T-cell development follows a defined set of stage-specific differentiation steps. However, molecular and cellular events occurring at early stages of human T-cell development remain to be fully elucidated. To address this, human umbilical cord blood (UCB) hematopoietic stem cells (HSCs) were induced to differentiate to the T lineage in OP9-DL1 cocultures. A developmental program involving a sequential and temporally discrete expression of key differentiation markers was revealed. Quantitative clonal analyses demonstrated that CD34+CD38− and CD34+CD38lo subsets of UCB contain a similarly high T-lineage progenitor frequency, whereas the frequency in CD34+CD38+/hi cells was 5-fold lower. Delta-like/Notch-induced signals increased the T-cell progenitor frequency of CD34+CD38−/lo cells differentiated on OP9-DL1, and 2 distinct progenitor subsets, CD34+CD45RA+CD7++CD5−CD1a− (proT1) and CD34+CD45RA+CD7++CD5+CD1a− (proT2), were identified and their thymus engrafting capacity was examined, with proT2 cells showing a 3-fold enhanced reconstituting capacity compared with the proT1 subset. Furthermore, in vitro–generated CD34+CD7++ progenitors effectively engrafted the thymus of immunodeficient mice, which was enhanced by the addition of an IL-7/IL-7 antibody complex. Taken together, the identification of T-progenitor subsets readily generated in vitro may offer important avenues to improve cellular-based immune-reconstitution approaches.
Introduction
T cells develop within the thymus from bone marrow–derived hematopoietic progenitors, and follow a series of stage-specific differentiation events, which are broadly characterized by the developmentally coordinated expression of CD4 and CD8.1
The initial stages of human T-cell development include precursors that express the stem cell marker CD34,2,3 which is also present on hematopoietic stem cells (HSCs) and on multipotent or lineage-specified progenitor cells. Within the known hierarchy of T-cell development, the earliest precursor subset is further defined by the lack of CD3, CD4, CD8, and CD1a expression.4 Although immature stages of T-cell development are typically delineated as CD34+CD1a− and CD34+CD1a+ cells, these populations remain heterogeneous. Of note, CD7 expression is considered to be one of the earliest cell surface markers known to appear during T lymphopoiesis.2,5 Importantly, the transition from CD34+CD7+CD1a− to CD34+CD7+CD1a+ by early thymocytes is associated with T-cell commitment, as a small percentage (∼ 10%) of these cells show rearrangement at the T-cell receptor β-chain (TCRβ) locus.6 In addition, CD34+CD7+CD1a+ cells appear to be T-lineage restricted, as these cells show low precursor activity toward non–T-cell lineages.7
Current understanding of the aforementioned early stages has been obtained from analyses of human fetal or adult thymocyte subsets, and by analyzing T-cell development in vitro using xenogeneic engraftment of mouse fetal thymus organ cultures (FTOCs).8,9 Although these systems have provided important insight into T-cell development, the capacity to evaluate specific progenitor populations has remained difficult to assess given the requirement of human thymus tissue, and the limited number of progenitor T cells that can be readily analyzed.
Previous work from our laboratory established that human T-lineage differentiation can be induced from umbilical cord blood (UCB)–derived HSCs cocultured with OP9-DL1 cells.10 However, these studies were not performed using quantitative clonal analyses, and it was unresolved whether different UCB-CD34+ subsets could give rise to T-lineage cells and whether Delta-like/Notch signals influence the T-progenitor frequency of CD34+ UCB cells. Finally, our initial studies10 showed that during the early stages of HSC/OP9-DL1 differentiation a population of cells resembling T progenitors became apparent, however the potential of these cells to serve as effective T-cell progenitors was not addressed.
Here, we examined the early stages of human T-cell development in vitro, and performed limiting dilution and single-cell assays to address the T-cell progenitor frequency of various UCB-derived CD34+ stem/progenitor subsets. Our results showed a sequential and temporally discrete acquisition of T-lineage differentiation markers. We assessed the effect of Delta-like/Notch interactions in enhancing T-cell progenitor potential among Notch-signaled CD34+ subsets. Furthermore, using limiting dilution approaches in FTOC, our findings revealed that different in vitro–generated T-progenitor subsets vary in their thymus-engrafting effectiveness. Finally, we obtained evidence showing that the CD34+CD7++ subset possesses the cardinal properties of a T-cell progenitor as demonstrated by its ability to home, seed, and reconstitute the thymuses of Rag2−/−γc−/− and nonobese diabetic/severe combined immunodeficient (NOD/SCID)/γcnull mice.
Together, these findings support the use of this system for the generation and study of human T progenitors, and highlight the potential for their eventual use as a cellular-based therapy for the treatment of T-cell immunodeficiencies.
Methods
Umbilical cord blood samples
Human UCB samples were obtained by syringe extraction and collected in a blood-pack unit containing citrate phosphate dextrose anticoagulant (Baxter Healthcare) from consenting mothers after delivery at Women's College Hospital in accordance with approved guidelines established by the Research Ethics Board of Sunnybrook Health Sciences Center and the Declaration of Helsinki. Within 12 hours of collection, UCB mononuclear cells were isolated as previously described.10,11 For each experiment, frozen UCB was thawed and pre-enriched into lineage-negative (Lin−) and lineage-positive (Lin+) fractions with the autoMACS (Miltenyi Biotec) using the StemSep Human Progenitor Enrichment Kit (StemCell Technologies), which depletes for cells expressing the following lineage markers: CD2, CD3, CD14, CD16, CD19, CD24, CD56, CD66b, and glycophorin-A. To isolate human HSCs, Lin− cells were stained with antihuman CD38-APC and antihuman CD34-PE mAbs and subsequently sorted for CD34+CD38−/lo cells using a BD Biosciences FACSAria sorter. Sorted human HSCs were greater than 99% pure as determined by postsort analysis.
Mice
BALB/c Rag2−/−γc−/− mice were kindly provided by Drs Kees Weijer and Hergen Spits (University of Amsterdam). NOD.cg-PrkdcscidIL2rgtm/Wjl/Sz (NOD/SCID/γcnull) mice were purchased from The Jackson Laboratory and housed and bred in a pathogen-free facility. All animal procedures were approved by the Sunnybrook Health Science Center Animal Care Committee.
Human HSC and OP9-DL1 cell coculture
Human-mouse FTOC
FTOC8 was performed by isolating fetal thymuses from embryos of timed-pregnant CD1 mice at day 15 of gestation (The Jackson Laboratory). The thymuses were cultured for 5 days in the presence of 1.35 mM deoxyguanosine (dGuo). Sorted human proT subsets derived from day-13 HSC/OP9-DL1 cocultures, supplemented with rhFlt-3L, rhIL-7, and SCF (30 ng/mL) (Peprotech), were placed into hanging drops with dGuo-treated fetal thymuses in Terasaki wells for 24 hours, followed by FTOC for 7 to 21 days as indicated. OP9-media and cytokines (Flt-3L/IL-7) were replenished every 5 days. Cells were analyzed by crushing thymus lobes on a nylon mesh cell strainer to obtain single-cell suspensions.
Quantitative real-time reverse-transcriptase polymerase chain reaction
Total RNA was isolated in Trizol-reagent and reverse transcribed using Superscript-III and Oligo(dT)12-18 primers (Invitrogen). Diluted cDNA samples from total OP9-control cocultures, total OP9-DL1 cocultures, sorted T-lineage subsets from OP9-DL1 cocultures as indicated in the figures, UCB purified Lin+CD3+ and CD33+ cells, or bulk and Lin− human postnatal thymocytes (PNTs) were used as templates for quantitative real-time reverse-transcriptase polymerase chain reaction (QRT-PCR) reactions. Detection of the QRT-PCR was performed with the SYBR Green PCR master mix according to the manufacturer's instructions (QIAGEN or Bio-Rad) on the Applied Biosystems Sequence Detection System 7000. All transcript levels were normalized to human β-actin.
Flow cytometry
Fluorescein isothiocyanate (FITC)–, R-phycoerythrin (PE)–, allophycocyanin (APC)–, PE-Cy7–, peridinin chlorophyll protein (PerCP) PerCP-Cy5.5–, Alexa Fluor700–, Alexa Fluor750–, and Pacific Blue–conjugated antibodies were purchased commercially (BD Biosciences or eBioscience). Cell suspensions were FcRII blocked and stained, and analyzed with a FACSCalibur (BD Biosciences) or an LSR-II cytometer (BD Biosciences). Data analysis was performed using FlowJo software (TreeStar) by gating on live lymphocytes and lack of propidium iodide uptake. Within FACS plots and histograms, numbers in quadrant corners represent percentage of gated cells.
Precursor frequency analysis
HSC limiting dilution assay was performed by serial dilutions from different cell subsets of UCB. For human in vitro–derived progenitor T cells, limiting dilution assays were performed using sorted subsets. Additional details are provided in the supplemental methods (available on the Blood website; see the Supplemental Materials link at the top of the online article). Progenitor frequencies were determined by the method of maximum likelihood applied to the Poisson model.13
Generation and administration of human progenitor T cells into immunodeficient mice
Sorted CD34+CD38−/lo cells (3.0 × 105) from Lin− UCB were cultured on OP9-DL1 cells per individual well of a 6-well plate and cultured for 10 to 11 days in OP9-media containing rhFlt-3L, rhIL-7, and rhSCF (30 ng/mL). CD34+CD7++ cells were sorted and injected intrahepatically into day-4 to -6 BALB/c Rag2−/−γc−/− or NOD/SCID/γcnull neonates. Each mouse received 2.5 to 5.0 × 105 CD34+CD7++ cells mixed with or without rhIL-7 (0.5 μg/mouse) and anti–IL-7 mAb, clone M25 (2.5 μg/mouse), cocktail in a total volume of 30 μL in PBS. As controls, mice were injected with either PBS or UCB CD34+ cells (1.5-2.5 × 105). Mice were boosted with IL-7/M25 cocktail every 3 to 4 days. Four to 6 weeks after transfer, thymus single-cell suspensions were counted, stained, and analyzed with an LSR-II cytometer. Data analysis was performed using FlowJo software by gating on live lymphocytes by exclusion of 4′-6-diamidino-2-phenylindole (DAPI) uptake followed by gating on cells expressing human CD45.
Results
Cellular analysis of the sequential induction of T-cell development in vitro
An important step in the establishment of an effective in vitro system for human T lymphopoiesis is to fully characterize the early stages of T-cell development. To this end, we performed a temporal kinetic analysis of early developmental changes that occur when UCB-derived HSCs are induced to differentiate on OP9-DL1 cells. As expected, flow cytometric analysis of the starting stem cell population showed that sorted Lin− CD34+CD38−/lo cells do not express markers of early T-cell differentiation, such as CD7, CD5, CD1a, and CD10, nor markers of late T-cell differentiation such as CD2, CD4, CD8, and CD3 (Figure 1A).
Developmental progression of human T-lineage cells from Lin− CD34+CD38−/lo HSCs cultured on OP9-DL1 cells. (A) Flow cytometric analysis for the cell surface expression of CD34, CD5, CD1a, CD10, CD2, CD4, CD8, CD3, and CD7 from purified human CD34+CD38−/lo HSCs before coculture with OP9-DL1 cells. (B-C) HSC/OP9-DL1 cocultures were harvested and analyzed by flow cytometry at the indicated time points for the expression of the markers as shown. The CD7 costaining with CD2 was performed with a different fluorophore-conjugated anti-CD7 mAb. Data are representative of at least 5 independent cocultures. Numbers in plots indicate percentage of cells within each quadrant.
Developmental progression of human T-lineage cells from Lin− CD34+CD38−/lo HSCs cultured on OP9-DL1 cells. (A) Flow cytometric analysis for the cell surface expression of CD34, CD5, CD1a, CD10, CD2, CD4, CD8, CD3, and CD7 from purified human CD34+CD38−/lo HSCs before coculture with OP9-DL1 cells. (B-C) HSC/OP9-DL1 cocultures were harvested and analyzed by flow cytometry at the indicated time points for the expression of the markers as shown. The CD7 costaining with CD2 was performed with a different fluorophore-conjugated anti-CD7 mAb. Data are representative of at least 5 independent cocultures. Numbers in plots indicate percentage of cells within each quadrant.
We made use of CD7 surface expression as a common marker for the temporal analysis of T-cell differentiation.1,10 Analysis of CD7 expression in early HSC/OP9-DL1 cocultures revealed that this approach recapitulates early and late stages of T-cell development, in which CD7 expression is first detected at day 4 on CD34+ cells, followed by high-level expression on CD34+ cells by days 6 to 8, and then slightly decreasing on a subset of CD34− cells at later time points (beyond day 14; Figure 1B).
During the initial week of coculture, as CD34+ cells rapidly acquire CD7 expression, the overall cell numbers showed a slight increase (supplemental Figure 1), and the cells remain negative for the expression of CD5, CD1a, CD2, and CD4. By day 8 of culture, CD5 expression is first detected on CD34+CD7++ cells, which remain CD1a− (Figure 1B). CD1a+ cells begin to be detected by day 10, and present on approximately 15% of the CD7++ cells, which correspond to cells that have also started to down-regulate CD34 expression. By day 14, expression of CD5 is observed on nearly all of the CD7++ cells, with CD1a being expressed on the majority of these cells. Day 14 also corresponds to when cellular expansion begins (supplemental Figure 1).
At later time points, CD7++ and CD7+ populations expressing CD2 and CD4 begin to predominate (Figure 1C). In addition, a population of CD7+CD1a++ cells continues to expand, and eventually accounted for nearly 90% of the CD7-expressing cells by day 48. A similar pattern can be observed for human postnatal thymocytes (supplemental Figure 2). In contrast to early time points (days 8-10), in which CD2 expression is low on CD7++ cells, by day 48 nearly 50% of the cells express high levels of CD2 (Figure 1C). The expression of CD4 on CD7++ cells emerges as early as day 12 (Figure 1B) and continues to increase, eventually accounting for approximately 75% of the CD7-expressing cells by day 48 (Figure 1C). Although a small percentage of CD1a+ or CD4+ cells that lack CD7 expression was detected, we have previously reported that these cells belong to the myeloid lineage.10
Molecular analysis of the sequential induction of T-cell development in vitro
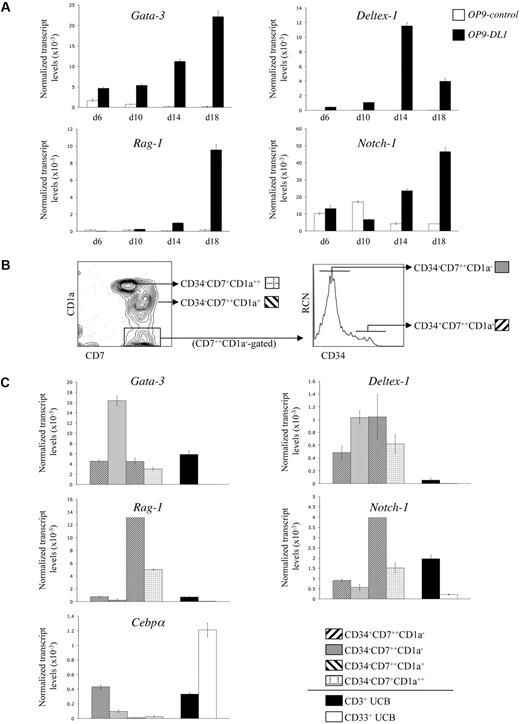
Although human HSC/OP9-DL1 cocultures exhibited a cellular expression pattern consistent with stages of T-cell development observed in the thymus, the precise temporal kinetics of Notch-dependent gene expression during early T-cell differentiation14,,–17 are undefined. We examined the expression of Gata-3, Deltex-1, Rag-1, and Notch-1 transcripts from HSCs cocultured with OP9-control (GFP-only) or OP9-DL1 cells. As shown in Figure 2A, expression of Gata-3, Deltex-1, Rag-1, and Notch-1 showed a general trend toward elevated transcript levels in OP9-DL1 compared with OP9-control cocultures, with a clear difference starting at around day 14. Consistent with its role in early T-cell specification and commitment,18 Gata-3 expression was differentially induced early and steadily increased over time in OP9-DL1 cocultures. Deltex-1, a known Notch-induced target gene,18 was also specifically up-regulated as early as day 6 in OP9-DL1 cocultures. Rag-1, an essential gene for TCR gene rearrangements,19 became differentially up-regulated in OP9-DL1 cocultures by day 14. Finally, expression of Notch-1 was observed throughout in both cocultures, but was clearly up-regulated as a consequence of Delta-like–induced signaling.18
Gene expression analysis of CD34+CD38−/lo HSCs cultured on OP9-DL1 cells. (A) Temporal kinetics of gene expression by quantitative real-time QRT-PCR analysis from human CD34+CD38−/lo HSCs cultured on either OP9-control or OP9-DL1 cells for 6, 10, 14, and 18 days, as indicated. (B) Flow cytometric analysis for the cell surface expression of CD7 and CD1a from a day-50 HSC/OP9-DL1 coculture, with CD34 expression shown for cells gated as CD7+ CD1a−. (C) Gene expression analysis by QRT-PCR from the coculture-derived subsets as indicated in panel B: CD34+CD7++CD1a−, CD34−CD7++CD1a−, CD34−CD7++CD1a+, CD34−CD7+CD1a++; see figure key. CD3+ T cells and CD33+ myeloid cells were purified from the Lin+ fraction of UCB samples and served as controls. Transcript levels for the indicated genes were normalized to human β-actin, and these data are representative of 3 independent experiments, with the STD error bars shown corresponding to values obtained from triplicate wells within an individual experiment.
Gene expression analysis of CD34+CD38−/lo HSCs cultured on OP9-DL1 cells. (A) Temporal kinetics of gene expression by quantitative real-time QRT-PCR analysis from human CD34+CD38−/lo HSCs cultured on either OP9-control or OP9-DL1 cells for 6, 10, 14, and 18 days, as indicated. (B) Flow cytometric analysis for the cell surface expression of CD7 and CD1a from a day-50 HSC/OP9-DL1 coculture, with CD34 expression shown for cells gated as CD7+ CD1a−. (C) Gene expression analysis by QRT-PCR from the coculture-derived subsets as indicated in panel B: CD34+CD7++CD1a−, CD34−CD7++CD1a−, CD34−CD7++CD1a+, CD34−CD7+CD1a++; see figure key. CD3+ T cells and CD33+ myeloid cells were purified from the Lin+ fraction of UCB samples and served as controls. Transcript levels for the indicated genes were normalized to human β-actin, and these data are representative of 3 independent experiments, with the STD error bars shown corresponding to values obtained from triplicate wells within an individual experiment.
Although the gene expression kinetics shown in Figure 2A are consistent with the induction of T-lineage differentiation by Notch/Delta-like interactions, we sought to more precisely characterize the changes in gene expression occurring at specific T-cell differentiation stages. To this end, subsets of CD7-expressing cells, with each subset representing a distinct and sequential stage of T-cell development, were analyzed. As shown in Figure 2B, the developmental progression of CD7-expressing cells, from a day-50 OP9-DL1 coculture, can be ordered sequentially based on the loss of CD34 and the gain of CD1a expression into 4 stages: CD34+CD7++CD1a−, CD34−CD7++CD1a−, CD34−CD7++CD1a++, and finally CD34−CD7+CD1a++. These subsets, as well as T cells (CD3+) and myeloid cells (CD33+) sorted from UCB as lineage controls, were then examined for the expression of Gata-3, Deltex-1, Rag-1, Notch-1, as well as the myeloid-specific gene Cebpα20 (Figure 2C). Up-regulation of Gata-3 transcripts became apparent as CD7++ cells lose CD34 surface expression, which is then reduced at later stages, a finding that is in keeping with previous observations.18 Deltex-1 was up-regulated in each of the CD7-expressing subsets compared with either CD3+ mature T cells or CD33+ myeloid cells. Of note, Rag-1 and Notch-1 transcript up-regulation was most pronounced at the CD34−CD7++CD1a+ stage, consistent with the role of these genes in the generation and functional outcomes of the pre-TCR complex.21 As expected, when CD34 expression is extinguished, CD7-expressing cells become more restricted to the T-cell lineage, which parallels the observed loss of Cebpα expression within these subsets.
Taken together, human HSC/OP9-DL1 cocultures display stage- and temporal-specific cellular and molecular signatures, which not only recapitulates key hallmarks of T lymphopoiesis but also provides a simple and effective way to further dissect the developmental program of human T cells.
CD34+CD38− and CD34+CD38lo UCB cells exhibit high T-lymphopoietic potential
Several studies have provided evidence that the UCB-CD34+ stem cell pool is heterogeneous in terms of its repopulation, differentiation, and renewal potential.22,23 Indeed, the CD34+ population can be subfractionated into distinct subsets based on CD38 expression.22,–24 The CD38− subfraction contains primitive precursors capable of long-term reconstitution with slower engraftment kinetics.23 Conversely, UCB cells from the CD34+CD38lo or CD38+/hi subsets exhibit different characteristics, giving rise to rapid myeloid-erythroid differentiation with short-term repopulating ability.22,–24 However, these studies did not address the frequency of progenitors with T-lineage potential among these different CD34+ subsets. To determine the T-progenitor frequency of the various UCB-CD34+CD7− subsets, CD34+CD38−, CD34+CD38lo, and CD34+CD38+/hi cells were sorted (supplemental Figure 3) and placed at limiting cell numbers into wells containing OP9-DL1 cells. As shown in Table 1, CD34+CD38− or CD34+CD38lo cells gave rise to T-lineage cells with similar overlapping frequencies of 1 in 4.8 and 1 in 3.9, respectively, whereas the CD34+CD38+/hi subset possessed a nearly 5-fold diminished T-lineage progenitor frequency of 1 in 19. Thus, the CD34+CD38− and CD34+CD38lo fractions contain a greater frequency of cells that can give rise to T-lineage cells.
Progenitor frequency analysis of human hematopoietic stem cell subsets
| HSC subset* . | Progenitor frequency−1 (95% confidence limits)† . |
|---|---|
| CD34+CD38− | 4.76 (3.66-6.21) |
| CD34+CD38lo | 3.85 (2.94-5.06) |
| CD34+CD38+/hi | 19.30 (14.77-25.22) |
| HSC subset* . | Progenitor frequency−1 (95% confidence limits)† . |
|---|---|
| CD34+CD38− | 4.76 (3.66-6.21) |
| CD34+CD38lo | 3.85 (2.94-5.06) |
| CD34+CD38+/hi | 19.30 (14.77-25.22) |
CD34+CD38−, CD34+CD38lo, and CD34+CD38+/hi HSCs were placed at limiting numbers in wells of a 96-well/plate containing OP9-DL1 cells, and cultured for 11 days before harvesting for flow cytometric analysis.
Individual wells were scored for the presence of T cells based on CD45+CD7++ staining. Statistical analysis was performed via the method of maximum likelihood applied to the Poisson model.13
In vitro–generated proT cells show thymus-reconstituting ability in vivo
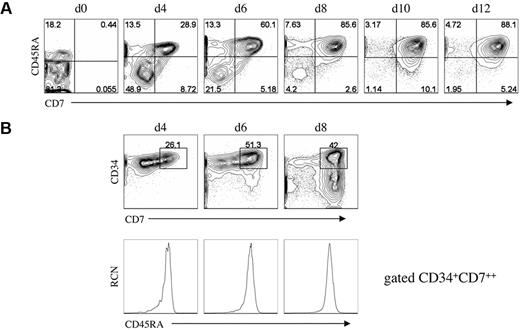
Although rare, thymus-seeding cells, identified as CD34+CD45RAhiCD7+, have been shown to be present in UCB25 or fetal bone marrow.2 To determine whether a similar population can be generated in vitro, we looked for cells bearing this phenotype at early coculture time points. Of note, the starting UCB-HSC population of CD34+CD38−/lo Lin− cells contained a subset that expressed the CD45RA isoform at low levels,3,26 however the majority of these cells were CD7− (Figure 3A). Our analysis showed that CD45RA expression is up-regulated within the first 4 days on CD34+ cells, and by day 6 nearly all CD34+CD7++ cells express CD45RA (Figure 3B). Thus, a population of CD34+CD7++CD45RA+ cells displaying a thymic-colonizing phenotype, as seen in vivo,2,25 is present in vitro and may also possess thymus-reconstituting potential.
Analysis for the presence of T-cell progenitors generated in vitro. (A) Flow cytometric analysis for the expression of CD7 and CD45RA from HSC/OP9-DL1 cocultures harvested at the indicated time points, including at day 0 before the start of coculture. (B) Flow cytometric analysis of CD7 and CD34 expression from HSC/OP9-DL1 cocultures harvested at days 4, 6, and 8 (top row), with CD45RA expression shown for cells gated as CD34+CD7++ (bottom row). Data are representative from 3 independent cocultures. Numbers in plots indicate percentage of cells within each quadrant; RCN, relative cell number.
Analysis for the presence of T-cell progenitors generated in vitro. (A) Flow cytometric analysis for the expression of CD7 and CD45RA from HSC/OP9-DL1 cocultures harvested at the indicated time points, including at day 0 before the start of coculture. (B) Flow cytometric analysis of CD7 and CD34 expression from HSC/OP9-DL1 cocultures harvested at days 4, 6, and 8 (top row), with CD45RA expression shown for cells gated as CD34+CD7++ (bottom row). Data are representative from 3 independent cocultures. Numbers in plots indicate percentage of cells within each quadrant; RCN, relative cell number.
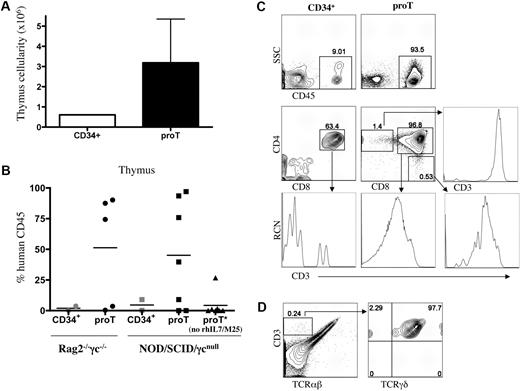
To evaluate this, CD34+CD7++CD45RA+ (proT) cells sorted from a day-11 HSC/OP9-DL1 coculture were injected intrahepatically into nonirradiated newborn NOD/SCID/γcnull or BALB/c Rag2−/−γc−/− mice,27,,,–31 and analyzed 4 to 6 weeks later. UCB-derived CD34+ HSCs were similarly injected into littermates for comparison. In light of the findings showing that the addition of IL-7 facilitates immune engraftment by human cells in immunodeficient mice,29 we made use of a cocktail of rhIL-7 and anti–IL-7 mAb, which has been shown to increase the activity of IL-7 in vivo.32 Figure 4A shows the thymus cellularity obtained from NOD/SCID/γcnull mice receiving either CD34+ HSCs or in vitro–generated proT cells. We observed a 6-fold increase in thymus cellularity in the mice receiving proT cells compared with mice injected with CD34+ cells.
Analysis of engraftment and differentiation of in vitro–derived progenitor T cells in immunodeficient mice. Human UCB CD34+CD38−/lo cells were differentiated on OP9-DL1 cells for 10 to 12 days, and CD34+CD7++ (proT) cells were sorted by flow cytometry. Neonatal NOD/SCID/γcnull or BALB/c Rag2−/−γc−/− mice were injected intrahepatically with 2.5 to 5.0 × 105 proT cells or 1.5 to 2.5 × 105 UCB-derived CD34+ cells. (A) Thymuses were harvested 4 to 6 weeks after transplantation, single-cell suspensions were obtained, and total cellularity was determined. The cellularity shown is for NOD/SCID/γcnull mice that received a transplant of proT cells and that displayed higher than 70% human CD45+ chimerism. (B) The percentage of human CD45+ cells present in the thymus of individual BALB/c Rag2−/−γc−/− and NOD/SCID/γcnull mice that received a transplant of UCB CD34+ or in vitro–generated proT cells is shown, with the mean percentage indicated by a horizontal bar. (C) Flow cytometric analysis for cell surface expression of human CD45 of thymocytes from a representative UCB CD34+- and proT-injected mouse (top row); middle row panels show human CD45+-gated cells analyzed for CD4 and CD8 cell surface expression, and CD3 expression is shown for either DP-gated cells (bottom row) or SP-gated cells (middle and bottom rows), as indicated by arrows. (D) Flow cytometric analysis of human CD45+-gated thymocytes from a mouse injected with in vitro–generated proT cells is shown for CD3 and TCRαβ expression (left); and CD3 and TCRγδ expression on CD3+TCRαβ−-gated cells, as indicated by arrow (right).
Analysis of engraftment and differentiation of in vitro–derived progenitor T cells in immunodeficient mice. Human UCB CD34+CD38−/lo cells were differentiated on OP9-DL1 cells for 10 to 12 days, and CD34+CD7++ (proT) cells were sorted by flow cytometry. Neonatal NOD/SCID/γcnull or BALB/c Rag2−/−γc−/− mice were injected intrahepatically with 2.5 to 5.0 × 105 proT cells or 1.5 to 2.5 × 105 UCB-derived CD34+ cells. (A) Thymuses were harvested 4 to 6 weeks after transplantation, single-cell suspensions were obtained, and total cellularity was determined. The cellularity shown is for NOD/SCID/γcnull mice that received a transplant of proT cells and that displayed higher than 70% human CD45+ chimerism. (B) The percentage of human CD45+ cells present in the thymus of individual BALB/c Rag2−/−γc−/− and NOD/SCID/γcnull mice that received a transplant of UCB CD34+ or in vitro–generated proT cells is shown, with the mean percentage indicated by a horizontal bar. (C) Flow cytometric analysis for cell surface expression of human CD45 of thymocytes from a representative UCB CD34+- and proT-injected mouse (top row); middle row panels show human CD45+-gated cells analyzed for CD4 and CD8 cell surface expression, and CD3 expression is shown for either DP-gated cells (bottom row) or SP-gated cells (middle and bottom rows), as indicated by arrows. (D) Flow cytometric analysis of human CD45+-gated thymocytes from a mouse injected with in vitro–generated proT cells is shown for CD3 and TCRαβ expression (left); and CD3 and TCRγδ expression on CD3+TCRαβ−-gated cells, as indicated by arrow (right).
As shown in Figure 4B, NOD/SCID/γcnull, as well as BALB/c Rag2−/−γc−/−, mice displayed a high frequency of human CD45+ cells in the thymus (45% and 51% average, respectively) when receiving proT cells. In contrast, a 15-fold lower level of human chimerism was observed when UCB CD34+ cells are transplanted into nonirradiated neonatal host mice. Although some thymus engraftment from the in vitro–generated proT cells could be observed when mice were not treated with the rhIL-7/M25 cocktail, the levels of human CD45+ chimerism (4% average) were significantly lower than in mice receiving IL-7, and reached the levels observed in mice treated with UCB CD34+ cells in the presence of the rhIL-7/M25 cocktail.
A representative flow cytometric analysis for the presence of human cells, by cell surface staining for human CD45 expression, demonstrated greater than 90% human chimerism in the thymus of a proT-injected mouse, which was in contrast to the typically low frequency of human cells (9%) present in a mouse that received a CD34+ transplant (Figure 4C). Further examination of the cell surface phenotype on CD45-gated cells from the CD34+ cells and proT-injected mice revealed that the majority of human cells had progressed along the T-lineage pathway and differentiated to express CD5, CD1a and lacked CD34 expression (data not shown). As shown in Figure 4C, CD4+CD8+ (double-positive [DP]) cells comprised the majority of the human thymocytes present in the engrafted mice. Notably, at this time point, only the DP cells present in the proT-engrafted mice showed increased levels of CD3 surface expression, indicating that the proT-derived cells were more developmentally advanced. In addition, the generation of CD4 single-positive (SP) and CD8 SP T cells was observed only in the proT-injected mouse, and gating on these SP populations demonstrated the presence of high levels of CD3 expression. Further analysis of the mice receiving proT cells revealed that most of the CD3+ T cells belonged to the αβ-lineage, whereas a small, but discrete population of CD3+ cells that lack TCRαβ (0.24%) corresponded to TCRγδ-bearing T cells (Figure 4D). A similar pattern of human engraftment and T-lineage differentiation was observed in BALB/c Rag2−/−γc−/− mice (data not shown).
These data clearly demonstrate that CD34+CD7++CD45RA+ T-lineage progenitors (proT), generated in vitro from CD34+CD38−/lo HSCs, exhibit key properties of being able to home to the thymus and effectively reconstitute the thymus of immune-deficient mice.
Identification of 2 distinct CD34+CD7++CD45RA+ progenitor subsets by CD5 expression
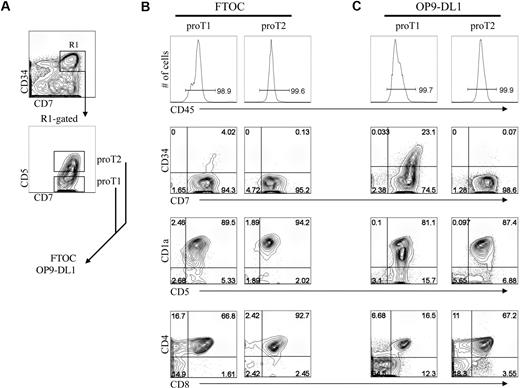
As the results shown in Figure 4 indicated that in vitro–generated CD34+CD7++ proT cells are capable of engrafting a host mouse thymus, we wanted to determine whether this ability resided uniformly or belonged to distinct subsets. To address this, we further dissected the CD34+CD45RA+CD7++CD1a− progenitor (proT) population based on the presence or absence of CD5 expression, and, as shown in Figure 5A, CD5 is expressed on approximately 45% of these cells. Of note, analysis of human postnatal thymocytes demonstrates that CD5 expression can also serve to identify 2 subsets of cells within the rare CD34+CD7++ progenitor population, with the majority of these expressing CD5 (supplemental Figure 2B). To determine whether these T-progenitor subsets have a differential ability to reconstitute and differentiate within a host thymus in vitro,2,8,33 CD34+CD45RA+CD7++CD1a− cells that are either CD5− or CD5+ (hereafter referred to as proT1 and proT2, respectively) were sorted from a day-13 HSC/OP9-DL1 coculture, and placed in FTOC (Figure 5B) or for comparison placed back onto OP9-DL1 cells (Figure 5C).
Analysis of engraftment and differentiation by in vitro–derived progenitor T-cell subsets in FTOC. Human UCB CD34+CD38−/lo HSCs were differentiated for 13 days on OP9-DL1 cells and CD34+CD7++CD5− (proT1) and CD34+CD7++CD5+ (proT2) subsets were sorted by flow cytometry as indicated in panel A, and placed into FTOC (B) or placed back onto OP9-DL1 cells (C) for 19 days. Cells were harvested and analyzed for cell surface expression of CD45, CD7, CD34, CD5, CD1a, CD8, and CD4. Total cellularity of fetal thymic lobes after reconstitution ranged from 6 to 8 × 103 or 5 to 12 × 103 with proT1 cells or proT2 cells, respectively. Data are representative of 3 independent experiments in which 1.5 × 104 sorted proT subsets were placed either into fetal thymus lobe pairs or in wells containing OP9-DL1 cells.
Analysis of engraftment and differentiation by in vitro–derived progenitor T-cell subsets in FTOC. Human UCB CD34+CD38−/lo HSCs were differentiated for 13 days on OP9-DL1 cells and CD34+CD7++CD5− (proT1) and CD34+CD7++CD5+ (proT2) subsets were sorted by flow cytometry as indicated in panel A, and placed into FTOC (B) or placed back onto OP9-DL1 cells (C) for 19 days. Cells were harvested and analyzed for cell surface expression of CD45, CD7, CD34, CD5, CD1a, CD8, and CD4. Total cellularity of fetal thymic lobes after reconstitution ranged from 6 to 8 × 103 or 5 to 12 × 103 with proT1 cells or proT2 cells, respectively. Data are representative of 3 independent experiments in which 1.5 × 104 sorted proT subsets were placed either into fetal thymus lobe pairs or in wells containing OP9-DL1 cells.
Shown in Figure 5B, both proT1 and proT2 subsets successfully reconstituted the FTOCs, and their progeny accounted for nearly all of the cells present in the lobes, based on human CD45 expression (≥ 95%). In addition, the reconstituted FTOCs contained T cells that were derived from either the proT1 or proT2 subsets. Although the input proT1 cells were initially CD34+CD7++CD5−, nearly all of the cells within the colonized lobes had differentiated into CD34−CD5+CD1a+ T-lineage cells, with 67% also coexpressing CD4 and CD8 and 2% to 16% expressing either CD8 or CD4, with the majority of these being CD4 intermediate single positive (data not shown). Similarly, the proT2 cells, which initially expressed CD5, also gave rise to T cells, however with an increase in the frequency of DP cells (93%). This difference may relate to the later differentiation state of the proT2 cells, which as demonstrated in supplemental Figure 4, purified proT1 cells gave rise to cells with the proT2 phenotype within 24 hours and the majority of these cells reached the next stage by 48 hours. In addition, purified proT2 cells did not give rise to cells with the proT1 phenotype. The precursor-product relationship from proT1 to proT2 is further highlighted by the presence of a small fraction (4%) of CD34+CD7++ cells remaining in FTOC seeded with proT1 but not proT2 cells. Keeping with this, proT1 cells placed back onto OP9-DL1 cells also showed the presence of a CD34+CD7++ progenitor population, which was absent in proT2 cultures (Figure 5C). Nevertheless, both proT1 and proT2 cells showed a similar ability to continue to differentiate along the T-lineage pathway in these cocultures, giving rise to CD1a+- and CD4/CD8-expressing cells.
Because proT1 and proT2 cells share a similar progenitor phenotype with cells found in the human thymus that have been shown to also possess natural killer (NK)–lineage potential, we addressed whether NK cells could also be generated from these subsets. Consistent with this, we confirmed that in vitro–derived proT1 and proT2 cells give rise to NK cells when cultured on OP9-control cells, supplemented with IL-15 (supplemental Figure 5). These results are consistent with studies demonstrating the presence of cells with dual T/NK potential within the CD34+CD7++ thymocyte subset.34,35 Of note, both proT1 and proT2 cells when cultured on OP9-DL1 cells did not give rise to NK cells, rather they continued to differentiate along the T-cell pathway (supplemental Figure 5), which is consistent with the known role of Notch signaling in maintaining commitment to the T-lineage while inhibiting alternate lineage outcomes.36
Although both proT1 and proT2 cells can give rise to T cells, it remained unclear whether these subsets contained a similar progenitor frequency to reconstitute a host thymus. To address this, sorted proT1 and proT2 cells were placed in limiting cell numbers in FTOCs or on OP9-DL1 cells for 7 days, and analyzed by flow cytometry for the presence of human T-lineage cells. The results shown in Table 2 demonstrate that the proT2 subset displayed a 3-fold higher T-lineage reconstituting frequency than that of the proT1 cells (1:400 and 1:1400, respectively). To further examine whether this difference was cell intrinsic, the T-progenitor frequency of these subsets was determined in a limiting dilution assay with OP9-DL1 cells. Of note, and in contrast to the progenitor frequencies observed in FTOCs, the results from the cocultures revealed that both proT subsets possess a similar and high (∼ 1:2) progenitor frequency (Table 2).
Progenitor frequency analysis of progenitor T-cell subsets
| ProT subset* . | Culture system† . | Progenitor frequency−1 (95% confidence limits)‡ . |
|---|---|---|
| ProT1, CD34+CD7+CD5− | FTOC | 1384.72 (979-1959) |
| ProT2, CD34+CD7+CD5+ | FTOC | 411.74 (256-663) |
| ProT1, CD34+CD7+CD5− | OP9-DL1 | 2.52 (1.75-3.63) |
| ProT2, CD34+CD7+CD5+ | OP9-DL1 | 1.95 (1.35-2.80) |
| ProT subset* . | Culture system† . | Progenitor frequency−1 (95% confidence limits)‡ . |
|---|---|---|
| ProT1, CD34+CD7+CD5− | FTOC | 1384.72 (979-1959) |
| ProT2, CD34+CD7+CD5+ | FTOC | 411.74 (256-663) |
| ProT1, CD34+CD7+CD5− | OP9-DL1 | 2.52 (1.75-3.63) |
| ProT2, CD34+CD7+CD5+ | OP9-DL1 | 1.95 (1.35-2.80) |
CD34+ CD38−/lo UCB-derived cells were cultured on OP9-DL1 cells for 12 to 14 days and proT1/proT2 cells, with the indicated phenotypes, were obtained by flow cytometric cell sorting.
ProT subsets were placed in limiting numbers in FTOC or in wells of a 96-well/plate containing OP9-DL1 cells and cultured for 7 days before harvesting for flow cytometric analysis.
Individual lobes or wells were scored for the presence of T cells based on CD45+CD7++ or CD7++CD1a−/+ staining, respectively. Statistical analysis was performed via the method of maximum likelihood applied to the Poisson model.13
In light of these findings, it would appear that human proT cells, which otherwise display a similarly high T-cell progenitor frequency when assayed on the OP9-DL1 monolayer, possess a differential capability to reconstitute a mouse thymus lobe in vitro, which may relate to differences in the expression of molecules important for entry across the thymus lobe outer membrane or niche occupancy within the thymus. To determine a potential mechanism for the observed difference in this ability, we analyzed by QRT-PCR for the expression of genes associated with thymus homing or entry.37,,,,–42 Figure 6A shows that proT2 cells express higher transcript levels of CCR9 (CD199), PSGL-1 (CD162), CD49b (α2 integrin), CD49d (α4 integrin), and CD49e (α5 integrin). A similar trend of elevated expression was observed for CD29 (β1 integrin) in proT2 cells. In addition, flow cytometric analysis of these subsets confirmed that proT2 cells express higher levels of CD49d than proT1 cells (Figure 6B). These data are consistent with previous findings41,42 that point to the CD49d/CD29 heterodimer, which binds to VCAM-1 (CD106) expressed on thymus stromal cells, as well as CCR9 and PSGL-1 as important players in facilitating thymus entry by the proT2 cell subset.
Gene expression analysis of CD34+CD7++CD5− and CD34+CD7++CD5+ proT cell subsets. (A) QRT-PCR analysis of CD34+CD7++CD5− (proT1) and CD34+CD7++CD5+ (proT2) cells purified by flow cytometric cell sorting from a day-14 HSC/OP9-DL1 coculture. Thymocytes obtained from the Lin− fraction of a human postnatal thymus (PNT) served as a control sample. Transcript levels for the indicated genes (Ccr9 [CD199], Selplg1 [PSGL-1, CD162], Itga2 [α2, CD49b], Itga4 [α4, CD49d], Itga5 [α5, CD49e], and Itgb1 [β1, CD29]) were normalized to human β-actin. These data are representative of 3 independent experiments, with the STD error bars shown corresponding to values obtained from triplicate wells within an individual experiment. (B) Flow cytometric analysis for cell surface expression of CD49d on gated CD34+CD7++CD5− (proT1, open) and CD34+CD7++CD5+ (proT2, shaded) cells from a day-11 HSC/OP9-DL1 coculture.
Gene expression analysis of CD34+CD7++CD5− and CD34+CD7++CD5+ proT cell subsets. (A) QRT-PCR analysis of CD34+CD7++CD5− (proT1) and CD34+CD7++CD5+ (proT2) cells purified by flow cytometric cell sorting from a day-14 HSC/OP9-DL1 coculture. Thymocytes obtained from the Lin− fraction of a human postnatal thymus (PNT) served as a control sample. Transcript levels for the indicated genes (Ccr9 [CD199], Selplg1 [PSGL-1, CD162], Itga2 [α2, CD49b], Itga4 [α4, CD49d], Itga5 [α5, CD49e], and Itgb1 [β1, CD29]) were normalized to human β-actin. These data are representative of 3 independent experiments, with the STD error bars shown corresponding to values obtained from triplicate wells within an individual experiment. (B) Flow cytometric analysis for cell surface expression of CD49d on gated CD34+CD7++CD5− (proT1, open) and CD34+CD7++CD5+ (proT2, shaded) cells from a day-11 HSC/OP9-DL1 coculture.
Discussion
The early stages of human T-cell development have been broadly defined by several investigators.1,43 Here, we have taken advantage of a simple and powerful in vitro system to further refine this view by examining the differentiation of human UCB-HSCs cultured with OP9-DL1 cells, in which the early stages of T-cell development can be readily characterized. The temporal kinetic analysis of early and late time points allowed us to discern an ordered pattern of developmental stages, which is highlighted by the sequential cell surface expression of CD34, CD45RA, CD7, CD5, CD1a, CD2, CD4, CD8, and CD3.
Although the OP9-DL1 system recapitulates most stages of human thymocyte differentiation, we noted one difference regarding the expression of CD2, which has been reported to be expressed on some of the early CD34+ thymocytes as well as in CD34+ cells found in the bone marrow.5,44,45 We observed CD2 expression only at low levels on cells that were down-regulating CD34 expression, and high expression of CD2 was seen only at later developmental stages. One possibility for these differences could be an accumulation of CD34+ cells with this early phenotype within the thymus or that the signals that normally induce the expression of CD2 on all thymocytes may be lacking in vitro.
Initial findings by our laboratory and others have clearly shown that UCB-CD34+ cells can be induced to differentiate to the T-cell fate upon coculture with stromal cells ectopically expressing Dll1.10,46 However, several groups have demonstrated that the CD34+ population is heterogeneous in regard to its self-renewal ability, engraftment, and lineage potential.22,24,47 With this in mind, we examined whether specific CD34+ subsets differed in their ability to serve as T-cell progenitors. In addition, Hogan et al suggested that the CD34+CD38− pool contains a higher frequency of cells with T-cell potential, because NOD/SCID mice engrafted with this fraction showed greater thymus repopulation compared with animals receiving CD34+CD38lo or CD38+/hi cells.23 In keeping with this, our results indicated that the CD38+/hi fraction, which is also known to contain myeloid progenitors, has a significantly 5-fold lower T-cell potential than the more primitive CD38− or CD38lo subsets, which surprisingly showed similar T-progenitor frequencies. The comparable progenitor frequencies by these CD34+CD38− or CD38lo cells may be accounted for by a report suggesting that CD38 is reversibly expressed between negative and low levels.48
The critical role of Notch signals for inducing T lymphopoiesis is now well established.49 Here, we identified the stages of T-cell development in which the induction of Notch target genes is first up-regulated. These stages corresponded to when CD34+ cells begin to express CD7 at high levels, with some further induction after the loss of CD34 expression. Our findings are supported by several studies demonstrating that UCB-CD34+CD7+-expressing cells are strongly biased to the lymphoid lineage with very little myeloid potential.3,25,33 These observations are consistent with the notion that T-cell specification occurs early, within the first week, and therefore these Notch-induced CD34+CD7++ cells would likely show an increased T-progenitor frequency. Indeed, our results indicated that after Notch/Delta-like interactions, CD34+CD7++ cells show a 2-fold higher T-progenitor frequency than the initial UCB-CD34+CD7− cells. These findings suggested that the HSC/OP9-DL1 cocultures readily support the generation of T-cell progenitors, which could be akin to thymus-colonizing cells.
It is well established that the thymus is continuously seeded with blood-borne progenitors, as the thymus-resident progenitors do not possess self-renewing potential.50 Although a recent study has demonstrated the existence of an intrathymic CD7− population having lymphomyeloid potential,51 the study by Haddad et al2 proposed that thymus-colonizing cells express CD34+CD7++CD45RA+. Cells with a similar CD7++ phenotype are detected in HSC/OP9-DL1 cocultures, and here we show that these in vitro–generated cells were able to serve as thymus-colonizing cells in vivo. In addition, we noticed the presence of 2 distinct progenitor subsets within the CD34+CD7++CD1a− population, termed proT1 (CD5−) and proT2 cells (CD5+). Both subsets are capable of thymus reconstitution in vitro, however, when used in limiting dilution assays, we observed dramatic differences in their ability to engraft a host thymus in vitro, with the more mature proT2 cells showing a 3-fold higher progenitor frequency than proT1 cells. In contrast, when assayed on OP9-DL1 cells, both proT subsets exhibited statistically similar progenitor frequencies, which were also dramatically higher (200-600×) than those observed in FTOC. These findings suggest that human proT cells, which otherwise possess high T-cell potential, are affected by xenogeneic entry barriers present in the mouse FTOC system, which severely lowers their engraftment effectiveness. In addition, we noted that these proT subsets differed in the expression of CCR9, PSGL-1, and multiple integrins, which serve to provide a potential mechanism for the enhanced engrafting ability demonstrated by proT2 cells. The higher expression levels of these molecules by proT2 cells was specific, in that the transcript levels of Cebpα and Gata-2 were higher in the proT1 subset, which is consistent with its more immature status (supplemental Figure 7).
The identification of a human T-cell progenitor capable of effectively trafficking to and reconstituting a host thymus in immunodeficient mice has yet to be reported. Here, we demonstrate the generation of cells with phenotypic, molecular, and functional characteristics of thymus-colonizing progenitors.2,37,39 These in vitro–generated proT cells showed a high level (40%-50%) of engraftment in BALB/c Rag2−/−γc−/− and NOD/SCID/γcnull mice, whereas a much lower frequency of chimerism was observed in the thymus of mice injected with UCB CD34+. Multiple studies have demonstrated high levels of human cell engraftment when CD34+ HSCs are administered intrahepatically into irradiated neonates52 ; however, it should be noted that we assessed the engraftment potential of in vitro–derived proT cells and UCB CD34+ using a more stringent nonirradiation setting. In addition, many of these studies performed HSC transplantations on neonates less than 48 hours old, which typically gives rise to greater than 80% human engraftment in peripheral blood, whereas this chimerism is drastically reduced in 1-week-old animals.52 Both the lack of irradiation and the age of the injected neonates (4 to 6 days old) serve as likely explanations for the low-level of engraftment we observed in UCB CD34+-treated mice, and may have also contributed to some of the low-level engraftments observed with the proT-injected mice. Nevertheless, our observed increase in thymus engraftment with in vitro–derived proT cells upon coadministration of rhIL7/anti-IL7 mAb32 is consistent with work from Shultz et al whereby treatment of HSC-engrafted mice with an Fc-IL7 fusion protein exhibited a higher frequency of human T cells in the host thymus.29
Our findings demonstrate the presence of mature SP T cells in the thymus of mice that underwent transplantation. As in the normal human thymus, T cells of both the αβ and γδ lineage were generated, indicating that the in vitro–generated CD34+CD7++ cells injected into these mice serve as a common precursor stage before lineage bifurcation of these T-lineage subsets.53 Furthermore, the early appearance of SP T cells indicates that selection events and thymic education have been established and are ongoing. This selection may be attributable to the expression of MHC class I, and MHC class II, on mouse thymic epithelial cells. Of note, the small chance exists that positive selection may be occurring on hematopoietic cells of human origin.28,54 Indeed, Manz and coworkers have results supporting the notion of dual restriction in humanized mice.28 The use of NOD/SCID/γcnull mice deficient in mouse MHC, and transgenic for the expression of human HLA molecules, may facilitate human TCR/MHC-selection and -restriction events. Although we observed robust engraftment of proT cells in the thymus after 6 weeks, human T cells were not readily detected in the periphery. This may be attributable to the length of time between cell transfer and analysis, and we are undertaking experiments evaluating later time points (> 6 weeks) to assess for the appearance of peripheral T cells and to further examine their function and TCR repertoire.
We demonstrate the ability to generate T-cell progenitors from human CD34+CD38−/lo UCB HSCs in vitro. CD34+CD7++ T progenitors generated in vitro colonized FTOC with the CD5+ subset possessing a higher progenitor potential in its ability to engraft FTOCs than its CD5− counterpart. Of higher significance, in vitro–generated proT cells seeded and effectively reconstituted the thymus of immunodeficient mice. Taken together, the insight obtained from this study makes the CD5+ proT2 cells an attractive progenitor subset for further studies aimed at enhancing the effectiveness of human T-cell reconstitution in mice. The use of in vitro–derived progenitor T cells may prove more therapeutically relevant over mature T cells by avoiding issues such as graft-versus-host disease, as these cells would undergo positive and negative selection within the host thymus.55 It is possible then to speculate that in vitro–generated T-progenitor cells may eventually serve as a viable option for cell-based therapies,9,55 as these cells can be generated in large numbers, allowing for novel strategies to be developed for the restoration of adaptive immunity in immunocompromised individuals.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to Gisele Knowles and Arian Khandani for their expert assistance with flow cytometric cell sorting, Roxanne Holmes for technical support, and Dr Rose Kung and the staff of the Perinatal and Gynaecology Program of Women's College Hospital and Sunnybrook Health Sciences Center for their ongoing support in providing material for our research. We also thank Dr Priscilla Chiu (University Health Network) for providing us with human postnatal thymocyte cell suspensions. We are also grateful to Drs Pablo Serra Devecchi and Divya K. Shah for helpful advice in the preparation of this paper.
This study was supported by grants to J.C.Z.-P. from the Canadian Institutes of Health Research (CIHR) HOP-83070, The Terry Fox Foundation, the Ontario HIV Treatment Network (OHTN), and The Krembil Foundation; and to J.E.D. from the Terry Fox Foundation. G.A. is supported by a Studentship from the OHTN. R.N.L.M.-M. was supported in part by a Postdoctoral Fellowship from the OHTN, and J.C.Z.-P. is a recipient of the Canada Research Chair in Developmental Immunology.
Authorship
Contribution: G.A. performed and designed the research, analyzed data, and wrote the paper; E.H. contributed vital reagents; C.D.S. contributed vital reagents and provided critical experimental design ideas; J.E.D. designed experimental approach; R.N.L.M.-M. performed research, analyzed data, and wrote the paper; and J.C.Z.-P. designed the research and wrote the paper.
Conflict-of-interest disclosure: C.D.S. is a stockholder in Nascent Biologics Inc. The remaining authors declare no competing financial interests.
Correspondence: Juan Carlos Zúñiga-Pflücker or Ross N. La Motte-Moh, Sunnybrook Research Institute, 2075 Bayview Ave, Rm A3-31, Toronto, ON M4N 3M5, Canada; e-mail: jczp@sri.utoronto.ca or ross.lamotte.mohs@utoronto.ca.


![Figure 6. Gene expression analysis of CD34+CD7++CD5− and CD34+CD7++CD5+ proT cell subsets. (A) QRT-PCR analysis of CD34+CD7++CD5− (proT1) and CD34+CD7++CD5+ (proT2) cells purified by flow cytometric cell sorting from a day-14 HSC/OP9-DL1 coculture. Thymocytes obtained from the Lin− fraction of a human postnatal thymus (PNT) served as a control sample. Transcript levels for the indicated genes (Ccr9 [CD199], Selplg1 [PSGL-1, CD162], Itga2 [α2, CD49b], Itga4 [α4, CD49d], Itga5 [α5, CD49e], and Itgb1 [β1, CD29]) were normalized to human β-actin. These data are representative of 3 independent experiments, with the STD error bars shown corresponding to values obtained from triplicate wells within an individual experiment. (B) Flow cytometric analysis for cell surface expression of CD49d on gated CD34+CD7++CD5− (proT1, open) and CD34+CD7++CD5+ (proT2, shaded) cells from a day-11 HSC/OP9-DL1 coculture.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/5/10.1182_blood-2008-10-187013/5/m_zh89990939800006.jpeg?Expires=1767767873&Signature=rynZvBIwN-fKLPCoIOoNe4PMjRzqFuPnTQcgbPgQVhJIQS-AlYiM2LZZqyxnindnOlYnik7~3Ao7NI2G5SYN4utBzkorGjoJHBr7H3VCbq5dsMY9GLiY5EuizS0bdqNXhcKVMb-9-xOWPslb~r96yZFej1LmsnfRWMqOcYxQ3GIOym85ysjGYYQrj0pJaXvr0ZfvvPkRCw2MgDPf1WZohAKXC4Vs3o6zhUjSE6cyt9zkPceWtCHJxKc61xSmH~N3yLfpFx8Ig09MX0rxz84QisYMsGIPIC~NIGJFOZNYYqxSOyHzZ1OF7UmqZ6~uDwUiC1yDU-RrODI-VtFBpY1LXA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal