t test (2 groups) were used to compare various experimental groups. Significance was set at a P value less than .05.
Results
RARα2 expression is associated with inferior survival and myeloma progression
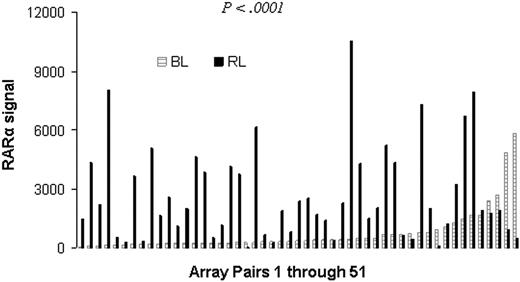
In an analysis of genes differentially expressed in paired MM cell samples collected at baseline (at diagnosis) and at relapse after transplantation, we discovered that RARα expression was significantly higher in myeloma cells at relapse (P < .001). Among the 51 paired arrays, 37 showed higher RARα gene expression at relapse (Figure 1). This result indicates that high RARα expression is associated with more aggressive MM.
Increased RARα expression in relapsed MM. Paired CD138+ MM cell samples were obtained at diagnosis and relapse from 51 patients diagnosed with MM. Gene expression profiling analysis were performed on these samples. Shown is the RARα signal at diagnosis (BL) and relapse (RL). Note that RARα expression is significantly increased at relapse compared with this at diagnosis (paired t test, P < .001).
Increased RARα expression in relapsed MM. Paired CD138+ MM cell samples were obtained at diagnosis and relapse from 51 patients diagnosed with MM. Gene expression profiling analysis were performed on these samples. Shown is the RARα signal at diagnosis (BL) and relapse (RL). Note that RARα expression is significantly increased at relapse compared with this at diagnosis (paired t test, P < .001).
To investigate gene expression patterns of RARα1 and RARα2 in MM, we used RT-PCR to examine their mRNA levels in MM cells obtained from newly diagnosed patients and in MM cell lines (Figure 2A-B). Samples from a total of 80 patients enrolled on TT2 and TT3 clinical trials were examined, and RARα1 mRNA was detected in all of the samples, while RARα2 was detected in only 26 (32.5%; see supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). A total of 36 MM cell lines were examined. Again, RARα1 mRNA was detected in all of the cell lines; while RARα2 was expressed in only 10 (28%; see supplemental Table 2). These results demonstrate that RARα2 was expressed in only a fraction of patients with MM (∼30%).
RARα2 expression is associated with poor prognosis in MM. RT-PCR detected the expression of RARα1, RARα2, and GAPDH in MM cells from patients (P1-P4; A) and MM cell lines (B). Representative results are shown. (C) Kaplan-Meier analysis of OS of patients newly diagnosed with RARα2+ (n = 26) and RARα2− (n = 54) MM in TT2 and TT3 trials. (D) RT-PCR was used to examine RARα2 expression in RARα2 knockdown MM cells (R2−). Wild-type (WT) and nontargeting scramble shRNA (SCR)–transfected cells were used as controls; RARα2 knockdown MM cells were cultured for 4 days, live cells were counted to calculate cell proliferation (E), and dead cells were determined by positive trypan blue staining, from which the dead-cell fraction was calculated (F). Results are expressed as means plus or minus SD of 3 independent experiments. *P < .05. (G) Western blot analysis of caspase-3, caspase-8, and caspase-9 (cleaved and uncleaved proteins) in WT, SCR, and R2− cells of ARK, KMS11, and OPM2 MM cells. Cells were collected at day 3 in the cultures. β-actin was used as loading control. Shown are the representative results of 3 independent experiments.
RARα2 expression is associated with poor prognosis in MM. RT-PCR detected the expression of RARα1, RARα2, and GAPDH in MM cells from patients (P1-P4; A) and MM cell lines (B). Representative results are shown. (C) Kaplan-Meier analysis of OS of patients newly diagnosed with RARα2+ (n = 26) and RARα2− (n = 54) MM in TT2 and TT3 trials. (D) RT-PCR was used to examine RARα2 expression in RARα2 knockdown MM cells (R2−). Wild-type (WT) and nontargeting scramble shRNA (SCR)–transfected cells were used as controls; RARα2 knockdown MM cells were cultured for 4 days, live cells were counted to calculate cell proliferation (E), and dead cells were determined by positive trypan blue staining, from which the dead-cell fraction was calculated (F). Results are expressed as means plus or minus SD of 3 independent experiments. *P < .05. (G) Western blot analysis of caspase-3, caspase-8, and caspase-9 (cleaved and uncleaved proteins) in WT, SCR, and R2− cells of ARK, KMS11, and OPM2 MM cells. Cells were collected at day 3 in the cultures. β-actin was used as loading control. Shown are the representative results of 3 independent experiments.
To validate the function of RARα2, we compared clinical outcomes of RARα2+ and RARα2− cases in 80 patients enrolled on our TT2 and TT3 clinical trials. As shown in Figure 2C, patients with RARα2 expression had a shorter OS than those without RARα2 expression (P = .0038). Patients with RARα2 expression also had other clinical and biological features associated with more aggressive myeloma, such as higher levels of lactate dehydrogenase (P = .043), more cytogenetic abnormalities (P = .004), lower TP53 expression (P < .001), and higher risk score defined by the 70-gene model (P < .001; Table 1). On multivariate analysis, however, RARα2 gene expression remained an independent factor associated with poor prognosis for both event-free survival (EFS) and OS (Table 1).
Univariate and multivariate analysis of clinical characteristics affecting EFS and OS by RARα 2 expression in TT2 and TT3
| Variable . | n/N (%) . | EFS . | OS . | ||
|---|---|---|---|---|---|
| HR (95% CI) . | P . | HR (95% CI) . | P . | ||
| Univariate | |||||
| Age ≥ 65 y | 0.75 (0.29, 1.97) | .562 | 0.93 (0.31, 2.76) | .889 | |
| LDH ≥ 190 U/L | 22/80 (28) | 2.27 (1.11, 4.64) | .024 | 2.38 (1.03, 5.52) | .043 |
| B2M ≥ 0.3 μM | 1.32 (0.66, 2.65) | .433 | 1.48 (0.64, 3.41) | .362 | |
| B2M > 0.47 μM | 1.21 (0.50, 2.94) | .678 | 1.26 (0.43, 3.73) | .674 | |
| Albumin < 35 g/dL | 0.70 (0.25, 2.01) | .512 | 0.85 (0.25, 2.87) | .793 | |
| Creatinine ≥ 176.8 μM | 1.53 (0.53, 4.40) | .427 | 1.14 (0.27, 4.91) | .859 | |
| CRP ≥ 8.0 mg/L | 1.32 (0.62, 2.83) | .473 | 1.29 (0.52, 3.19) | .588 | |
| Hb < 100 g/L | 1.60 (0.76, 3.37) | .213 | 1.31 (0.52, 3.28) | .561 | |
| Cytogenetic abnormalities | 29/79 (37) | 2.25 (1.09, 4.65) | .028 | 3.62 (1.51, 8.68) | .004 |
| RARα2+ | 26/80 (33) | 2.13 (1.06, 4.27) | .034 | 3.33 (1.42, 7.81) | .006 |
| 210764_at < 738 | 10/80 (13) | 3.80 (1.61, 8.94) | .002 | 4.74 (1.79, 12.52) | .002 |
| 70-gene–defined GEP high risk | 5/80 (6) | 5.26 (1.73, 15.97) | .003 | 7.59 (2.36, 24.43) | < .001 |
| Multivariate | |||||
| Cytogenetic abnormalities | 29/79 (37) | 2.67 (1.26, 5.64) | .01 | 4.41 (1.75,11.10) | .002 |
| RARα2+ | 26/79 (33) | 2.22 (1.07, 4.58) | .032 | 3.19 (1.32, 7.68) | .01 |
| 210764_at < 738 | 10/79 (13) | 3.66 (1.50, 8.91) | .004 | 3.93 (1.43, 10.84) | .008 |
| Variable . | n/N (%) . | EFS . | OS . | ||
|---|---|---|---|---|---|
| HR (95% CI) . | P . | HR (95% CI) . | P . | ||
| Univariate | |||||
| Age ≥ 65 y | 0.75 (0.29, 1.97) | .562 | 0.93 (0.31, 2.76) | .889 | |
| LDH ≥ 190 U/L | 22/80 (28) | 2.27 (1.11, 4.64) | .024 | 2.38 (1.03, 5.52) | .043 |
| B2M ≥ 0.3 μM | 1.32 (0.66, 2.65) | .433 | 1.48 (0.64, 3.41) | .362 | |
| B2M > 0.47 μM | 1.21 (0.50, 2.94) | .678 | 1.26 (0.43, 3.73) | .674 | |
| Albumin < 35 g/dL | 0.70 (0.25, 2.01) | .512 | 0.85 (0.25, 2.87) | .793 | |
| Creatinine ≥ 176.8 μM | 1.53 (0.53, 4.40) | .427 | 1.14 (0.27, 4.91) | .859 | |
| CRP ≥ 8.0 mg/L | 1.32 (0.62, 2.83) | .473 | 1.29 (0.52, 3.19) | .588 | |
| Hb < 100 g/L | 1.60 (0.76, 3.37) | .213 | 1.31 (0.52, 3.28) | .561 | |
| Cytogenetic abnormalities | 29/79 (37) | 2.25 (1.09, 4.65) | .028 | 3.62 (1.51, 8.68) | .004 |
| RARα2+ | 26/80 (33) | 2.13 (1.06, 4.27) | .034 | 3.33 (1.42, 7.81) | .006 |
| 210764_at < 738 | 10/80 (13) | 3.80 (1.61, 8.94) | .002 | 4.74 (1.79, 12.52) | .002 |
| 70-gene–defined GEP high risk | 5/80 (6) | 5.26 (1.73, 15.97) | .003 | 7.59 (2.36, 24.43) | < .001 |
| Multivariate | |||||
| Cytogenetic abnormalities | 29/79 (37) | 2.67 (1.26, 5.64) | .01 | 4.41 (1.75,11.10) | .002 |
| RARα2+ | 26/79 (33) | 2.22 (1.07, 4.58) | .032 | 3.19 (1.32, 7.68) | .01 |
| 210764_at < 738 | 10/79 (13) | 3.66 (1.50, 8.91) | .004 | 3.93 (1.43, 10.84) | .008 |
P values are from the Wald χ2 test in Cox regression. All univariate P values are reported regardless of significance. Multivariate model uses stepwise selection with entry level .1, and variable remains if it meets the .05 level. A multivariate P value greater than .05 indicates variable forced into model with significant variables chosen using stepwise selection.
HR indicates hazard ratio; 95% CI, 95% confidence interval; LDH, lactate dehydrogenase; B2M, β2-microglobulin; CRP, C-reactive protein; Hb, hemoglobin; and GEP, gene expression profiling.
To examine the function of both RARα1 and RARα2 in MM cells, specific shRNAs (using the lentivirus shRNA expression vector system) were used to knock down RARα1 or RARα2 in MM cells. RARα2+ (ARK, KMS11, and OPM2) and RARα2− (KMS28PE and XG1) MM cell lines were transfected with RARα2-specific shRNAs. RARα2 gene expression was confirmed by RT-PCR (Figure 2D); cell proliferation and cell death were examined in the cultures at day 4. RARα2 knockdown induced significant cell growth inhibition (Figure 2E) and cell death (Figure 2F) in RARα2+ ARK, KMS11, and OPM2 cells, but not in RARα2-deficient KMS28PE and XG1 cells (Figure 2E-F). However, knockdown of RARα1 in myeloma cells (including RARα2+ and RARα2− cells) did not induce cell death or proliferation inhibition (data not shown). To examine the apoptotic mechanism by which RARα2 knockdown induced cell death, RARα2-silenced ARK, KMS11, and OPM2 cells were cultured for 3 days. As shown in Figure 2G, increased levels of cleaved caspase-3, caspase-8, and caspase-9 were observed by Western blots in RARα2-knockdown MM cells compared with wild-type and scrambled-transfected control cells, suggesting both death receptor–dependent and –independent apoptotic pathways may be involved. These results demonstrate that RARα2, but not RARα1, plays an important role in MM cell growth and survival.
Identification of molecular pathways associated with RARα2 expression
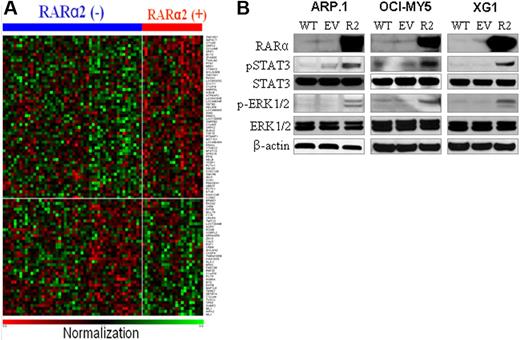
To investigate RARα2-induced signals in MM cells, we performed gene expression profiling analysis of MM cell samples from the 80 patients (including 26 RARα2+ and 54 RARα2− samples). A total of 96 genes were significantly differentially expressed in RARα2+ cell samples compared with RARα2− samples (P < .05), including 56 up-regulated and 40 down-regulated genes (Figure 3A, supplemental Table 3). Among the down-regulated genes, TP53 is a well-known tumor-suppressor gene, the low expression of which was associated with MM progression.22 Among the up-regulated genes, SPP1 was reported to enhance MM cell survival, migration, angiogenesis, and bone destruction.23-26 Melanoma 2 (AIM2) was recently shown to be 1 of the 70 high-risk signature genes in MM,11 and transcription factor Dp-1 (TFDP1) was reported to facilitate cell-cycle progression.27-30 High expression of RAS p21 protein activator 1 (RASA1) in myeloma cells from patients with RARα2+ myeloma suggested that Ras-Raf–mitogen-activated protein kinase kinase (MEK)–extracellular signal–regulated kinase (ERK) signaling pathways may be the RARα2-mediated downstream signaling pathway. Recently, investigation reported that activation of Ras induced activation of STAT3 signaling pathway.31,32 To confirm the results, we overexpressed RARα2 in RARα2-deficient cells (ARP.1, OCI-MY5, and XG1) by lentivirus-mediated RARα2 cDNA transfection (Figure 3B), and Western blots were applied to evaluate the protein levels of the main molecules in MEK/ERK and STAT3 signaling pathways. As shown in Figure 3B, increased phosphorylated (p) ERK1/2 and pSTAT3 were detected in RARα2-overexpressing myeloma cells compared with wild-type and EV-transfected control cells, demonstrating that both MEK/ERK and STAT3 signaling pathways were involved. It is well-documented that JAK/STAT3 and MEK/ERK signaling pathways play a crucial role in myeloma tumor cell survival, tumor development, and progression.33-36 Thus, these results strongly suggest that STAT3 and MEK/ERK signaling pathways are the target signaling pathways associated with high RARα2-induced MM cell growth and survival.
Identification of RARα2-associated signals. (A) Heat maps of the 96 differentially expressed genes in RARα2− (n = 54) and RARα2+ (n = 26) MM cell samples from the 80 patients. Gene symbols are plotted at the right side, and samples are plotted along the top horizontal axis. Red indicates up-regulation; and blue indicates down-regulation. (B) Western blots examined pSTAT3 and STAT3 and p-RK and ERK in RARα2-overexpressing (R2) cells of ARP.1, OCI-MY5, and XG1 cells. Wild-type (WT) and EV cells were used as controls. β-actin was used as loading control. Shown are the representative results of 3 independent experiments.
Identification of RARα2-associated signals. (A) Heat maps of the 96 differentially expressed genes in RARα2− (n = 54) and RARα2+ (n = 26) MM cell samples from the 80 patients. Gene symbols are plotted at the right side, and samples are plotted along the top horizontal axis. Red indicates up-regulation; and blue indicates down-regulation. (B) Western blots examined pSTAT3 and STAT3 and p-RK and ERK in RARα2-overexpressing (R2) cells of ARP.1, OCI-MY5, and XG1 cells. Wild-type (WT) and EV cells were used as controls. β-actin was used as loading control. Shown are the representative results of 3 independent experiments.
ATRA treatment induces cell death and growth inhibition selectively in RARα2+ MM cells
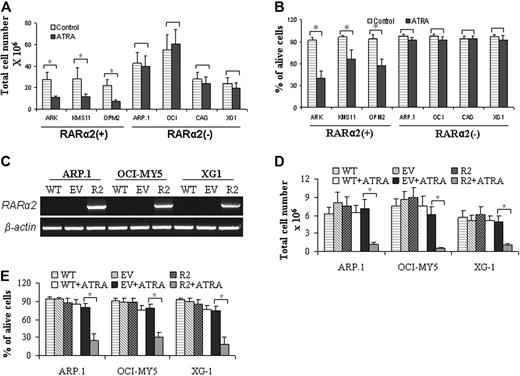
We subsequently evaluated the effects of ATRA treatment on the growth of RARα2+ and RARα2− MM cells. RARα2+ (ARK, KMS11, and OPM2) and RARα2− (ARP1, OCI-MY5, CAG, and XG-1) MM cells were treated for 4 days with ATRA (10−6 M) in the cultures. Cell proliferation and death were evaluated. ATRA treatment induced significant cell growth inhibition (Figure 4A; P < .05) and cell death (Figure 4B; P < .05) of RARα2+ MM cell lines when compared with untreated control cells. In contrast, however, ATRA treatment exhibited minimal or no effects on cell proliferation inhibition and cell death in RARα2− MM cells (Figure 4A-B; P > .05). These results suggest that RARα2 plays a crucial role in ATRA-induced MM cell death and growth inhibition.
ATRA treatment selectively induced cell growth inhibition and death in RARα2+ MM cells. RARα2+ ARK, KMS11, and OPM2 cells, and RARα2− ARP.1, OCI-MY5, CAG, and XG1 cells were treated with ATRA (10−6 M) for 4 days in the cultures. Cell proliferation (A) and death (B) were evaluated. Untreated cells were used as controls. Results are expressed as means plus SD of 3 independent experiments. *P < .05. (C) RT-PCR examined RARα2 mRNA in RARα2-overexpressing cells (R2). Wild-type (WT) and EV-transfected cells were used as controls; RARα2-overexpressing ARP.1, OCI-MY5, and XG1 cells were treated with ATRA (10−6 M) for 4 days in the cultures. Cell proliferation (D) and cell death (E) were evaluated. WT and EV cells with or without ATRA treatment were used as controls. R2 cells without ATRA treatment were also used as control. Results are expressed as means SD of 3 independent experiments. *P < .05.
ATRA treatment selectively induced cell growth inhibition and death in RARα2+ MM cells. RARα2+ ARK, KMS11, and OPM2 cells, and RARα2− ARP.1, OCI-MY5, CAG, and XG1 cells were treated with ATRA (10−6 M) for 4 days in the cultures. Cell proliferation (A) and death (B) were evaluated. Untreated cells were used as controls. Results are expressed as means plus SD of 3 independent experiments. *P < .05. (C) RT-PCR examined RARα2 mRNA in RARα2-overexpressing cells (R2). Wild-type (WT) and EV-transfected cells were used as controls; RARα2-overexpressing ARP.1, OCI-MY5, and XG1 cells were treated with ATRA (10−6 M) for 4 days in the cultures. Cell proliferation (D) and cell death (E) were evaluated. WT and EV cells with or without ATRA treatment were used as controls. R2 cells without ATRA treatment were also used as control. Results are expressed as means SD of 3 independent experiments. *P < .05.
To determine the role of RARα2 in ATRA-induced MM cell growth inhibition and death, RARα2 overexpression in ARP.1, OCI-MY5, and XG1 cells were used for ATRA treatment. RT-PCR confirmed the RARα2 overexpression (Figure 4C). RARα2-overexpressing cells were treated with ATRA for 4 days in the cultures. ATRA treatment potently inhibited cell growth (Figure 4D) and induced cell death (Figure 4E) in the RARα2-transfected cells, but not in wild-type or EV-transfected cells (P < .05). These results provide direct evidence that RARα2 plays a crucial role in ATRA-induced MM cell death and growth inhibition.
Identification of ATRA-induced signaling pathways in RARα2+ MM cells
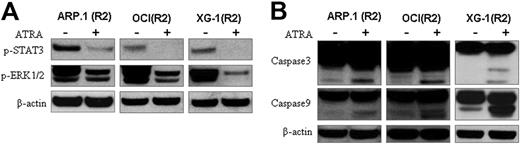
We next examined the effects of ATRA treatment on the activation of the potential RARα2-mediated signaling pathways. RARα2-overexpressing ARP.1, OCI-MY5, and XG-1 cells were treated with ATRA for 24 hours; cell lysate was prepared and Western blots were used. As shown in Figure 5A, ATRA treatment decreased pSTAT3 and pERK1/2, indicating that ATRA treatment inhibited the JAK/STAT3 and MEK/ERK signaling pathways. To examine apoptotic pathways associated with ATRA-induced cell death of RARα2+ myeloma cells, RARα2-overexpressing ARP.1, OCI-MY5, and XG-1 cells were treated with ATRA for 3 days. As shown in Figure 5B, increased levels of cleaved caspase-3 and caspase-9 were observed in ATRA-treated MM cells compared with untreated cells, indicating that caspase-dependent apoptotic signaling pathways were involved in ATRA-induced myeloma cell death in RARα2+ MM cells.
Identification of ATRA-induced signals in RARα2+ MM cells. (A) Western blots examined pSTAT3 and pERK1/2 in RARα2 (R2)–overexpressing ARP.1, OCI-MY5 (OCI), and XG-1 cells with (+) or without (−) ATRA treatment. (B) Western blot analysis of caspase-3 and caspase-9 in R2-overexpressing ARP.1, OCI, and XG-1 cells with (+) or without (−) ATRA treatment. β-actin was used as loading control. Shown are the representative results of 3 independent experiments.
Identification of ATRA-induced signals in RARα2+ MM cells. (A) Western blots examined pSTAT3 and pERK1/2 in RARα2 (R2)–overexpressing ARP.1, OCI-MY5 (OCI), and XG-1 cells with (+) or without (−) ATRA treatment. (B) Western blot analysis of caspase-3 and caspase-9 in R2-overexpressing ARP.1, OCI, and XG-1 cells with (+) or without (−) ATRA treatment. β-actin was used as loading control. Shown are the representative results of 3 independent experiments.
ATRA treatment inhibited the growth of xenograft tumors developed from RARα2-overexpressing myeloma cells in SCID mice
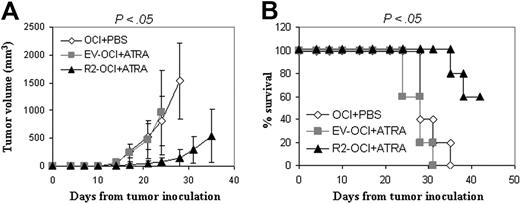
We examined the effects of ATRA treatment on the growth of RARα2+ myeloma tumors in the SCID mouse model. RARα2-overexpressing OCI-MY5 myeloma cells were injected subcutaneously into mice (n = 5); tumors were allowed to develop for 10 days, and ATRA treatment (10 mg/kg subcutaneously) was given 3 times per week for 3 weeks. As shown in Figure 6A, ATRA treatment significantly inhibited the growth of RARα2+ myeloma tumors (P < .05) compared with untreated OCI-MY5 control tumors, whereas ATRA treatment induced minimal or no inhibitory effects on EV-transfected myeloma tumors (P > .05). ATRA treatment significantly improved the survival of mice with RARα2+ myeloma tumors compared with untreated mice (P < .05), whereas no significant difference was observed between mice with ATRA-treated EV-transfected tumors and untreated wild-type tumors (P > .05; Figure 6B). These results demonstrated the inhibitory effects of RA-based reagents on RARα2+ MM diseases.
Efficacy of ATRA in treating the RARα2-overexpressing MM tumors in SCID mice. Mice (n = 5 in each group) were challenged with 2 × 106 OCI-MY5 (OCI), EV, or RARα2 (R2)–transfected cells. At 10 days following tumor inoculation, ATRA treatment was given to EV-transfected (EV-OCI + ATRA) and R2-transfected (R2-OCI + ATRA) MM tumors 3 times per week for 3 weeks; OCI tumors receiving PBS (OCI + PBS) were used as controls. (A) Tumor volumes of mice following tumor injection. Results are expressed as means plus SD. (B) Survival curve of mice following tumor injection. Shown are the representative results of 2 independent experiments.
Efficacy of ATRA in treating the RARα2-overexpressing MM tumors in SCID mice. Mice (n = 5 in each group) were challenged with 2 × 106 OCI-MY5 (OCI), EV, or RARα2 (R2)–transfected cells. At 10 days following tumor inoculation, ATRA treatment was given to EV-transfected (EV-OCI + ATRA) and R2-transfected (R2-OCI + ATRA) MM tumors 3 times per week for 3 weeks; OCI tumors receiving PBS (OCI + PBS) were used as controls. (A) Tumor volumes of mice following tumor injection. Results are expressed as means plus SD. (B) Survival curve of mice following tumor injection. Shown are the representative results of 2 independent experiments.
Discussion
Our study demonstrates the crucial role of RARα2 in myeloma cell growth, survival, and disease progression, and the surprising effects of ATRA on RARα2+ myeloma. Using paired array analysis, we showed that RARα expression significantly increased at relapse compared with diagnosis. RARα encodes 2 major isoforms: RARα1 and RARα2. RT-PCR detected ubiquitous RARα1 expression in MM cells, but RARα2 in cells of approximately 30% of newly diagnosed patients (N = 80) and MM cell lines (N = 36) analyzed. Patients with RARα2 expression had a shorter OS with identical treatments. Knockdown of RARα2 but not RARα1 induced potent MM cell death and growth inhibition. Overexpressing RARα2 in MM cells activated STAT3 and MEK/ERK signaling pathways. These results demonstrate the crucial role of RARα2, but not RARα1, in MM cell growth and disease progression. Interestingly, ATRA treatment induced potent cell death and growth inhibition in RARα2+ but not RARα2− MM cells, and overexpressing RARα2 in RARα2-deficient MM cells restored their sensitivity to ATRA treatment. Furthermore, ATRA treatment significantly inhibited the growth of xenograft tumors in SCID mice developed from RARα2-overexpressing MM cells. Therefore, our study provides a rationale for RA-based therapy in aggressive RARα2+ MM diseases.
The major RARα isoforms, RARα1 and RARα2, differ in the N-terminal AF-1 functional domains.18 However, the specific functional roles of RARα1 and RARα2 in cell growth, differentiation, and survival are defined neither in normal nor in tumor cells.18 In this study, we found that RARα1 was ubiquitously expressed in all MM cells, and RARα2 was expressed in cells from about 30% of patients with MM. Our findings are consistent with the previous investigation showing the distinct expression patterns of RARα1 and RARα2 in normal tissues.18 We also showed that patients with RARα2 expression had shorter OS in clinical trials and that knockdown of RARα2 but not RARα1 induced potent MM cell death and growth inhibition; furthermore, overexpressing RARα2 in MM cells activated STAT3 and MEK/ERK signaling pathways, which are well documented as having a crucial role in tumor cell survival, tumor development, and progression.33-36 Interestingly, a recent investigation also showed that RARα was associated with oncogenic activities in a mouse model.37 Based on these observations, especially the distinct expression patterns of RARα1 and RARα2 in MM cells, it is plausible to conclude that it is RARα2 but not RARα1 that may play crucial roles in MM cell growth and survival, and MM development and progression.
We found that RARα2 expression activates both JAK/STAT and MEK/ERK signaling pathways in MM cells; the underlying mechanisms still remain unclear. However, it was previously reported that IL-6–induced STAT3 activation and surfactant protein expression required the protein-protein interaction between STAT3 and RARα in the primary alveolar type II epithelial cells in the lung tissue.38 Fusion proteins of PML-RARα, STAT5b-RARα, and NuMa-RARα were shown to enhance STAT3 transcriptional activity in acute promyelocytic leukemia tumor cells.39-41 These observations suggested that RARα2 might be related to the activation of STAT3 signaling in other cells. Although ATRA was shown to inhibit MEK/ERK pathways,42,43 the ligand-independent effects of RARα1 and/or RARα2 on MEK/ERK signaling pathways were not reported previously. Therefore, further investigations are necessary to define the biological functions of RARα2 in MM and other tumor malignancies.
Both RARα1 and RARα2 are receptors for RA-based differentiating reagents, including ATRA. However, the specific functional roles of RARα1 and RARα2 in mediating RA-induced cell growth and differentiation are still not defined. In this study, we found that ATRA treatment selectively killed RARα2+ MM cells with minimal or no effects on RARα2− MM cells; furthermore, overexpressing RARα2 in RARα2− MM cells restore their sensitivity to ATRA treatment; importantly, ATRA treatment significantly inhibited the growth of xenograft tumors developed from RARα2-overexpressing MM cells in SCID mice. These results were consistent with previous investigations showing that ATRA treatment induced potent inhibition on OPM-2 MM cell growth,44,45 but no significant effects on U266 and L363 cells, which were detected as RARα2− cells in our study.46 Based on these observations, we conclude that RARα2 expression played a crucial role in mediating ATRA-induced MM cell death and growth inhibition.
In this study, we found that RARα2 expression was highly related to poor prognosis in TT2 and TT3 clinical trials. Nonetheless, our previous investigation reported 70 high-risk genes in MM in which RARα2 was not included.11 Since the 70 high-risk genes were discovered by gene array analysis, and the gene array can only detect the total signal of RARα (including RARα1 and RARα2), it is plausible to assume that the ubiquitous expression of RARα1 in MM cells has masked the signal of RARα2 in the gene array analysis and prevented RARα2 from being detected as a high-risk gene in MM diseases.
Even though ATRA treatment induced significant inhibition on RARα2-overexpressing MM tumors in the mouse model, this inhibition is relatively lower compared with the dramatic antimyeloma activities in vitro. Reasons may include the following: (1) since xenograft MM tumors are solid tumors, it is difficult for ATRA to infiltrate into the tumor microenvironment to get enough drug concentration for the induction of antitumor activities; (2) we found that ATRA treatment significantly increased cytochrome P450 enzymes (data not shown), which suggest increased degradation of ATRA in cells; and (3) we also found that ATRA treatment decreased the protein levels of RARα2 in MM cells (data not shown), suggesting decreased sensitivities of MM cells to the ATRA treatment. Therefore, further screening of more stable and more effective RA-based reagents and further investigation of new strategies to increase the sensitivities of MM cells to the treatment of RA-based reagents are needed for the clinical uses of RA-based reagents in MM therapy.
In conclusion, this study demonstrates that RARα2 played a crucial role in MM cell growth and survival and disease progression and, more importantly, in mediating ATRA-induced antimyeloma activities. We believe that this study is the first to show that RARα2 plays a crucial role in MM biology and in mediating ATRA-induced antimyeloma activities. Further preclinical studies are needed to examine the potential and efficacy of RA-based differentiating agents in the therapy of RARα2+ MM diseases, which do not benefit from current therapeutic strategies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health grants PO1 CA55819 (J.D.S., F.Z., B.B.) and RO1 CA115399 (G.T., F.Z.), institutional start-up funds from the University of Utah (F.Z.), and a Senior Award from the Multiple Myeloma Research Foundation (F.Z.).
National Institutes of Health
Authorship
Contribution: F.Z., J.D.S., and S.W. designed the study; S.W., F.Z., and G.T. wrote the paper; S.W., F.Z., L.S., W.X., Z.Z., and H.X. performed the research and analyzed the data; and B.B., G.T., and M.Z. provided patient samples and critical suggestions.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Fenghuang Zhan, Division of Hematology/BMT/Myeloma, University of Utah School of Medicine, Huntsman Cancer Institute, University of Utah, 30 North 1900 East, Salt Lake City, UT 84132; e-mail: fenghuang.zhan@hsc.utah.edu; or John D. Shaughnessy Jr, Myeloma Institute for Research and Therapy, University of Arkansas for Medical Sciences, 4301 W Markham, Little Rock, AR; e-mail: shaughnessyjohn@uams.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal